Abstract
This paper represents the first survey on the application of AI techniques for the analysis of biomedical images with forensic human identification purposes. Human identification is of great relevance in today’s society and, in particular, in medico-legal contexts. As consequence, all technological advances that are introduced in this field can contribute to the increasing necessity for accurate and robust tools that allow for establishing and verifying human identity. We first describe the importance and applicability of forensic anthropology in many identification scenarios. Later, we present the main trends related to the application of computer vision, machine learning and soft computing techniques to the estimation of the biological profile, the identification through comparative radiography and craniofacial superimposition, traumatism and pathology analysis, as well as facial reconstruction. The potentialities and limitations of the employed approaches are described, and we conclude with a discussion about methodological issues and future research.
1. Introduction
Forensic Sciences are the set of disciplines whose common objective is the materialization of the evidence for legal purposes through a scientific methodology. In this sense, any science becomes ‘forensic’ when it serves the judicial procedure. Human identification (ID) [1], often the main task that forensic sciences have to face, is crucial in a multitude of contexts of great importance in our society: from the identification of missing persons and the estimation of the age of unaccompanied migrants (whose rights could be otherwise violated) to crime analysis and massive disaster victim ID scenarios. In all these cases, personal identity is associated with the preservation and defense of Human Rights and is a tool to repair the violation of these rights.
The most commonly used methods for human ID are DNA testing and fingerprint comparison systems (AFIS), mostly due to their high accuracy (over 99%). These methods are expensive (AFIS costs can run in the millions of dollars depending on the agencies participating and the complexity of the system [2]) and time-consuming (weeks for DNA test from bone). However, their main drawback is their limited applicability: both require prior records, a trustable baseline and preserved material for the DNA extraction or fingerprint comparison. In other words, the application of these methods fails when there is not enough ante-mortem (AM) or post-mortem (PM) information available due to the lack of data (second DNA sample) or to the state of preservation of the corpse. While the skeleton usually survives both natural and non-natural decomposition processes (fire, salt, water, etc.), the soft tissue progressively degrades, being eventually lost. Therefore, techniques like DNA or fingerprint comparison are not suitable in cases where such records do not exist or in scenarios with poorly preserved bodies. To carry out the ID when the circumstances are not favorable (as is the case of skeletonized, burned or degraded individuals, mixed or disconnected remains, mass graves, etc.), methods based on Forensic Anthropology (FA) represent the main alternative at our disposal. FA studies the skeleton for its application to medico-legal issues [3], and encompasses techniques for skeleton-based forensic identification (SFI) such as craniofacial superimposition or comparative radiography. In fact, the experience of several practitioners in certain scenarios suggests the poorer effectiveness of DNA analysis (around 3% of the IDs) and dactyloscopy (15–25%) against SFI techniques (70–80%) [4]. SFI methods employed by forensic anthropologists, odontologists, and pathologists represent, in many cases, the victim’s last chance for ID.
In the last decades, artificial intelligence (AI) has allowed to automate repetitive or tedious tasks for human beings (e.g., the automation of industrial processes or cleaning tasks), as well as to surpass humans’ capacity in performing complex tasks (e.g., processing massive amounts of data to extract new knowledge or overcome human champions playing Chess or Go). Recently, advances related to machine learning (ML), under the terminological umbrella of deep learning (DL), have provided astonishing advances in image recognition, image restoration, image generation, speech recognition, and machine translation, among others. The medical field has not been an exception: AI has provided tremendously useful tools for practitioners in parameter estimation, image segmentation, pathology classification, or image enhancement, just to name a few of representative scenarios. It is, however, surprising how FA has largely remained apart from these advances and, still today, in general terms, it is an essentially manual and precarious discipline at the technological level. This lack of technological development and hybridization of AI with FA is noteworthy since, among other reasons, the human ID field, to which FA belongs, has a remarkable and increasing social and economic importance: the global human ID market reached $43.0 billion in 2019 and should reach $83.9 billion by 2024 (https://www.bccresearch.com/market-research/biotechnology/human-identification-forensics-genealogy-and-security-applications-market-report.html).
Very few surveys exist partly or totally focused on the application of AI techniques to particular SFI tasks and methodologies [5,6]. In this sense, up to the authors’ knowledge, this is the first paper to tackle the broad subject of AI-based biomedical image analysis for FA-based human ID. From this perspective, we will not tackle other forensic ID techniques like identification from biomolecular evidence (DNA), identification from latent prints (fingerprints, earprints), identification from methods of communication (e.g., handwriting), identification from podiatry and walking, or identification from personal effects. The important field of facial recognition and identification [7,8] is not addressed either, despite its great importance and technological development, because facial images are not considered biomedical images (in the sense that they are not visual representations of the interior of a human body, and they are not generally acquired for diagnostic or therapeutic purposes). In addition, there already exist exhaustive surveys [9,10,11] focusing on human ID based on facial images.
In this manuscript, we try to cover both the ID of deceased and living individuals employing hard tissues (bones) displayed using different biomedical imaging modalities (see Figure 1). The most popular biomedical image modalities employed for ID are X-ray images and CTs. The reason for this can be twofold. First, X-ray images are the most commonly acquired medical imaging modality worldwide. Just as an example, 2.02 million chest X-ray images were performed in 2015/16 by the National Health Service of United Kingdom [12], and 150 million are annually acquired in US [13]. In the forensic domain, CT imaging is increasingly being performed in forensic examinations, whereas most reports that exist on PM MRI are generally based on small case samples [14]. Second, even if PM CT and MRI have proven to be useful diagnostic tools in forensic medicine, some studies remark that CT is superior to MRI in the visualization of osseous and ligamentous injuries after trauma [15].
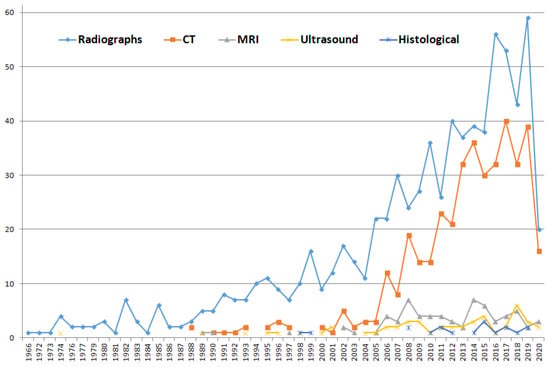
Figure 1.
Papers published using different biomedical image modalities for forensic ID. This figure was obtained by introducing the following queries in Scopus (search performed the 11th of May 2020): (TITLE-ABS-KEY(forensic identification radiograph) OR (TITLE-ABS-KEY(forensic identification X-ray image))); (TITLE-ABS-KEY(forensic identification CT)); (TITLE-ABS-KEY(forensic identification MRI)); (TITLE-ABS-KEY(forensic identification ultrasound)); and TITLE-ABS-KEY (forensic identification histological image). Those searches provided 771, 398, 68, 49 and 18 papers, respectively. The most commonly employed biomedical image modalities in this domain are X-ray images and computed tomographies (CTs).
After an introductory section devoted to the methodological background (Section 2), the paper focuses on the SFI-based approaches that employ biomedical images (such as radiographs, CTs, MRIs, or 3D bone scans) to the ID of living and deceased individuals. In Section 3 the different families of forensic ID methods and AI-based techniques used in the literature are specified. The manuscript concludes with Section 4, where some conclusions, recommendations and future research lines are discussed.
2. Methodological Background
2.1. Forensic Anthropology and Human Identification
FA addresses the application of physical anthropology (devoted to the study of human body) to legal cases, usually with a focus on the human skeleton. FA methods represent an alternative and counterpart with a much broader range of applicability than other ID approaches (like fingerprints and DNA). According to the Scientific Working Group for Forensic Anthropology (SWGANTH), forensic anthropologists contribute to identification at two levels. The first level is through methods that establish positive identifications. The second level is through methods that contribute to the identification by limiting potential matches to the analyzed individual. Among the former group, the SWGANTH includes comparative radiography and the comparison of surgical implants, while it leaves for the second level craniofacial superimposition, biological profiling, medical and/or dental records, abnormalities and pathological conditions, and comparative photography. While surgical implant identification simply involves locating the manufacturer’s symbol along with the device’s unique serial number, the remaining methods are largely complex to apply. The overall pipeline of SFI is described in Figure 2. Below, we briefly describe the main FA methodologies for human ID that are relevant to this review.
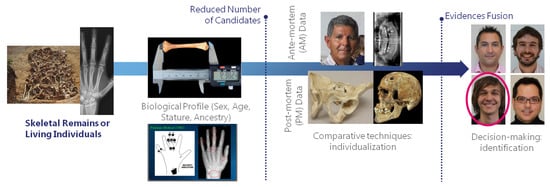
Figure 2.
Skeleton-based forensic identification (SFI) pipeline. These techniques can be applied for positive identification and potential candidates reduction (or shortlisting) of both living and dead individuals. They are commonly employed when other ID techniques (e.g., DNA or fingerprint comparison) are not applicable, or in combination with them.
The estimation of the biological profile (BP) has been studied for more than 300 years, and nowadays it plays a crucial role in narrowing the range of potential matches during the ID process. BP involves the study of skeletal remains with the aim of finding characteristic traits that support determining the identity of the individual. It is a sequential process in which sex, age, stature and ancestry, in this particular order, are estimated. These traits include:
- Sex estimation of adult individuals. It is recommended to employ the pelvic and skull morphologic traits [16]. When this is not possible, the discriminant formulas for postcranial skeleton proposed by [17] are recommended. Sex is the first characteristic to be estimated, since many formulas in remaining steps vary depending on sex.
- Sex estimation of subadults (i.e., children). Sexual characteristics are not fully developed and discriminant until puberty has been passed. For this reason, estimating the sex of children is one of the main difficulties faced by FA when studying subadult individuals. However, there are some approaches that have shown a high potential, like the analysis of morphological features of the ilium, mainly the sciatic notch [18].
- Age estimation of dead adult individuals. The analysis of the degenerative processes of the pubic symphysis is recommended, preferably following the method proposed in [19]. This method should be combined with the analysis of canine root transparency (presented in [20]) according to the two-step procedure proposed in [21]. When this is not possible, the recommended methods are: that proposed in [22] for the analysis of the coxal atrial veneer; that proposed in [23] for the sternal end of the fourth rib; and finally the analysis of the processes of obliteration of the cranial sutures proposed in [24], this being the least reliable method but the only one available on many occasions.
- Age estimation of dead children. For individuals who have not yet reached maturity (subadults), the methods proposed by Scheuer and Black in 2004 [25] are recommended.
- Age estimation of the living. The hand and wrist development atlas introduced in [26], the method for dental development proposed in [27], and the analysis of ossification status of the sternal epiphysis of the clavicle presented in [28] are recommended for different age ranges. Age estimation of the living acquires special relevance when determining the legal age of the person being scrutinized (i.e., to determine if the person is 18 years old to carry out age-dependent legal procedures appropriately in accordance with the rule of law).
- Stature estimation is performed employing long bones. When the remains are contemporary and of Mediterranean origin, it is recommended to use the formulas proposed in [29] for the femur and humerus, and those proposed in [30] for the tibia. When the remains are from the North American population, the formulas proposed in [31] as well as the FORDISC computer program [32] will preferably be applied.
- The estimation of population ancestry is the most inaccurate element of the BP given the low reliability of its results. It should only be used as an orientation criterion when there is a good agreement between the human study groups and skeletal biology, giving preference to morphological criteria of the skull.
Comparative radiography (CR) involves the direct comparison of AM radiographs, generally acquired for medical reasons, with PM radiographs acquired only for ID purposes, using specific and individualizing structures. Both radiographs are visually compared and evaluated for similarity in osseous shapes and densities, to determine whether they belong to the same subject [33]. Several bones and cavities have been reported as useful for candidate short-listing or positive identification based on their individuality and uniqueness. In particular, the most widely recognized as useful and reliable methods of identification are teeth, frontal cranium bones, vertebras, and clavicles, although dental-based ID is the most employed and discriminative technique. Also, the most commonly AM and PM images employed with the CR technique include radiographs [34], CT images [35], and 3D surface models [36]. The application of CR requires the superimposition of the AM and PM data for their visual comparison by producing PM radiographs simulating the AM ones in scope and projection. This is a time-consuming and error-prone trial-and-error process that relies completely on the skills and experience of the analyst. CR requires a prior record of clinical images not always available, but if present, this technique can be extremely accurate, reaching 100% reliability for certain bones [37]. The whole CR-based ID process is depicted in Figure 3.
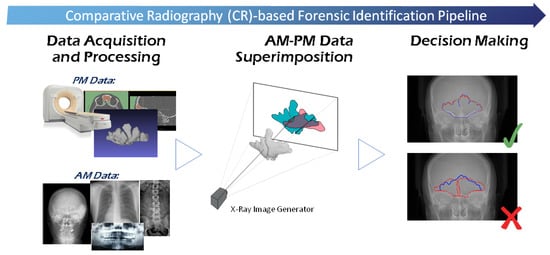
Figure 3.
General comparative radiography (CR)-based ID process. After data acquisition and processing (e.g., anatomical regions of interest segmentation), and prior to decision making, the ante-mortem (AM) and post-mortem (PM) materials are registered, so their overlap is maximized. In this figure, this superimposition process is depicted using frontal sinuses in X-Ray images.
Craniofacial Superimposition (CFS) is probably the most challenging SFI method [38,39]. It involves the superimposition of an image of a skull with a number of AM face images of an individual and the analysis of their morphological correspondence. This skull-face overlay (SFO) process is usually done by corresponding anatomical (anthropometrical) landmarks located in the skull (craniometric) and the face (cephalometric). Thus, differently to CR, two objects of different nature are compared (a face and a skull). CFS has been used for one century, yet it is not a mature and fully accepted technique due to the absence of solid scientific approaches, significant reliability studies, and international standards. On the other hand, this technique is widely employed in developing countries because its application is inexpensive and the only required AM data is one or more photographs of the face. In the manner of CR, the most recent comprehensive surveys of the CFS field differentiate three consecutive stages (Figure 4) for the whole CFS process [40,41]: (1) the acquisition and processing of the materials, i.e., skull (or skull 3D model) and the AM facial photographs with the corresponding location of landmarks on both; (2) the SFO process, which deals with accomplishing the best possible superimposition of the skull on a single AM photograph of a missing person. This process is repeated for each available photograph, obtaining different overlays; (3) the decision making which evaluate the degree of support of being the same person or not (exclusion) based on the previous SFOs. This decision is influenced by the morphological correlation between the skull and the face, the matching between the corresponding landmarks according to the soft tissue depth, and the consistency between asymmetries. These criteria can vary depending on the region and the pose [42].
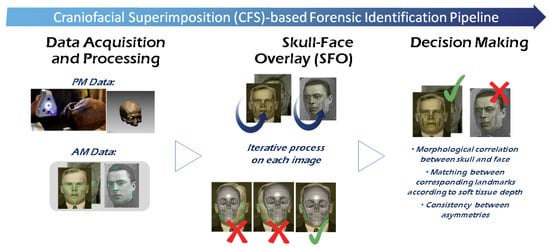
Figure 4.
General Craniofacial Superimposition (CFS)-based ID process.
Forensic facial approximation, also called facial reconstruction, is an ID technique for unknown skeletal remains, or corpses encountered in criminal investigations, based on the estimation of a face from a skull with the aim to obtain information about the deceased person’s identity. In [9] four different approaches are distinguished: (1) two-dimensional (2D) representation of the face over a photograph of the skull [43,44]; (2) three-dimensional (3D) manual construction of the face in clay or mastic over the skull or skull cast [43,45]; (3) computerized sculpting of the face using haptic feedback devices and a 3D scan of the skull [46,47]; and (4) computerized construction of the face using more complex computer-automated 3D routines [48,49,50,51]. All these approaches have in common the dependence of soft tissue thickness measurements of the face.
2.2. Artificial Intelligence and Forensic Human Identification
In the FA domain, AI methods can allow modeling and structuring human experts’ knowledge, as well as they can be used to extract new knowledge from massive databases and shortening ID times through the automation of certain tasks. They can also reduce human subjectivity and errors; and can contribute to provide a greater scientific basis that favors the admissibility of expert evidence, given that courtroom forensic testimony is often criticized by defense lawyers as lacking a scientific basis. In this sense, the Daubert criteria [52] determine whether evidence is admissible in a court of law. An identification method fulfills the Daubert criteria when: (1) it is testable and peer reviewed; (2) it possesses known potential error rates; and (3) it is accepted by the forensic community. Finally and overall, AI-based forensic ID approaches can help to tackle the currently unapproachable number of open ID cases worldwide.
AI is a broad interdisciplinary field including ML, knowledge representation, automatic reasoning, natural language processing, robotics, and computer vision (CV), among others. Within this vast research field, the most commonly employed AI tools applied to FA-based ID problems belong to the subfields of CV, ML, and soft computing (SC). CV is the scientific discipline that deals with the automatic interpretation of images [53]. ML is the branch of AI developing techniques that allow computers to learn directly from data [54]. The performance of ML-based AI systems (including DL approaches [55]) is reaching or even exceeding the human level on an increasing number of complex tasks. SC, or computational intelligence, techniques [56] are widely used as they exploit the tolerance for imprecision and uncertainty to achieve tractability, robustness, and low computational cost when solving real-world problems. SC focuses on the design of hybrid intelligent systems that combine nature-inspired computational approaches to appropriately handle vague and incomplete data. Within SC, fuzzy sets and fuzzy systems are aimed for reasoning and knowledge representation under imprecision and uncertainty [57] while evolutionary algorithms and metaheuristics provide single- and multi-objective methods for optimization, search and ML, yielding high-quality solutions in a reasonable time [58].
3. Forensic Human Identification through the Analysis of Biomedical Images
3.1. AI-Based Approaches for Biological Profile Estimation
The most common scenario in FA is to have direct access to the bones; bones that are extracted, cleaned and manipulated by the forensic anthropologist. However, radiological imaging, mainly CT scanning, is gaining popularity for BP since it is a non-invasive approach (in the sense that the human expert do not need to have direct visual access to the bone), and it allows better possibilities for observation and metric calculations. Estimation of BP from X-ray images has been approached from three different scientific communities with three different purposes. In clinical medicine, the biological age is important when determining endocrinological diseases in adolescents or for optimally planning the time-point of paediatric orthopaedic surgery interventions. Nevertheless, in legal medicine, is used to approximate unknown chronological age, when determining age in cases of criminal investigations or for asylum-seeking procedures, where identification documents of children or adolescents are missing. Finally, physical and forensic anthropologists are interested in determining sex, age, stature and ancestry of any human remain.
The bigger international efforts have been focused on the development of accurate and objective automatic approaches for assessing whether living individuals have reached the threshold age that implies legal adulthood. There are different computer-based proposals to assist the assessment procedure by using radiological imaging methods. Usually, MRI or X-ray images of the hand/wrist are employed in some AI-based proposals using SC techniques such as fuzzy decision trees [59], random forests [60], or neural networks [61]. Recently, the proliferation of convolutional neural networks (ConvNets) has attracted interest in medical image analysis. DL techniques overcome many of these limitations by allowing to automatically learn the suitable features for image interpretation without any direct human intervention during the training process.
Nevertheless, to our knowledge, there is no automatic approach facing a complete forensic BP assessment considering different stages and a whole age range. Most published automatic methods operate only with X-ray scans of Caucasian subjects younger than 10 years, with a few approaches dealing with subjects less than 18 years old. The automation of age estimation in adults is much less technologically developed.
The following subsections will focus on sex and age estimation. Table 1 and Table 2 include an overview of the main AI-based approaches employed in the literature for sex and age estimation, respectively. There are some cases (e.g., Pinto et al. [62], Abdullah et al. [63] or Pietka et al. [64,65,66]) that, probably, fit more the image/signal processing domain than the AI research field. However, we consider those works relevant and related at a computational level and, thus, they are also mentioned in the corresponding table or subsection. It is also important to highlight that there are other remarkable approximations (like [67]) that, despite their use of advanced intelligent systems, are not tackled in this paper, since they do not actually use biomedical images (in the case of [67], the author uses ConvNets to estimate the sex of individuals by detecting biometric tracts in photographs of hands). Finally, it is worth mentioning that there are also previous studies using AI-based approaches in BP estimation, like ML for sex estimation [68,69], but these employ measurements manually taken by forensic anthropologists and not images, so again they are not included in this survey.

Table 1.
An overview of the literature on artificial intelligence (AI)-based approaches for sex estimation from skeletal data. References are ordered according to their publication date.

Table 2.
An overview of the literature on AI-based approaches for age estimation from skeletal data. References are ordered according to their publication date.
3.1.1. Sex Estimation from Skeletal Structures
Sex estimation is a fundamental pillar of the BP. If sex estimation is incorrect, then the identification may be delayed. Current methods are mainly based on morphometric or morphological criteria [70,71]. The morphometric approach involves the measurements of hands, feet, and the extremities corresponding to the upper, lower and long bones. On the other hand, the morphological approach finds its foundation in the sexual dimorphism, presented in certain degrees in most of the bones in human body. In particular, the most common and popular parts of human body for sex estimation are skull and the pelvis area [62]. However, these methods are subject to human biases, require a high degree of expertise, are complex, time-consuming [72], and are not always suitable, mainly due to the presence of a significant damage (chemical and/or physical) on the skeletal remains.
AI techniques, especially DL approaches, offer a flexible and powerful solution to the sex estimation from skeletal structures. Some researchers have tried different DL techniques for sex estimation [62,63,73,74], mainly focusing on adults, since the adult’s body is mature enough to point out significant clues which can help to distinguish its sex, but may not be suitable for sex estimation in children [75]. Table 1 offers an overview of the main AI-based approaches employed in the literature for sex estimation.
Darmawan et al. [73] used a Hybrid Particle Swarm Artificial Neural Network technique for sex estimation of individuals. They used a dataset of left hand X-ray images of Asian population. Their data set was small and their results suggest a different accuracy for different age groups. Pinto et al. [62] introduced a methodology for objective quantification of sexually dimorphic features on images of skull and pelvis using the Wavelet Transform, as a multi-scale mathematical tool that allows for measurements of shape variations that are hidden at different scales of resolution. This information can be used by experts to improve the accuracy of BP assessment, and to describe the geographic and temporal variations within and among populations. In [63], the authors presented an automated Haversian canal detection system based on the histomorphology, which uses only bone fragments to estimate age and sex. They divided their detection system into two parts. In the first part, they manually analyzed and observed differences in the parameters of male and female sample of bones. In the second part, they applied microstructural image processing techniques to identify the gender. Bewes et al. [74] addressed the problem of sex estimation of skeletal remains by training a ConvNet with images of 900 skulls virtually reconstructed from hospital CT scans. When tested on previously unseen images of skulls, the deep network showed 95% accuracy at sex estimation. In [77], the authors employ an ensemble of shallow multilayer perceptrons to perform sex estimation from six cranial measurements (cranial sagittal arc, cranial sagittal chord, apical sagittal arc, apical sagittal chord, occipital sagittal arc, and Occipital sagittal chord). They tested their approach on 267 whole-skull CT scans (153 females and 114 males) from the Uighur ethnic group in the north of China (females aged 18–88 and males aged 20–84). An accuracy >94% was reported in all cases.
On the other hand, other authors such as Kaloi and He [75], have focused on the estimation of sex in children. They proposed a technique called GDCNN (Gender Determination with ConvNets), where left hand radiographs of the children between a wide range of ages (ranging from 1 month to 18 years old) are examined to determine the gender. To identify the area of attention (part of the hand) they used Class Activation Maps, discovering that the lower part of the hand around carpals (wrist) is more important than other factors for child sex estimation. They obtained an accuracy of 98%, identifying the gender of a child even with half of the lower part of the hand, which is impressive considering the incompletely grown skeleton of the children. In another contribution [78], the authors examined the possibilities offered by 3D descriptors on sex identification accuracy, and tested their multi-region based representation on 100 head PM CT scans (54 male and 46 female subjects between the ages of 5 to 85 years, from south east Asia). The authors yield comparable results to the commonly reported sex prediction range (70–90%) using morphometric or morphological assessment by forensic anthropologists.
As conclusion, most works for sex estimation employ either morphological methods (that rely on the visual assessment of sexually dimorphic traits) or metric methods (based on the variability in male and female dimensions, mostly utilizing different statistical methods to derive models/equations). This field has not been particularly explored using AI approaches and, generally, when tackled, the different presented approaches have been focused on sex estimation in adult individuals.
3.1.2. Age Estimation from Skeletal Structures
In legal medicine, when identification documents of children or adolescents are missing, as may be the case in asylum seeking procedures or in criminal investigations, estimation of physical maturation is used as an approximation to assess unknown chronological age. Some of the established radiological methods estimating unknown age in children and adolescents are based on visual examinations of bone ossification in X-ray images of the hand [26,79], even if some proposals to automatically analyze the sternal end of the fourth rib in CTs have also been presented [80]. Ossification is best followed in the hand due to the large number of assessable bones that are visible in X-ray images from this anatomical region, together with the fact that aging progress is not simultaneous for all hand bones. From the level of ossification assessed by the radiologist, the most common methods for the estimation of physical maturation of an individual are the GP [81] method and the TW2 [82,83] method. The GP method is the approach used by the majority of radiologists due to its simplicity and speed. This method is based on the comparison between the X-ray image of the hand and an atlas at various chronological ages. The patient’s radiograph is compared to the suitable image in the atlas. The TW2 method analyzes specific bones, instead of the whole hand as in the GP method. In particular, this method takes into account a set of specific ROIs divided into epiphysis/metaphysis ROIs and carpal ROIs. Very recently, ConvNets have shown to be successful for bone age estimation and there are some published applications based on the GP method [84,85,86,87,88] and on the TW2 method [59,89,90,91]. Table 2 includes an overview of the main AI-based approaches employed in the literature for age estimation.
Larson et al. [85] used a ConvNet over a total of 14,036 clinical hand radiographs and their corresponding reports, obtained from two children’s hospitals, to train and validate the model. The RMS of a second test set composed of 1377 examinations from the publicly available GP Digital Hand Atlas [95,98] was compared with an existing automatic model, termed BoneXpert (BoneXpert automatically reconstructs the borders of 15 bones (including metacarpal and phalangeal bones, the distal radius, and the ulna) from radiographs of hands using a generative model (active appearance model), and then estimates the age from the shape, intensity, and texture scores derived from principal component analysis for each bone. However, it is important to remark that this software is used in clinical settings, to estimate skeleton maturity and detect abnormalities/diseases, and it is not used in forensic settings for age estimation.) [115]. The estimates of the model, the clinical report, and three human reviewers were within the 95% limits of agreement. RMS for the Digital Hand Atlas data set was 0.73 years, compared with 0.61 years of a previously reported model. Kim et al. [84] used a GP method-based DL technique to develop an automatic software system for bone age estimation. Using that software, they estimated the bone age from left-hand radiographs of 200 patients (3–17 years old) using first-rank bone age (only the software), computer-assisted bone age (two radiologists with software assistance), and GP atlas-assisted bone age (two radiologists with GP atlas assistance). The reference bone age was determined by the consensus of two experienced radiologists. The first-rank bone ages determined by the automatic software system showed a 69.5% of concordance rate and significant correlations with the reference bone age (r = 0.992; p < 0.001). The concordance rates also increased with the use of the automatic software for both reviewers, and the X-ray images evaluation time required by the radiologists were reduced between 18.0% and 40.0%. Their results suggested that automatic software system reliably showed accurate bone age estimations, and appeared to enhance efficiency by reducing evaluation times without compromising diagnostic accuracy. In [86], the authors created a DL system to automatically detect and segment the hand and wrist. They perform an automated bone age assessment with a fine-tuned ConvNet over a set of 4278 female and 4047 male radiographs (with chronological age of 5–18 years old). Their model achieves 57.32% and 61.40% accuracy for the female and male cohorts on held-out test images. Female test radiographs were assigned a bone age within 1 year in 90.39% of cases and within 2 years in 98.11% of cases. Male test radiographs were assigned in 94.18% of cases within 1 year and in 99.0% of cases within 2 years. Attention maps were created which reveal what features the trained model uses to perform bone age assessment. Lee et al. [87] presented a way to use DL for age estimation from a subject’s hand X-ray images, employing a set of feature points on the hand. These points have to be defined to serve as a reference to crop a certain region that is informative in terms of aging-induced morphological changes. Mutasa et al. [88] using their proposed customized neural network architecture trained on 10,289 images of different skeletal age, achieved a test set MAE of 0.637 and 0.536, respectively. Their results support the hypothesis that purpose-built neural networks provide improved performance over networks derived from pre-trained imaging data sets.
Automated approaches reproducing the TW2 method can be mainly classified based on whether they use image processing or knowledge-based techniques, and a thorough review can be found at Mansourvar et al. [61]. The majority of the image processing-based methods date back to the 2000s. These methods use hand radiographs of living individuals as knowledge source for training classifiers. In [59], a computing-with-words-based classifier for skeletal maturity assessment is proposed. In [89] the proposal is based on a neural network and a fuzzy filter output. In [90], a fuzzy inference system is used for age assessment. More recently, Spampinato et al. [91] proposed and tested several DL approaches. In particular, several existing pre-trained ConvNets are employed to assess skeletal bone age automatically, based on the TW2 method and using a dataset with about 1400 X-ray images. The results showed an average discrepancy between the manual and the automatic evaluation of about 0.8 years. They also designed and trained from scratch a ConvNet, which proved to be the most effective and robust solution in assessing bone age across ethnic groups, age ranges and gender. Furthermore, this was the first automated skeletal bone age assessment work tested on a public dataset.
The advent and proliferation of ConvNets using X-ray hand radiographs have facilitated new applications to the evaluation of age using other bones. The iliac crest apophysis provides an excellent subject for the application of forensic age diagnostics to the living, particularly for determining age thresholds of 14, 16 and 18 years. For this reason, Li et al. [113] developed a DL system to perform automatic bone age estimation based on 1875 clinical X-ray pelvic radiological images, particularly for individuals between 10 and 25 years old. It can handle all possible cases of automated skeletal bone age assessment, even for samples from individuals of 19, 20, and 21 years old. However, it may not be practical in determining ages over 22 years due to the little change in the mean score of ossification. Compared to the existing cubic regression model, their ConvNet model achieves better average performance (MAE = 0.89 and RMSE = 1.21). These results also improve the DL architectures based on left-hand X-ray images, where the MAE values range from 0.54 to 0.80 years [84,85,88,91]. However, although their statistical analysis indicates a high positive correlation between the estimated and real age (r = 0.916; p < 0.05), this number is less accurate than hand X-ray radiographic images methods (r = 0.992; p < 0.001).
As an alternative to the age estimation methods based on X-ray images, research in age estimation using MRI has gained tremendous interest in recent years. The interest in developing automatic MRI-based methods for age estimation is determined by addressing the problems of exposure to ionizing radiation, the necessity to define new MRI specific staging systems, and the subjective influence of the examiner [114]. In Stern et al. [60], they used random forests to separately regress chronological age from intensity-based features extracted from 11 selected hand bones of adolescent subjects. A decision tree excluding metacarpal and phalanx information from older subjects served as a heuristic fusion strategy for age estimation, making this method ad hoc and dependent on parameter tuning. In Stern and Urschler [109], the capability of RFs for information fusion was explored by allowing it to internally decide from which bones to learn a subject’s chronological age. Thus, they treated aging as a global developmental process without the necessity for heuristic fusion schemes, as in [60], or predefined nonlinear functions, as in [115]. Following the current research trend of replacing handcrafted features in random forests with automatically learned ones, in [109] they proposed a ConvNet architecture to combine age information from individual bones in an automatic fashion by letting the architecture learn directly the most relevant features for age estimation. More recently, in [114] the authors present a solution for automatic age estimation from 3D MRI scans of the hand. They evaluate ML methods, as RFs and ConvNets, with different variants of the image information used as input for learning. Trained on a dataset of 328 MRI images, they compare the performance of the different input strategies and demonstrate unprecedented results achieving the state-of-the-art accuracy compared with previous MRI-based methods. For estimating biological age, they obtained a mean absolute error of 0.37 ± 0.51 years for the age range of subjects ≤18 years, i.e., where bone ossification has not yet saturated. Finally, they adapted their best performing method to 2D images, and applied it to a dataset of X-ray images in order to validate their findings, showing that their method is in line with the state-of-the-art methods developed specifically for X-ray data.
As conclusion, bone age assessment is one of the most important topics in FA and, specially, in the evaluation of biological maturity of children. It is usually performed by comparing an X-ray of left hand-wrist with an atlas of known sample bones. With the rise of DL, most works currently employ ConvNets to tackle the problem. Age estimation in adults remains a challenge, as well as the development of approaches that integrate hybrid evidence for achieving more confident results (such as X-ray of the left hand and teeth, and physical or psychological examination, to ensure that the subject has reached the legal age).
3.2. AI-Based Approaches for Traumatism and Pathology Analysis
Over the recent years, the success of ML in general, and DL in particular, to classify images has caused great interest in its application to medical image analysis in several relevant fields, including the detection of skin cancer [116], gastrointestinal lesions [117], diabetic retinopathy [118], mammographic lesions [119] or lung nodules [120]. However, a representative pathological example that is of high interest, but also rare, is bone lesions [121]. To our knowledge, there are just a few works in the field of orthopaedics related to the application of DL to detect bone lesions or pathologies in X-ray images.
Olczack et al. [122] extracted 256,000 wrists, hand, and ankle radiographs from Danderyd’s Hospital and identified 4 classes: fracture, laterality, body part, and exam view. Then, they evaluated the diagnostic accuracy of 5 openly available deep networks adapted to this task. All networks exhibited an accuracy of at least 90% when identifying laterality, body part, and exam view. The final accuracy for fractures was estimated at 83% for the best performing network.
Chung et al. [123] evaluated the ability of AI techniques for detecting and classifying proximal humerus fractures using plain anteroposterior shoulder radiographs. The evaluated dataset was composed of 1891 images (1 image per person) of normal shoulders (n = 515) and 4 proximal humerus fracture types (greater tuberosity, 346; surgical neck, 514; 3-part, 269; 4 part, 247) classified by 3 specialists. They trained a ConvNet after augmenting the training dataset. The ability of the ConvNet was measured by top-1 accuracy in comparison with humans (28 general physicians, 11 general orthopedists, and 19 orthopedists specialized in the shoulder) to detect and classify proximal humerus fractures. Their results showed 96% accuracy, for distinguishing normal shoulders from proximal humerus fractures, and 65–86% accuracy for classifying the fracture type.
In Gupta et al. [124], the authors address the problem of classifying bone lesions from X-ray images by increasing the small number of positive samples in the training set. They propose a generative data augmentation approach based on a cycle-consistent generative adversarial network that synthesizes bone lesions on images without pathology. They pose the generative task as an image-patch translation problem that they optimize specifically for distinct bones (humerus, tibia, and femur). In experimental results, they confirm that the described method mitigates the class imbalance problem in the binary classification task of bone lesion detection. They show that the augmented training sets enable the training of superior classifiers achieving better performance on a held-out test set. Additionally, they demonstrate the feasibility of transfer learning and apply a generative model that was trained on one body part to another.
3.3. AI-Based Approaches for Comparative Radiography
Methodological approaches for performing CR-based identification are divided into three groups according to the dimensionality of the employed data: 2D-2D (radiograph-radiograph), 2D-3D (radiograph-CT or 3D surface image) and 3D-3D (CT-CT or 3D model). The greater the dimensionality, the greater the accuracy and robustness of the methods. Within each of these groups, methods can be further classified into manual approaches and semi-automatic approaches. In this section, we will focus on the semi-automatic methods, those in which some tasks of the identification process are automated by means of AI techniques.
3.3.1. 2D-2D Approaches for Comparative Radiography
The comparison of AM and PM radiographs is the most extended approach in forensic literature. In order to highlight the applicability of CR-based forensic ID, it is important to remark that X-ray images represent the most commonly employed medical imaging modality [125]. In particular, chest X-rays are the most commonly performed radiology examination world-wide [126] because they are able to produce images of the heart, lungs, airways, blood, vessels, spine, and chest [127], and because of their diagnosis and treatment potential [126,128].
There are several works that semi-automatically compare different skeletal structures between AM and PM radiographs. These skeletal structures include frontal sinus [129,130], cranial vault [131], and teeth [132,133,134]. These methods are based on the comparison of the silhouettes of skeletal structures in radiographs using geometric morphometric techniques. The elliptical Fourier analysis [135] is used to compare AM and PM silhouettes obtaining a shortlist of the most probable PM matches for each AM case. The segmentation of the skeletal structures in AM and PM radiographs is required in all these methods. Related to that, there are a few computational approaches that automate the manual segmentations using ad-hoc rule-based segmentation methods, as the automated dental identification system (ADIS) [136,137] for teeth comparison [138], or [139] for frontal sinuses segmentation, or via the direct comparison of the intensities as the computer-assisted decedent identification (CADI) [138] for vertebrae comparison. However, the latter approach suffers from the elapsed time between the AM and PM radiographs, and the consequent change in the intensities of the skeletal structures. CADI reduces its impact via the manual selection of a region of interest around each vertebra, the equalization of the pixels within these areas (e.g., with a histogram equalization filter), and lastly the comparison of AM and PM vertebrae using the Jaccard similarity metric.
3.3.2. 3D-2D Approaches for Comparative Radiography
In the manual approach, the comparison methodology requires the acquisition of PM simulated radiographs from a CT trying to simulate the AM radiographs [35,140,141], instead of real radiographs. The acquisition of these simulated radiographs is a time-consuming, error-prone, and subjective task.
However, there are just a few automatic approaches for the comparison of AM radiographs and PM 3D images [33,142,143]. These approaches are based on the use of 3D laser range scanners for the acquisition of 3D surface models of the skeletal structures, clavicles in [33,143], and patellae in [142]. They follow a procedure where a set of 2D projected images are obtained from these PM 3D surface models through the 3D model rotation. These 2D projections only contain the silhouette of the target skeletal structure. Finally, this set of PM projections is automatically compared to the manually segmented silhouette of the skeletal structure in the AM radiographs using elliptical Fourier analysis descriptors. However, the limitation of these methods lies in the set of predefined 2D projections, and in the assumption that the parameters that modulate the perspective distortions are known.
Alternatively, Gómez et al. [144] developed an evolutionary image registration method which successfully solved this image comparison problem (see Figure 5). The AM data are clinical radiographies (2D images) of a particular bone that have to be compared against the actual PM bone (a 3D model). The promising results that were achieved led them to recently design a methodology to fully automate the ID process by CR (see Figure 3). This method was tested with frontal sinuses, clavicles and patellae obtaining a superb performance. However, the method showed the following drawbacks: (1) none of the considered projective projections reproduced the perspective distortion of radiographs where the X-ray generator was not perpendicular to the image receptor (e.g., in the Water’s projection of radiographs of frontal sinuses [145]); (2) the robustness of the evolutionary algorithm employed, Differential Evolution (DE), especially with clavicles and patellae, that in some runs led to bad superimpositions due to the stochastic nature of DE and the highly multimodal search space tackled; and (3) the large amount of time required to obtain a superimposition with DE (1800 s on average). This large time is motivated by the high computational cost required by each evaluation of this evolutionary computation technique (on average, it takes 0.25 s) uncovering the computationally expensive optimization nature of the CR problem, as well as the high number of evaluations required by the optimizer to converge. They also tackled the problem of segmenting multiple organs (hearts, lungs and clavicles) in chest X-ray images using ConvNets [146]. They proposed several new deep architectures to deal with this complex problem. Their best performing proposal obtained better results than the competitor methods on clavicles with 0.0884, 0.939 and 18.022 for Jaccard Index, Dice Similarity Coefficient, and Hausdorff Distance metrics, respectively. This performance is in line with the ability of humans to accurately delimit the contour of clavicles in X-ray images. The same authors also presented the first system for the automatic segmentation of frontal sinuses in skull radiographs [147] (see Figure 6), as well as the first complete, but preliminary, computer-aided CR-based ID support system using frontal sinuses [148]. This system, that integrates automatic segmentation and registration, is able to rank the candidates in a way that 70% times the true positive ID case is ranked in first or second position, as well as is able to filter out 50% of candidates while always keeping the true positive ID case within the sample.
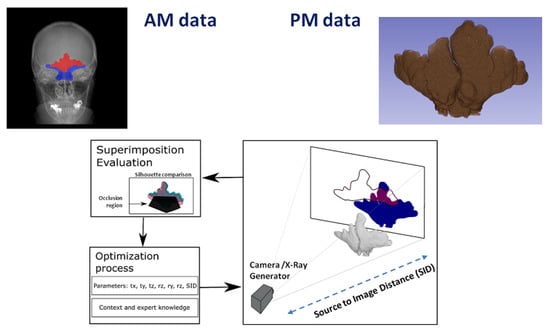
Figure 5.
Schematic representation of the automatic comparison of anatomical structures in radiographic materials. This figure uses the frontal sinuses as an example. Once AM (X-Ray) and PM (CT) data are segmented (either automatically or manually), both are superimposed using an evolutionary image registration algorithm. Three main interconnected blocks are represented: (i) The transformation to obtain a projection of the 3D model; in this example, the geometric transformation includes translation (tx, ty, and tz), rotation (rx, ry, and rz), and source to image distance (SID). (ii) The similarity metric that compares the PM projection and the AM segmentation considering an occlusion region (i.e., where the frontal sinuses are occluded or not clearly defined). (iii) The optimization process to estimate the nine parameters of the transformation that are only limited by the context and expert knowledge from the X-ray acquisition protocol.
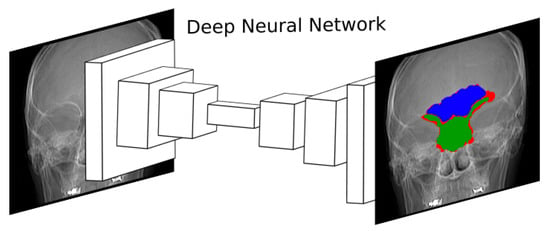
Figure 6.
Diagram showing the approximation followed in [147]. A deep ConvNet is trained to segment frontal sinuses in skull radiographs. A real outcome of the network is displayed on the right, where the frontal sinus, the occlusion region, and the segmentation error are displayed in blue, green and red, respectively. The frontal sinus is an anatomical region of extremely diffuse boundaries and complex morphology. In fact, its lower limit overlaps other anatomical structures, hardening the segmentation process.
3.3.3. 3D-3D Approaches for Comparative Radiography
The CT-CT comparison approach is the most reliable one, since the 3D shapes can be directly compared [149,150,151,152]. A few computerized approaches have been proposed for the comparison of AM and PM 3D data of different skeletal structures such as teeth [153,154,155], frontal sinuses [156], or lumbar vertebrae [157]. The segmentation of the 3D skeletal structures in both the AM and PM CTs (although the PM data could be acquired with a 3D laser range scanner), their automatic registration and the measuring of the quality of the match are required for the application of these methods. However, the availability of 3D AM data (such as CT) is scarce compared to the number of AM radiographs available reducing significantly their applicability.
In conclusion, in recent years there has been a breakthrough in automating forensic ID issues using CR. However, the automation by integrating all available information about a forensic case, using multiple superimposition procedures (with the same or different dimensionality) of multiple skeletal structures, is still a future goal. Automatic segmentation of certain structures, such as clavicles and frontal sinuses in radiographs, or the automatic superimposition of radiographs and 3D models have already been addressed. However, there are still no automatic solutions for the analysis and segmentation of any type of skeletal structure in any type of image (radiographs or CTs). There are also no automatic tools for the automatic location and classification of morphological patterns in radiographs and CTs.
3.3.4. Virtopsy
Virtopsy (or virtual autopsy) [158] is a minimally invasive procedure to perform an autopsy, that employs radiological imaging methods routinely used in clinical medicine, such as CT and MRI, to find the reason for death. Virtopsy is a multi-disciplinary technology that combines forensic medicine and pathology, roentgenology, computer graphics, biomechanics, and physics.
Some contributions presented preliminary studies about the possible application of ML to virtopsy [159,160,161], highlighting the importance and suitability of interactive ML methods [162], while others present simple image processing techniques to detect forensically relevant information in the images (e.g., the detection of metal objects embedded in a cadaver in [163]). However, in [164], the authors actually employ DL methods to detect and segment a hemopericardium (i.e., blood in the pericardial sac of the heart) in post-mortem CTs to better identify cases with a possibly non-natural cause of death (in particular, the presence of hemopericardium often leads to the diagnosis of pericardial tamponade as a cause of death). Their best performing deep network classified all cases of hemopericardium from the validation images correctly with only a few false positives, while most segmentation networks tended to underestimate the amount of blood in the pericardium. Also, in [165], a semi-supervised DL pipeline is employed to localize and classify orthopedic implants in the lower extremities (specifically the femur) on a large database of whole-body post-mortem CT scans. For the localization component, Dice scores of 0.99, 0.96, and 0.98 and mean absolute errors of 3.2, 7.1, and 4.2 mm were obtained in the axial, coronal, and sagittal views, respectively. For the classification component, test cases were properly labeled with an accuracy >97% (the recall for two of the classes was 1.00, but fell to 0.82 and 0.65 for the other two). Despite these examples, we conclude that this field remains largely unexplored.
3.4. AI-Based Approaches for Craniofacial Superimposition
The following subsections review the main existing computer-aided CFS approaches for each of the three commonly identified CFS stages (see Figure 4), highlighting the automatic methods developed over the last 15 years. Additionally, Table 3 summarizes the main AI-based contributions to the CFS problem for each particular stage.

Table 3.
An overview of the literature on AI-based approaches to the CFS problem. The stage of the process, i.e., Acquisition and Processing of the Materials (APM), skull-face overlay (SFO) and SFO assessment and decision making (ADM), that is addressed using an AI method is indicated with a ✔.
3.4.1. Acquisition and Processing of the Materials
The computerized systems developed for the first stage of CFS are related to face enhancement and skull modelling procedures. Skull 2D images, skull live images (video superimposition), and more frequently nowadays skull 3D models can be used in CFS. With the use of scanning devices, like laser range scanners, the forensic anthropologist can get a skull 3D model with a precision lower than one millimetre in a reasonable time [187]. The use of a 3D model instead of a 2D image is recommended, because it is a more accurate representation of the real skull.
Since the first proposal to use a skull 3D model to tackle the CFS problem [166], 3D image reconstruction software is necessary to construct the 3D model by aligning the views in a common coordinate frame. Such image registration process consists of finding the best 3D rigid transformation (composed of a rotation and a translation) to align the acquired views of the object. In this sense, a method [170,171] was proposed based on evolutionary algorithms for the automatic alignment of skull range images. Different views of the skull to be modeled were acquired by using a laser range scanner, and a two-step pair-wise range image registration technique was successfully applied to such images. The method is able to reconstruct the skull 3D model even if there is no turntable and the views are wrongly scanned. Today’s technology, i.e., current 3D acquisition devices and corresponding software, automatically solve the alignment of the different acquisition views without the necessity of turn-tables o any additional device.
Finally, a different related task was presented in [175]. The authors proposed a new algorithm to deal with the 3D open model mesh simplification problem from an evolutionary multi-objective point of view. An open model refers to a surface with open ends. The problem is based on the location of a certain number of points in order to approximate a mesh as accurately as possible to the initial surface. The algorithm considers two conflicting objectives, the accuracy and the simplicity of a simplified 3D mesh.
3.4.2. Skull-Face Overlay
Several proposals have been presented in the literature to perform the SFO task. The most natural way to deal with the SFO problem is to replicate the original scenario of the AM photograph in which the living person was in a given pose somewhere inside the camera’s field. Regarding computer-aided automatic methods, the task of replicating on a skull the pose and the remaining acquisition parameters of a given facial photograph is the main goal of the SFO process. This is similar to the classic CV problem of replicating the pose of a 3D object from a photo based on some reference points. Technically, we are given n points having 3D positions , …, and target 2D positions , …, . The goal is to find a projection P so that every P() is as close as possible to .
The first computer-aided approach for the SFO task was proposed by Nickerson et al. in 1991 [166]. The landmarks of the 3D skull mesh were selected to calculate the affine and perspective transformation (rotation, scaling and translation) to map with the landmarks of the 2D face image using real-coded genetic algorithms. Another automatic approach was presented in [169], in which they considered two different neural networks for the implementation of an objective assessment of the symmetry between two nearly frontal 2D images (skull and facial image). In the last decade, several works have tackled the SFO automation using evolutionary algorithms and fuzzy sets [173,174,177] (see Figure 7). These approaches are based on overlaying a 3D model of the skull over a facial photograph by minimizing the distance among pairs of landmarks while handling the imprecision introduced by the facial landmark location [40,188]. The minimization process involves the search for a specific projection of the skull model leading to the best possible matching between corresponding landmarks. More recently, Valsecchi et al. [184] proposed a novel automatic SFO algorithm called POSEST-SFO. Unlike prior approaches, POSEST-SFO algorithm solves a system of polynomial equations relating the distances between the points before and after the projection. This latter algorithm was tested on a synthetic data set comprised of 9 CBCTs from 9 different subjects and 60 simulated photographs, i.e., 540 SFOs. This method is extremely fast, since it lasts 78 milliseconds to automatically perform a single SFO. In the most realistic scenario, considering soft tissue thickness (the mean distance was employed) and ±5 pixels of error in facial landmarks, the mean back-projection error was 2.0 mm and 3.2 mm in frontal and lateral photos, respectively. However, this algorithm, contrary to previous publications [177,179], does not address the sources of uncertainty, i.e., the articulation of the mandible, the estimation of the soft tissue thickness, and the intra- and inter-error in landmark location.
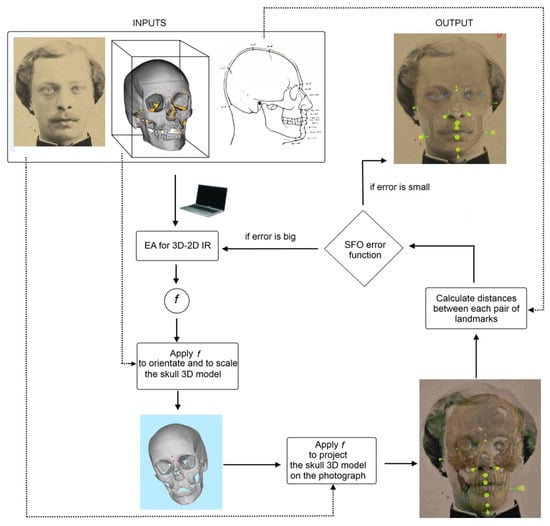
Figure 7.
Diagram showing the approximation followed in [173,174,177]. An evolutionary algorithm (EA) iteratively guides the search for the transformation parameters of an image registration (IR) process. The goal is to get the best possible skull-face overlay (SFO) for craniofacial identification. More specifically, the forensic expert marks fuzzy landmarks on the facial image and then marks crisp/precise landmarks on the skull (3D model that can be rotated and manipulated with ease and precision). Those crisp landmarks are fuzzified taking into account facial soft tissue depth population studies. Finally, these 3D fuzzy landmarks are projected onto the 2D image, overlap measurements between two fuzzy sets are calculated, and all this information is integrated into the EA fitness function.
3.4.3. Skull-Face Overlay Assessment and Decision Making
The forensic expert has to make a decision to determine the degree of support that the skull and facial photographs belong to the same person or not. This decision is made through the analysis of the previous SFOs and influenced by several criteria assessing the skull-face anatomical correspondence. This way, different authors have defined and classified those criteria in four different families [168,189,190]: (1) Analysis of the consistency of the bony and facial outlines/morphological curves; (2) Assessment of the anatomical consistency by positional relationship; (3) Line location and comparison to analyze anatomical consistency; and (4) Evaluation of the consistency of the soft tissue thickness between corresponding cranial and facial landmarks. This is a subjective process that relies on the forensic expert’s skills and the quantity and quality of the used materials.
There are just a few works tackling the automation of the analysis of craniofacial correspondences within the framework of CFS ID [168,191,192]. Most of the existing literature was published more than 20 years ago and the works are very basic and limited. In addition, they do not consider the use of either skull 3D models or computer techniques to perform the SFO. Besides, the employed technique for the shape analysis implies manual interaction. They provide a value that does not take into account the actual spatial relation between skull and face since the employed methods are invariant to translation, scale and rotation. Finally, these systems only implement a single group of the criteria to assess the craniofacial correspondence.
Recently, in [178,182,183], the authors present a hierarchical system to evaluate the anatomical consistence of morphological criteria between the face and the skull and give support to the forensic expert decision-making process. From a series of SFOs of the same individual, the computer-assisted decision support system (CADSS) provides to the forensic expert a quantitative output value that is indicative of the morphological matching consistency of a given CFS problem. This quantitative value is based on the use of several skull-face anatomical criteria combined at different levels by means of fuzzy aggregation functions considering the evaluation of the anatomical correspondence between the skull and the face at three different levels: criterion evaluation (level 3), SFO evaluation (level 2), and CFS evaluation (level 1). The sources of uncertainty and degrees of confidence involved in the process (bone preservation and 3D model quality, image quality, discriminatory power of each individual criterion, BP influence) were modeled and considered at each level of the system [183]. With the aim of comparing the performance of real forensic practitioners with this CFS CADSS, the authors applied their CADSS to the same experimental dataset of [193]. In that study, 26 participants from 17 different institutions were asked to deal with 14 identification scenarios, some of them involving the comparison of multiple candidates and unknown skulls. A total number of 60 SFO problems were tackled. The mean value of the results of the 26 experts, the result of the three best experts, and the outcomes of their automatic CADSS are shown in Table 4. The designed CADSS can be considered the first automatic tool for classifying couples of unknown faces and skulls as positive or negative cases with accuracy similar to the best performing forensic experts [182]. However, one of the reached conclusions was that the identification results based on the performance of the CADSS could be strongly influenced by the poor quality of some SFOs.

Table 4.
The table shows the mean value of the results of the 26 experts, the results of the three best experts, and the outcome of the CADSS presented in [182]. Detailed performance indicators are shown, such as the percentage of correct decisions, the number of positive and negative decisions given in each case, and the corresponding rate of true and false positives and true and false negatives. Ground truth refers to the real nature of each CFS case (P = Positive, N = Negative).
A completely different approach from the previous ones is presented in [180]. The authors proposed a model for automated skull recognition without the necessity of superimposition. They claimed the use of a publicly available dataset, IdentifyMe, consisting of 464 skull images, along with semi-supervised and unsupervised transform learning models. In order to automate this process in [181], they proposed a Shared Transform Model for learning discriminative representations. The model learns robust features while reducing the intra-class variations between skulls and digital face images. Experimental evaluation on the IdentifyMe dataset showcases the efficacy of the proposed model by achieving improved performance for the two protocols given with the dataset.
3.4.4. 3D-3D Computer-Aided Approaches for Craniofacial Superimposition
Over the recent years, some authors have proposed a 3D skull-3D face approach for CFS unlike the existing traditional (image and video) and computer-aided 2D-3D approaches mentioned above.
Duan et al. [176] proposed a novel ID method based on the correlation measure between 3D skull and 3D face in morphology. The mapping between skull and face is obtained using canonical correlation analysis. Unlike existing techniques, this method does not need the accurate relationship between skull and face, and only measures the correlation between them. In order to measure the correlation between skull and face more reliably and improve the identification capability, a region fusion strategy is adopted. Experimental results validate the proposed method, and show that the region based method significantly boost the matching accuracy. The correct recognition rate reaches 100% using a CT dataset.
In [186] another 3D-3D superimposition approach was presented in an effort to contribute to computer-aided CFS. The proposed method emphasizes adherence to two important parameters: (1) maintaining the life-size of the face image in relation to the size of the skull, and (2) orienting the skull on an anthropological basis using selected feature points. The proposed method commences by reconstructing the 3D face model from a given 2D face image using a mean simplified generic elastic model, followed by registering the face model to a 3D skull along the jaw line using the analytical curvature B-spline (AC B-spline). The accuracy index of the registration is, then, evaluated to suggest the degree to which the face image corresponds to a skull. The superimpositions of positive and negative cases were conducted on a set of 3D skulls versus a set of 2D face images. The accuracy indices of the registration results suggest that the AC B-spline is more robust in 3D-3D superimposition compared to the other existing methods. The full experimental results have demonstrated the potential of the proposed method as an assistive tool to the forensic scientists for craniofacial identification.
As conclusion, CFS is arguably the SFI approach that has been more benefited from the use of AI techniques in the last years. However, the main limitation it currently presents is the lack of massive empirical evidence in favor of its use as forensic ID method. This prevents many forensic experts and institutions from using it for ID.
3.5. AI-Based Approaches for Facial Reconstruction
Forensic facial reconstruction (or forensic facial approximation) is the process of recreating the face of an individual (whose identity is often not known) from his/her skeletal remains (Figure 8). Several works on the automation of facial reconstruction have been proposed during the last 15 years, giving rise to completely computerized and largely automated methods, often using CT scans as training sets [50,51,194,195,196,197,198].

Figure 8.
Forensic facial reconstruction is probably the most subjective, and one of the most controversial, techniques in the Forensic Anthropology (FA) field. In addition to remains involved in criminal investigations, facial reconstructions are created for remains believed to be of historical value and for remains of prehistoric hominids and humans. In particular, this figure displays the facial reconstruction of the Homo Heidelbergensis. Image taken from Wikimedia Commons.
Vandermulen et al. [194] presented a fully automatic procedure for craniofacial reconstruction, using a reference database of head CT scans. All reference images are automatically segmented into head volumes (enclosed by the external skin surface) and bone/skull volumes, both represented by a signed distance transform (sDT) map. The reference skull sDTs are non-linearly warped to the target skull sDT and this warping is applied to all reference skin sDTs. A linear combination of the warped reference skin sDTs is proposed as the reconstruction of the external skin surface of the target subject. Results on a pilot reference database (N = 20) show the feasibility of this approach, although further investigations are required. First, metal streak artefacts need to be removed from the images, since they possibly distort the reconstructions to an unacceptable extent. Second, the warping procedure needs to be examined more carefully, paying attention, on one hand, to better fitting the reference to the target skull and, on the other hand, to provide a smooth extrapolation of the warping. Third, other linear combinations besides the mere average need to be explored. Finally, a more extensive quantitative validation framework for the reconstructions needs to be carried out.
Tu et al. [196] proposed the automation of the reconstruction process through a data-driven 3D generative model of the face that was constructed using a database of CT head scans for a given skull. The reconstruction can be constrained based on prior knowledge such as age and/or weight. To determine whether or not these reconstructions have merit, geometric methods for comparing reconstructions against a gallery of facial images are proposed. First, Active Shape Models, a specific type of deformable model [199], are used to automatically detect a set of facial landmarks on each image. These landmarks are associated with 3D points on the reconstruction. Direct comparison of the reconstruction is problematic since, in general, the camera geometry used for image capture is unknown and there are uncertainties associated with the reconstruction and landmark detection processes. The first method of comparison uses constrained optimization to determine the optimal projection of the reconstruction onto the image. Residuals are, then, analyzed resulting in a ranking of the gallery. The second method uses boosting to learn which points are both reliable and discriminating. This results in a match/no-match classifier.
Claes et al. [48] described the common pipeline of modern facial approximation software. First, it is necessary that an expert examines the unknown skull in order to determine the BP. Then, a virtual replica of the skull is produced and represented according to the modeling parameters. A craniofacial template encoding face, skull and soft tissue information are derived from a head database. Then, an admissible geometric transformation drives the adaptation of the craniofacial template onto the unknown skull, according to the “proximity” between the skulls. As result, the template face is deformed onto the predicted face associated with the unknown skull, linking together information coming from both the database and the examination of the unknown skull. Finally, a skin texture and hairiness are added to the reconstructed face.
Guyomarc’h et al. [50] developed a computerized method for estimating facial shape based on CT scans of 500 French individuals, Anthropological Facial Approximation in Three Dimensions. Facial soft tissue depths are estimated based on age, sex, corpulence, and craniometrics, and projected using reference planes to obtain the global facial appearance. Position and shape of the eyes, nose, mouth, and ears are inferred from cranial landmarks through geometric morphometrics. The 100 estimated cutaneous landmarks are then used to warp a generic face to the target facial approximation. A validation by re-sampling on a subsample demonstrated an average accuracy of ≈4 mm for the overall face. The resulting approximation is an objective probable facial shape, but is also synthetic (i.e., without texture), and therefore needs to be enhanced artistically prior to its use in forensic cases. This facial approximation approach is integrated in the TIVMI software and is available freely for further testing.
De Buhan and Nardoni [51] combine classical features as the use of a skulls/faces database, and more original aspects: (1) a shape matching method is used to link the unknown skull to the database templates; and (2) the final face is seen as an elastic 3D mask that is deformed and adapted onto the unknown skull. In this method, the skull is considered as a whole surface and not restricted to some anatomical landmarks, allowing a dense description of the skull/face relationship. Liu and Li [197] employed a database of portrait photos to create many face candidates, and then performed a superimposition. First, they build an effective autoencoder for image-based facial reconstruction and, second, they use a generative model for constrained face inpainting. Their experiments have demonstrated that the proposed pipeline was stable and accurate. Imaizumi et al. [198] developed a software solution for 3D facial approximation from the skull based on CT scans of the head obtained from 59 Japanese adult volunteers (40 males, 19 females). The positional relationship between the skull and head surface shape were analyzed by creating anatomically homologous shape models. Before modeling, skull shapes were simplified by concealing hollow structures of the skull. Surficial tissue thickness, represented by the distance between corresponding vertices of the simplified skull and head surface, was calculated for each individual, and averaged for each sex. Although the approximate head shapes of known individuals showed a relatively good resemblance in both the shape of the whole head and facial parts, some errors were identified, particularly in areas with thick superficial tissue at the cheek, and thicker tissue at the glabella, nose, mouth, and chin. Moreover, they created referential models for CFS from average models of the skull and head surface shape for each sex.
Recently developed computer-enabled tools have facilitated the estimation of face shape from genetic sequences [200,201]. Called “molecular photofitting” [202], these methods can supplement facial approximation methods to predict faces being especially useful for morphologies with limited tangible relationships to the skeletal structure. In terms of specific face traits, red hair color and blue/brown colors of the iris are regarded as accurately predictable from genes alone [203]. Approximately 70% accuracy has been recorded for red hair prediction [204] whereas positive predictive intervals of colours of the iris ranged from 66–100% for blue eyes and 70–100% for brown eyes [205,206,207,208,209]. Typically, positive predictive values for brown eyes were higher (>85%) than for blue (>75%), with a drastic reduction in the same statistic for the so-called intermediate eye colours [206]. Predictive models for skin color are also being investigated, tested, and validated [210,211].
4. Discussion and Conclusions
In this article, we have reviewed some of the main works applying AI techniques to different biomedical image modalities (mainly X-ray images, CTs and MRIs, but also 3D surface scans of PM materials, i.e., bone remains) with the purpose of contributing to forensic human identification of deceased and living individuals. SFI is one of the main tools at our disposal when techniques like DNA analysis or fingerprint comparison cannot be applied, because there is no second sample to compare with or because the materials are so degraded that soft tissues are not preserved, among other reasons.
AI techniques have been applied with remarkable success in many challenging tasks, including healthcare and medical imaging. From this point of view, it is highly surprising the residual presence that AI currently has in forensic anthropologists’ daily practice. Even if some integral tools start to arise [212], to date forensic experts have no AI-based tools available to automate SFI tasks.
Some of the solutions reviewed in this manuscript employ CV, SC and ML techniques to automate identification techniques such as CFS or CR, through the unbiased and accurate processing, analysis and comparison of AM data and PM data. Moreover, some of the presented solutions would enable fast multiple comparisons, offering efficient filtering tools that can largely reduce the candidates of a database in minutes instead of days. On the other hand, decisions could be supported on objective and reproducible results that, perhaps, would have a stronger impact in a court of law. The studies cited in this manuscript show that AI techniques, like neural networks, can be trained to estimate the BP, describe trauma- or pathological conditions, of an individual from skeletal remains or radiographic images. Other techniques are able to contribute to automatically perform the visual comparison of anatomical structures with high accuracy and have the potential to remove human bias in all these tasks.
We identify a set of limitations and opportunities in the field of AI-based solutions to FA problems involving biomedical images. Regarding the limitations, we could mention the following:
- The reduced number of multidisciplinary research groups including forensic researchers in anthropology and related sciences (odontology, forensic medicine or anatomy) and AI experts. This interaction is essential in order to establish a fluid and productive collaboration between different scientific disciplines. On one hand, it allows unifying terminology and facilitating the useful transfer of knowledge among scientists. On the other hand, it allows joining forces in a common direction towards challenging research projects. This interdisciplinary collaboration should bear fruit in the recognition that machines are not here to replace human beings, but to assist them and facilitate their work in those tasks that humans do not want perform or want to perform faster and easier.
- With few and commendable exceptions [213,214], there is a lack of large and open public data sets for research purposes in FA. Currently, data availability is the starting point and requirement for the validation and comparison of many proposed AI techniques (like DL approaches). This is specially true nowadays, in the DL era; since we need to remark one of the main characteristics, and at the same time limitations, of ML models (largely used nowadays in many of the described tasks, like classification, regression, or segmentation): the result will be as good as the data available, so if the dataset used to train these systems is not broad and diverse enough, the results will surely be suboptimal. Furthermore, to compare the performance of newly developed methods, a common forensic dataset of known case studies should be available. This is a common practice in other fields as close as clinical medicine and biomedical research, with representative examples such as classification and localization of common thorax diseases in chest X-ray [215], melanoma classification in skin photographs [216], the cancer imaging archive [217], the Allen mouse and human brain atlases for combining genomics and neuroanatomy [218], the MIMIC critical care database that contains health data associated with approximately 40,000 critical care patients [219], the Alzheimer’s Disease neuroimaging initiative [220], or the OpenNeuro initiative for sharing MRI and fMRI data [221], among others. It is also important to recognize the fact that it is not only about acquiring and storing data, but how these data are compiled in order to meet ethical requirements and avoid machine bias [222], among others. All this problematic, related to the absence of large and complete datasets, may also be at the root of the inadequacy found in how some methods calculate, express and interpret the obtained errors (for instance, in age estimation [223]).
Associated with those limitations, we find the following opportunities in this research field:
- Many of today’s most popular and successful AI techniques to tackle imaging problems are rarely used in FA applications. This fact will undoubtedly open a niche for its application. We can think about techniques of few-shot learning [224] (to train models from a few examples), generative adversarial networks [225] or non-adversarial generative models [226] (for example, to generate data when these are scarce), or Siamese networks [227] (to establish if two images provided as input belong to the same class). Similarly, the use of 3D ConvNets [228] to directly process three-dimensional information is non-existent, as well as the combined use of recurrent and convolutional networks [229] to carry out more complex tasks (such as the textual description of images automatically, i.e., image captioning). Even the automation of certain tasks, like landmarking [230] or head pose estimation [231], is seldom performed using state-of-the-art ML/CV approaches.
- As in many other medical applications, the accuracy of the results is not the only goal to be accomplished. The results are also intended to be understandable by human users. The recently introduced concept of explainable AI encompasses AI systems for opening black box models, for improving the understanding and comprehensibility of what the models have learned, and/or for explaining individual predictions [232,233]. FA demands solutions designed considering the required explainability of some particular tasks while achieving the required performance and accuracy, providing the designed (human-centric) models or decision support systems with the ability to trust the system output [234]. These types of approaches are very scarce within the research topic addressed in this article. However, its introduction into the field would be highly desirable, due to the necessity for the conclusions of a FA report to be understood by a medical examiner or a judge.
We end this article with some of the specific activities or interventions that we consider preferential in order to fill the technological gap in SFI. These represent a list of future works in the fields of CFS, CR and BP, the three main forensic identification methodologies addressed in this work:
- In relation to CFS, it would be especially relevant to carry out systematic and massive studies to verify its effectiveness as a forensic identification technique. AI techniques would be very useful in carrying out massive experiments and in automating and objectifying the sub-tasks involved in the CFS process. All this would contribute to increasing the scientific support of CFS, as well as considering CFS as a primary identification technique (instead of secondary) (Remember that primary identification methods are those that allow positive identification of a person. While secondary identification techniques are those that allow candidates to be discarded, i.e., they do not “tell” if the AM and PM materials correspond to the same person, but rather if they are not compatible and, therefore, do not correspond to the same person.). In practical terms, this massive validation would proceed under the hypothesis that the skull is like a fingerprint, which, in case this hypothesis is empirically validated, would serve to give more validity to CFS as evidence in a court of justice.
- CR already is a primary identification technique. The main task to be carried out would be the automation and integration of all stages of the CR process:
- -
- Automatic analysis and segmentation of any type of skeletal structure in any type of image, radiographs or CT scans.
- -
- Automatic location and classification of morphological patterns on radiographs and CT scans that can help with identification.
- -
- Automatic superimposition of AM and PM materials, regardless of whether they are radiographs, 3D surface models and/or CT scans.
- -
- Lastly, the development of a decision support system able to aggregate as much information as possible to help the forensic expert in the decision-making process.
- In relation to BP, there is also a real need (and urgent need in many cases (For instance, the age estimation of living individuals, that could be related to cases of pedophilia in pictures or videos, or the determination of the legal age in cases unaccompanied minor migrants.)) to incorporate tools that facilitate BP estimation.
- -
- The usual methodology in FA is generally limited in effectiveness. Many of these methods use very subjective criteria and, in many cases, have not been properly validated [223]. Some of the most commonly used BP methods were developed 50–100 years ago, as Todd’s ten-phase method to analyze the pubic symphysis to estimate age [235], the Greulich and Pyle method based on the atlas [26] from the late fifties, and that of Tanner-Whitehouse [79], developed in 1975. These were elaborated with a limited number of examples (a few hundred), normally biased by a common ancestry, and a limited age range compared to modern populations. In addition, the phases/patterns were observed and documented by one or a few researchers (based on their experience, knowledge and ability to recognize patterns), and a large group of methods is based on a series of linear measurements directly taken from the bone with the caliber or on radiographic images. From this point of view, it is necessary to develop identification methods with a higher degree of certainty and discriminant capability, which ensure compliance with scientific standards and, consequently, their admissibility as expert evidence.
- -
- Despite the publication of new and more sophisticated methods for BP estimation, these are not integrated into the usual practice of FA. Such daily practice is almost exclusively based on the use of manual measurement tools and spreadsheets, and thus practitioners usually perceive these new methods as more complex than traditional approaches. The use of specific software is thus limited to a small number of practitioners. The lack of tools and computer platforms that facilitate and integrate the use of more complex and effective methods causes anthropologists to continue using methods that are already obsolete, though easy to use.
- -
- In order to face the two former limitations, it is also essential to address the difficulty of obtaining appropriate data/case studies for the development and validation of BP methods. On the one hand, as already mentioned before, study samples are very scarce or difficult to achieve. We refer, for example, to skeletal human remains, access to corpses available for research, invasive clinical studies, etc. On the other hand, such samples must meet important requirements: absence of pathological conditions or trauma (main limitation in clinical studies), both sexes, age groups and different ancestries well represented, complete and reliable AM information from official records, etc.
Funding
This work was supported by the Spanish Ministry of Science, Innovation and Universities, and European Regional Development Funds (ERDF) under grants EXASOCO (PGC2018-101216-B-I00) and RTI2018-095894-B-I00; by the Regional Government of Andalusia under grant EXAISFI (P18-FR-4262); and by the Instituto de Salud Carlos III, Government of Spain and ERDF through the DTS18/00136 project. Pablo Mesejo is funded by the European Commission H2020-MSCA-IF-2016 through the Skeleton-ID Marie Curie Individual Fellowship [reference 746592]. Dr. Ibáñez’s work is funded by Spanish Ministry of Science, Innovation and Universities-CDTI, Neotec program 2019 [reference EXP-00122609/SNEO-20191236]. Also, this work has received financial support from the ERDF and the Xunta de Galicia, Centro de Investigación del Sistema Universitario de Galicia, Ref. ED431G 2019/01.
Conflicts of Interest
Dr. Mesejo, Dr. Martos and Dr. Ibáñez are partners of Panacea Cooperative Research, which holds and markets SkeletonID, an AI-based software tool to support the forensic expert in human identification tasks.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| AM | Ante-Mortem |
| BP | Biological Profile |
| CFS | Craniofacial Superimposition |
| ConvNet | Convolutional Neural Network |
| CR | Comparative Radiography |
| CT | Computed Tomography |
| CV | Computer Vision |
| DL | Deep Learning |
| FA | Forensic Anthropology |
| ID | Human Identification |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| PM | Post-Mortem |
| ROI | Region of Interest |
| SC | Soft Computing |
| SFI | Skeleton-based Forensic Identification |
| SFO | Skull-Face Overlay |
References
- Thompson, T.; Black, S. Forensic Human Identification: An Introduction; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Thibault, E.A.; Lynch, L.M.; McBride, R.B.; Walsh, G. Proactive Police Management; Prentice Hall: Upper Saddle River, NJ, USA, 2004. [Google Scholar]
- Ubelaker, D.H. Forensic anthropology: Methodology and diversity of applications. In Biological Anthropology of the Human Skeleton; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 41–69. [Google Scholar]
- Beauthier, J.P.; Valck, E.; Lefevre, P.; Winne, J.D. Mass disaster victim identification: The tsunami experience. Open Forensic Sci. J. 2009, 2. [Google Scholar] [CrossRef]
- Damas, S.; Cordón, O.; Ibáñez, O.; Santamaría, J.; Alemán, I.; Botella, M.; Navarro, F. Forensic identification by computer-aided craniofacial superimposition: A survey. ACM Comput. Surv. (CSUR) 2011, 43, 1–27. [Google Scholar] [CrossRef]
- Nissan, E. Computer Applications for Handling Legal Evidence, Police Investigation and Case Argumentation; Springer: Berlin/Heidelberg, Germany, 2012; Volume 5. [Google Scholar]
- Jain, A.K.; Li, S.Z. Handbook of Face Recognition; Springer: Berlin/Heidelberg, Germany, 2011; Volume 1. [Google Scholar]
- Valentine, T.; Davis, J.P. Forensic facial identification: A practical guide to best practice. In Forensic Facial Identification: Theory and Practice of Identification from Eyewitnesses, Composites and CCTV; John Wiley & Sons: Chichester, UK, 2015; pp. 323–347. [Google Scholar]
- Stephan, C.N.; Caple, J.M.; Guyomarc’h, P.; Claes, P. An overview of the latest developments in facial imaging. Forensic Sci. Res. 2019, 4, 10–28. [Google Scholar] [CrossRef]
- Zhao, W.; Chellappa, R.; Phillips, P.; Rosenfeld, A. Face recognition: A literature survey. ACM Comput. Surv. 2003, 35, 399–458. [Google Scholar] [CrossRef]
- Ding, C.; Tao, D. A comprehensive survey on pose-invariant face recognition. ACM Trans. Intell. Syst. Technol. 2016, 7, 1–42. [Google Scholar] [CrossRef]
- England, N.; Improvement, N. Diagnostic Imaging Dataset Statistical Release; Department of Health: London, UK, 2016; Volume 421.
- Laserson, J.; Lantsman, C.D.; Cohen-Sfady, M.; Tamir, I.; Goz, E.; Brestel, C.; Bar, S.; Atar, M.; Elnekave, E. Textray: Mining clinical reports to gain a broad understanding of chest x-rays. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 553–561. [Google Scholar]
- Yen, K.; Lövblad, K.O.; Scheurer, E.; Ozdoba, C.; Thali, M.J.; Aghayev, E.; Jackowski, C.; Anon, J.; Frickey, N.; Zwygart, K.; et al. Post-mortem forensic neuroimaging: Correlation of MSCT and MRI findings with autopsy results. Forensic Sci. Int. 2007, 173, 21–35. [Google Scholar] [CrossRef]
- Obenauer, S.; Herold, T.; Fischer, U.; Grabbe, E.; Fadjasch, G.; Saternus, K.; Koebke, J. Evaluation of injuries of the upper cervical spine in a postmortem study with digital radiography, CT and MRI. RoeFo-Fortschritte auf dem Gebiete der Roentgenstrahlen und der Neuen Bildgebenden Verfahren 1999, 171, 473–479. [Google Scholar]
- Ferembach, D. Recomendations for age and sex diagnosis of skeletons. J. Hum. Evolut. 1980, 9, 517–549. [Google Scholar]
- Aguilera, I.A. Determinación del Sexo en el Esqueleto Postcraneal. Estudio de una Población Mediterránea Actual. Ph.D. Thesis, Universidad de Granada, Granada, Spain, 1997. [Google Scholar]
- Olivares, J.I.; Aguilera, I.A. Validation of the sex estimation method elaborated by Schutkowski in the Granada Osteological Collection of identified infant and young children: Analysis of the controversy between the different ways of analyzing and interpreting the results. Int. J. Leg. Med. 2016, 130, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.; Suchey, J.M. Skeletal age determination based on the os pubis: A comparison of the Acsádi-Nemeskéri and Suchey-Brooks methods. Hum. Evolut. 1990, 5, 227–238. [Google Scholar] [CrossRef]
- Lamendin, H.; Baccino, E.; Humbert, J.; Tavernier, J.; Nossintchouk, R.; Zerilli, A. A simple technique for age estimation in adult corpses: The two criteria dental method. J. Forensic Sci. 1992, 37, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Baccino, E.; Zerilli, A. The two step strategy (TSS) or the right way to combine a dental (Lamendin) and an anthropological (Suchey–Brooks system) method for age determination. Proc. Am. Acad. Forensic Sci. 1997, 3, 150. [Google Scholar]
- Lovejoy, C.O.; Meindl, R.S.; Pryzbeck, T.R.; Mensforth, R.P. Chronological metamorphosis of the auricular surface of the ilium: A new method for the determination of adult skeletal age at death. Am. J. Phys. Anthropol. 1985, 68, 15–28. [Google Scholar] [CrossRef]
- Işcan, M.Y.; Loth, S.R.; Wright, R.K. Age estimation from the rib by phase analysis: White males. J. Forensic Sci. 1984, 29, 1094–1104. [Google Scholar]
- Meindl, R.S.; Lovejoy, C.O. Ectocranial suture closure: A revised method for the determination of skeletal age at death based on the lateral-anterior sutures. Am. J. Phys. Anthropol. 1985, 68, 57–66. [Google Scholar] [CrossRef]
- Scheuer, L.; Black, S. The Juvenile Skeleton; Elsevier: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Greulich, W.W.; Pyle, S.I. Radiographic Atlas of Skeletal Development of the Hand and Wrist; Stanford University Press: Redwood City, CA, USA, 1959. [Google Scholar]
- Demirjian, A.; Goldstein, H.; Tanner, J.M. A new system of dental age assessment. Hum. Biol. 1973, 45, 211–227. [Google Scholar] [PubMed]
- Kellinghaus, M.; Schulz, R.; Vieth, V.; Schmidt, S.; Schmeling, A. Forensic age estimation in living subjects based on the ossification status of the medial clavicular epiphysis as revealed by thin-slice multidetector computed tomography. Int. J. Leg. Med. 2010, 124, 149–154. [Google Scholar] [CrossRef]
- Nunes de Mendonça, M. Contribución para la identificación humana a partir del estudio de las estructuras óseas. In Determinacion de la Talla a Traves de la Longitud de los Huesos Largos; Universidad Complutense de Madrid: Madrid, Spain, 1998. [Google Scholar]
- Belmonte, M. Determinación de la Estatura a Través de la Tibia en Población Española Contemporánea. Ph.D. Thesis, Universidad de Granada, Granada, Spain, 2012. [Google Scholar]
- Trotter, M.; Gleser, G.C. A re-evaluation of estimation of stature based on measurements of stature taken during life and of long bones after death. Am. J. Phys. Anthropol. 1958, 16, 79–123. [Google Scholar] [CrossRef]
- Ousley, S.D.; Jantz, R.L. FORDISC 2.0: Personal Computer Forensic Discriminant Functions; University of Tennessee: Knoxville, TX, USA, 1996. [Google Scholar]
- Stephan, C.N.; Amidan, B.; Trease, H.; Guyomarc’h, P.; Pulsipher, T.; Byrd, J.E. Morphometric comparison of clavicle outlines from 3D bone scans and 2D chest radiographs: A shortlisting tool to assist radiographic identification of human skeletons. J. Forensic Sci. 2014, 59, 306–313. [Google Scholar] [CrossRef]
- Christensen, A.M.; Smith, M.A.; Gleiber, D.S.; Cunningham, D.L.; Wescott, D.J. The Use of X-ray Computed Tomography Technologies in Forensic Anthropology. Forensic Anthropol. 2018, 1, 124. [Google Scholar] [CrossRef]
- Hatch, G.M.; Dedouit, F.; Christensen, A.M.; Thali, M.J.; Ruder, T.D. RADid: A pictorial review of radiologic identification using postmortem CT. J. Forensic Radiol. Imaging 2014, 2, 52–59. [Google Scholar] [CrossRef]
- Thali, M.J.; Braun, M.; Dirnhofer, R. Optical 3D surface digitizing in forensic medicine: 3D documentation of skin and bone injuries. Forensic Sci. Int. 2003, 137, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Fleischman, J.M. Radiographic identification using midline medical sternotomy wires. J. Forensic Sci. 2015, 60, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Iscan, M.Y.; Helmer, R. Forensic Analysis of the Skull; Willey-Liss: New York, NY, USA, 1993. [Google Scholar]
- Stephan, C.N. Craniofacial identification: Techniques of facial approximation and craniofacial superimposition. In Handbook of Forensic Anthropology and Archaeology; Left Coast Press: Walnut Creek, CA, USA, 2009; Volume 25, pp. 304–321. [Google Scholar]
- Damas, S.; Wilkinson, C.; Kahana, T.; Veselovskaya, E.; Abramov, A.; Jankauskas, R.; Jayaprakash, P.; Ruiz, E.; Navarro, F.; Huete, M.; et al. Study on the performance of different craniofacial superimposition approaches (II): Best practices proposal. Forensic Sci. Int. 2015, 257, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Huete, M.; Kahana, T.; Ibáñez, O. Past, present, and future of CFS: Literature and international surveys. Leg. Med. 2015, 17, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Damas, S.; Cordón, O.; Ibáñez, O. Handbook on Craniofacial Superimposition: The MEPROCS Project; Springer Nature: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Stephan, C.N.; Henneberg, M. Building faces from dry skulls: Are they recognized above chance rates? J. Forensic Sci. 2001, 46, 432–440. [Google Scholar] [CrossRef]
- Taylor, R.; Craig, P. The wisdom of bones: Facial approximation on the skull. In Computer Graphic Facial Reconstruction; Academic Press: Boston, MA, USA, 2005; pp. 33–55. [Google Scholar]
- Wilkinson, C. Forensic Facial Reconstruction; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Wilkinson, C.; Rynn, C. Craniofacial Identification; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Lee, W.J.; Wilkinson, C.M.; Hwang, H.S. An accuracy assessment of forensic computerized facial reconstruction employing cone-beam computed tomography from live subjects. J. Forensic Sci. 2012, 57, 318–327. [Google Scholar] [CrossRef]
- Claes, P.; Vandermeulen, D.; De Greef, S.; Willems, G.; Clement, J.G.; Suetens, P. Computerized craniofacial reconstruction: Conceptual framework and review. Forensic Sci. Int. 2010, 201, 138–145. [Google Scholar] [CrossRef]
- Parks, C.L.; Richard, A.H.; Monson, K.L. Preliminary performance assessment of computer automated facial approximations using computed tomography scans of living individuals. Forensic Sci. Int. 2013, 233, 133–139. [Google Scholar] [CrossRef]
- Guyomarc’h, P.; Dutailly, B.; Charton, J.; Santos, F.; Desbarats, P.; Coqueugniot, H. Anthropological Facial Approximation in Three Dimensions (AFA3D): Computer-Assisted Estimation of the Facial Morphology Using Geometric Morphometrics. J. Forensic Sci. 2014, 59, 1502–1516. [Google Scholar] [CrossRef]
- de Buhan, M.; Nardoni, C. A facial reconstruction method based on new mesh deformation techniques. Forensic Sci. Res. 2018, 3, 256–273. [Google Scholar] [CrossRef]
- Foster, K.R.; Huber, P.W. Judging Science: Scientific Knowledge and the Federal Courts; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Forsyth, D.A.; Ponce, J. Computer Vision: A Modern Approach; Prentice Hall Professional Technical Reference: Upper Saddle River, NJ, USA, 2002. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Engelbrecht, A.P. Computational Intelligence: An Introduction; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Zadeh, L.A. Soft computing and fuzzy logic. In Fuzzy Sets, Fuzzy Logic, and Fuzzy Systems: Selected Papers by Lotfi a Zadeh; World Scientific: Singapore, 1996; pp. 796–804. [Google Scholar]
- Eiben, A.E.; Smith, J.E. Introduction to Evolutionary Computing; Springer: Berlin/Heidelberg, Germany, 2003; Volume 53. [Google Scholar]
- Aja-Fernández, S.; de Luis-Garcıa, R.; Martın-Fernández, M.A.; Alberola-López, C. A computational TW3 classifier for skeletal maturity assessment. A computing with words approach. J. Biomed. Inform. 2004, 37, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.; Ebner, T.; Bischof, H.; Grassegger, S.; Ehammer, T.; Urschler, M. Fully automatic bone age estimation from left hand MR images. Med. Image Comput. Comput. Assist. Interv. 2014, 17, 220–227. [Google Scholar] [PubMed]
- Mansourvar, M.; Ismail, M.A.; Herawan, T.; Gopal Raj, R.; Abdul Kareem, S.; Nasaruddin, F.H. Automated bone age assessment: Motivation, taxonomies, and challenges. Comput. Math. Methods Med. 2013, 2013, 391626. [Google Scholar] [CrossRef]
- Pinto, S.C.D.; Urbanová, P.; Cesar, R.M., Jr. Two-Dimensional Wavelet Analysis of Supraorbital Margins of the Human Skull for Characterizing Sexual Dimorphism. IEEE Trans. Inf. Forensics Secur. 2016, 11, 1542–1548. [Google Scholar] [CrossRef]
- Abdullah, H.; Jamil, M.M.A.; Nor, F.M. Automated Haversian Canal Detection for Histological Sex Determination. In Proceedings of the IEEE Symposium on Computer Applications & Industrial Electronics (ISCAIE), Langkawi, Malaysia, 24–25 April 2017; pp. 69–74. [Google Scholar]
- Pietka, E.; Gertych, A.; Pospiech, S.; Cao, F.; Huang, H.; Gilsanz, V. Computer-assisted bone age assessment: Image preprocessing and epiphyseal/metaphyseal ROI extraction. IEEE Trans. Med. Imaging 2001, 20, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Pietka, E. Computer-assisted bone age assessment—Database adjustment. Int. Congr. Ser. 2003, 1256, 87–92. [Google Scholar] [CrossRef]
- Pietka, E.; Gertych, A.; Pospiechâ, S.; Cao, F.; Huang, H.; Gilzanz, V. Computer-assisted bone age assessment: Graphical user interface for image processing and comparison. J. Digit. Imaging 2004, 17, 175–188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afifi, M. 11K Hands: Gender recognition and biometric identification using a large dataset of hand images. Multimedia Tools Appl. 2019, 78, 20835–20854. [Google Scholar] [CrossRef]
- du Jardin, P.; Ponsaillé, J.; Alunni-Perret, V.; Quatrehomme, G. A comparison between neural network and other metric methods to determine sex from the upper femur in a modern French population. Forensic Sci. Int. 2009, 192, 127.e1-6. [Google Scholar] [CrossRef] [PubMed]
- Navega, D.; Vicente, R.; Vieira, D.N.; Ross, A.H.; Cunha, E. Sex estimation from the tarsal bones in a Portuguese sample: A machine learning approach. Int. J. Leg. Med. 2015, 129, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Buikstra, J.E. Standards for data collection from human skeletal remains. Ark. Archaeol. Surv. Res. Ser. 1994, 44. [Google Scholar] [CrossRef]
- Krishan, K.; Chatterjee, P.M.; Kanchan, T.; Kaur, S.; Baryah, N.; Singh, R. A review of sex estimation techniques during examination of skeletal remains in forensic anthropology casework. Forensic Sci. Int. 2016, 261, 165.e1–165.e8. [Google Scholar] [CrossRef]
- Sierp, I.; Henneberg, M. The Difficulty of Sexing Skeletons from Unknown Populations. J. Anthropol. 2015, 2015, 1–13. [Google Scholar] [CrossRef]
- Darmawan, M.F.; Yusuf, S.M.; Rozi, M.A.; Haron, H. Hybrid PSOANN for sex estimation based on length of left hand bone. In Proceedings of the 2015 IEEE Student Conference on Research and Development (SCOReD), Kuala Lumpur, Malaysia, 13–14 December 2015; pp. 478–483. [Google Scholar]
- Bewes, J.; Low, A.; Morphett, A.; Pate, F.D.; Henneberg, M. Artificial intelligence for sex determination of skeletal remains: Application of a deep learning artificial neural network to human skulls. J. Forensic Leg. Med. 2019, 62, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Kaloi, M.A.; He, K. Child Gender Determination with Convolutional Neural Networks on Hand Radio-Graphs. arXiv 2018, arXiv:1811.05180. [Google Scholar]
- Mahfouz, M.; Badawi, A.; Merkl, B.; Fatah, E.E.A.; Pritchard, E.; Kesler, K.; Moore, M.; Jantz, R.; Jantz, L. Patella sex determination by 3D statistical shape models and nonlinear classifiers. Forensic Sci. Int. 2007, 173, 161–170. [Google Scholar] [CrossRef]
- Yang, W.; Liu, X.; Wang, K.; Hu, J.; Geng, G.; Feng, J. Sex determination of three-dimensional skull based on improved backpropagation neural network. Comput. Math. Methods Med. 2019, 2019, 9163547. [Google Scholar] [CrossRef]
- Arigbabu, O.A.; Liao, I.Y.; Abdullah, N.; Noor, M.H.M. Can computer vision techniques be applied to automated forensic examinations? A study on sex identification from human skulls using head CT scans. In Proceedings of the Asian Conference on Computer Vision, Taipei, Taiwan, 20–24 November 2016; pp. 342–359. [Google Scholar]
- Tanner, J.M.; Whitehouse, R.; Cameron, N.; Marshall, W.; Healy, M.; Goldstein, H. Assessment of Skeletal Maturity and Prediction of Adult Height (TW2 Method); Saunders: London, UK, 2001. [Google Scholar]
- Prieto, J.; Mihaila, S.; Hilaire, A.; Fanton, L.; Odet, C.; Revol-Muller, C. Age estimation from 3D X-ray CT images of human fourth ribs. In Proceedings of the International Conference on Image Processing, Computer Vision, and Pattern Recognition (IPCV), Las Vegas, NV, USA, 16–19 July 2012; p. 1. [Google Scholar]
- Breen, M.A.; Tsai, A.; Stamm, A.; Kleinman, P.K. Bone age assessment practices in infants and older children among Society for Pediatric Radiology members. Pediatr. Radiol. 2016, 46, 1269–1274. [Google Scholar] [CrossRef]
- Malina, R.M.; Beunen, G.P. Assessment of skeletal maturity and prediction of adult height (TW3 method). Am. J. Hum. Biol. 2002, 14, 788–789. [Google Scholar] [CrossRef]
- Pinchi, V.; De Luca, F.; Ricciardi, F.; Focardi, M.; Piredda, V.; Mazzeo, E.; Norelli, G.A. Skeletal age estimation for forensic purposes: A comparison of GP, TW2 and TW3 methods on an Italian sample. Forensic Sci. Int. 2014, 238, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Shim, W.H.; Yoon, H.M.; Hong, S.H.; Lee, J.S.; Cho, Y.A.; Kim, S. Computerized Bone Age Estimation Using Deep Learning Based Program: Evaluation of the Accuracy and Efficiency. Am. J. Roentgenol. 2017, 209, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.B.; Chen, M.C.; Lungren, M.P.; Halabi, S.S.; Stence, N.V.; Langlotz, C.P. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018, 287, 313–322. [Google Scholar] [CrossRef]
- Lee, H.; Tajmir, S.; Lee, J.; Zissen, M.; Yeshiwas, B.A.; Alkasab, T.K.; Choy, G.; Do, S. Fully automated deep learning system for bone age assessment. J. Digit. Imaging 2017, 30, 427–441. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, K.G. Applying Deep Learning in Medical Images: The Case of Bone Age Estimation. Healthc. Inform. Res. 2018, 24, 86–92. [Google Scholar] [CrossRef]
- Mutasa, S.; Chang, P.D.; Ruzal-Shapiro, C.; Ayyala, R. MABAL: A Novel Deep-Learning Architecture for Machine-Assisted Bone Age Labeling. J. Digit. Imaging 2018, 31, 513–519. [Google Scholar] [CrossRef]
- Hsieh, C.W.; Jong, T.L.; Chou, Y.H.; Tiu, C.M. Computerized geometric features of carpal bone for bone age estimation. Chin. Med. J. 2007, 1, 767–770. [Google Scholar] [CrossRef]
- Mansourvar, M.; Asemi, A.; Raj, R.G.; Kareem, S.A.; Antony, C.D.; Idris, N.; Baba, M.S. A fuzzy inference system for skeletal age assessment in living individual. Int. J. Fuzzy Syst. 2017, 19, 838–848. [Google Scholar] [CrossRef]
- Spampinato, C.; Palazzo, S.; Giordano, D.; Aldinucci, M.; Leonardi, R. Deep learning for automated skeletal bone age assessment in X-ray images. Med. Image Anal. 2017, 36, 41–51. [Google Scholar] [CrossRef]
- Rucci, M.; Coppini, G.; Nicoletti, I.; Cheli, D.; Valli, G. Automatic analysis of hand radiographs for the assessment of skeletal age: A subsymbolic approach. Comput. Biomed. Res. 1995, 28, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Gross, G.W.; Boone, J.M.; Bishop, D.M. Pediatric skeletal age: Determination with neural networks. Radiology 1995, 195, 689–695. [Google Scholar] [CrossRef]
- Mahmoodi, S.; Sharif, B.S.; Chester, E.G.; Owen, J.P.; Lee, R. Skeletal growth estimation using radiographic image processing and analysis. IEEE Trans. Inf. Technol. Biomed. 2000, 4, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Gertych, A.; Zhang, A.; Sayre, J.; Pospiech-Kurkowska, S.; Huang, H. Bone age assessment of children using a digital hand atlas. Comput. Med. Imaging Graph. 2007, 31, 322–331. [Google Scholar] [CrossRef]
- Gertych, A.; Piętka, E.; Liu, B.J. Segmentation of regions of interest and post-segmentation edge location improvement in computer-aided bone age assessment. Pattern Anal. Appl. 2007, 10, 115–123. [Google Scholar] [CrossRef]
- Hsieh, C.; Jong, T.; Tiu, C. Bone age estimation based on phalanx information with fuzzy constrain of carpals. Med. Biol. Eng. Comput. 2007, 45, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Gertych, A.; Liu, B.J. Automatic bone age assessment for young children from newborn to 7-year-old using carpal bones. Comput. Med. Imaging Graph. 2007, 31, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qi, J.; Liu, Z.; Ning, Q.; Luo, X. Automatic bone age assessment based on intelligent algorithms and comparison with TW3 method. Comput. Med. Imaging Graph. 2008, 32, 678–684. [Google Scholar] [CrossRef]
- Tristán-Vega, A.; Arribas, J.I. A radius and ulna TW3 bone age assessment system. IEEE Trans. Biomed. Eng. 2008, 55, 1463–1476. [Google Scholar] [CrossRef] [PubMed]
- Thodberg, H.H.; Kreiborg, S.; Juul, A.; Pedersen, K.D. The BoneXpert method for automated determination of skeletal maturity. IEEE Trans. Med. Imaging 2009, 28, 52–66. [Google Scholar] [CrossRef]
- Thodberg, H.H. An automated method for determination of bone age. J. Clin. Endocrinol. Metab. 2009, 94, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Spampinato, C.; Scarciofalo, G.; Leonardi, R. An Automatic System for Skeletal Bone Age Measurement by Robust Processing of Carpal and Epiphysial/Metaphysial Bones. IEEE Trans. Instrum. Meas. 2010, 59, 2539–2553. [Google Scholar] [CrossRef]
- Martin, D.D.; Meister, K.; Schweizer, R.; Ranke, M.B.; Thodberg, H.H.; Binder, G. Validation of automatic bone age rating in children with precocious and early puberty. J. Pediatr. Endocrinol. Metab. 2011, 24, 1009–1014. [Google Scholar] [CrossRef]
- Davis, L.M.; Theobald, B.; Bagnall, A.J. Automated Bone Age Assessment Using Feature Extraction. In Intelligent Data Engineering and Automated Learning (IDEAL); Yin, H., Costa, J.A.F., Barreto, G.D.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7435, pp. 43–51. [Google Scholar]
- Mansourvar, M.; Raj, R.G.; Ismail, M.A.; Kareem, S.A.; Shanmugam, S.; Wahid, S.; Mahmud, R.; Abdullah, R.H.; Nasaruddin, F.H.F.; Idris, N. Automated web based system for bone age assessment using histogram technique. Malays. J. Comput. Sci. 2012, 25, 107–121. [Google Scholar]
- Lin, H.; Shu, S.; Lin, Y.; Yu, S. Bone age cluster assessment and feature clustering analysis based on phalangeal image rough segmentation. Pattern Recognit. 2012, 45, 322–332. [Google Scholar] [CrossRef]
- Adeshina, S.A.; Lindner, C.; Cootes, T.F. Automatic segmentation of carpal area bones with random forest regression voting for estimating skeletal maturity in infants. In Proceedings of the 2014 11th International Conference on Electronics, Computer and Computation (ICECCO), Abuja, Nigeria, 29 September–1 October 2014; pp. 1–4. [Google Scholar]
- Stern, D.; Urschler, M. From individual hand bone age estimates to fully automated age estimation via learning-based information fusion. In Proceedings of the IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 150–154. [Google Scholar]
- Giordano, D.; Kavasidis, I.; Spampinato, C. Modeling skeletal bone development with hidden Markov models. Comput. Methods Progr. Biomed. 2016, 124, 138–147. [Google Scholar] [CrossRef]
- Kashif, M.; Deserno, T.M.; Haak, D.; Jonas, S.M. Feature description with SIFT, SURF, BRIEF, BRISK, or FREAK? A general question answered for bone age assessment. Comput. Biol. Med. 2016, 68, 67–75. [Google Scholar] [CrossRef]
- Seok, J.; Kasa-Vubu, J.; DiPietro, M.A.; Girard, A.R. Expert system for automated bone age determination. Expert Syst. Appl. 2016, 50, 75–88. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Z.; Dong, X.; Liang, W.; Xue, H.; Zhang, L.; Zhang, Y.; Deng, Z. Forensic age estimation for pelvic X-ray images using deep learning. Eur. Radiol. 2019, 29, 2322–2329. [Google Scholar] [CrossRef]
- Štern, D.; Payer, C.; Urschler, M. Automated age estimation from MRI volumes of the hand. Med. Image Anal. 2019, 58, 101538. [Google Scholar] [CrossRef]
- Thodberg, H.H.; Neuhof, J.; Ranke, M.B.; Jenni, O.G.; Martin, D.D. Validation of bone age methods by their ability to predict adult height. Horm. Res. Paediatr. 2010, 74, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Mesejo, P.; Pizarro, D.; Abergel, A.; Rouquette, O.; Beorchia, S.; Poincloux, L.; Bartoli, A. Computer-aided classification of gastrointestinal lesions in regular colonoscopy. IEEE Trans. Med. Imaging 2016, 35, 2051–2063. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. J. Am. Med. Assoc. 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.L.; Hsu, C.H.; Hidayati, S.C.; Cheng, W.H.; Chen, Y.J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Therapy 2015, 8, 2015–2022. [Google Scholar] [PubMed]
- Franchi, A. Epidemiology and classification of bone tumors. Clin. Cases Miner. Bone Metab. 2012, 9, 92. [Google Scholar]
- Olczak, J.; Fahlberg, N.; Maki, A.; Razavian, A.S.; Jilert, A.; Stark, A.; Sköldenberg, O.; Gordon, M. Artificial intelligence for analyzing orthopedic trauma radiographs: Deep learning algorithms—Are they on par with humans for diagnosing fractures? Acta Orthop. 2017, 88, 581–586. [Google Scholar] [CrossRef]
- Chung, S.W.; Han, S.S.; Lee, J.W.; Oh, K.S.; Kim, N.R.; Yoon, J.P.; Kim, J.Y.; Moon, S.H.; Kwon, J.; Lee, H.J.; et al. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop. 2018, 89, 468–473. [Google Scholar] [CrossRef]
- Gupta, A.; Venkatesh, S.; Chopra, S.; Ledig, C. Generative image translation for data augmentation of bone lesion pathology. arXiv 2019, arXiv:1902.02248. [Google Scholar]
- Sandström, S.; Ostensen, H.; Pettersson, H.; Åkerman, K. The WHO Manual of Diagnostic Imaging: Radiographic Technique and Projections; World Health Organization: Brussels, Belgium, 2003; Volume 2. [Google Scholar]
- Daffner, R.H.; Hartman, M. Clinical Radiology: The Essentials; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Rigby, D.; Hacking, L. Interpreting the chest radiograph. Anaesth Intensive Care 2018, 19, 50–54. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Romeny, B.T.H.; Viergever, M.A. Computer-aided diagnosis in chest radiography: A survey. IEEE Trans. Med. Imaging 2001, 20, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.M. Assessing the variation in individual frontal sinus outlines. Am. J. Phys. Anthropol. 2005, 127, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.M. Testing the reliability of frontal sinuses in positive identification. J. Forensic Sci. 2005, 50, 18–22. [Google Scholar] [CrossRef]
- Maxwell, A.B.; Ross, A.H. A radiographic study on the utility of cranial vault outlines for positive identifications. J. Forensic Sci. 2014, 59, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Chen, H. Matching of dental X-ray images for human identification. Pattern Recognit. 2004, 37, 1519–1532. [Google Scholar] [CrossRef]
- Chen, H.; Jain, A.K. Dental biometrics: Alignment and matching of dental radiographs. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Nomir, O.; Abdel-Mottaleb, M. Human identification from dental x-ray images based on the shape and appearance of the teeth. IEEE Trans. Inf. Forensics Secur. 2007, 2, 188–197. [Google Scholar] [CrossRef]
- Caple, J.; Byrd, J.; Stephan, C.N. Elliptical fourier analysis: Fundamentals, applications, and value for forensic anthropology. Int. J. Leg. Med. 2017, 131, 1675–1690. [Google Scholar] [CrossRef]
- Devi, P.; Thimmarasa, V.; Mehrotra, V.; Singla, V. Automated dental identification system: An aid to forensic odontology. J. Indian Acad. Oral Med. Radiol. 2011, 23, 360. [Google Scholar] [CrossRef]
- Anuja, P.; Doggalli, N. Software in forensic odontology. Indian J. Multidiscip. Dent. 2018, 8, 94. [Google Scholar]
- Derrick, S.M.; Hipp, J.A.; Goel, P. The Computer-Assisted Decedent Identification Method of Computer-Assisted Radiographic Identification. In New Perspectives in Forensic Human Skeletal Identification; Academic Press: Cambridge, MA, USA, 2018; pp. 265–276. [Google Scholar]
- Tabor, Z.; Karpisz, D.; Wojnar, L.; Kowalski, P. An automatic recognition of the frontal sinus in x-ray images of skull. IEEE Trans. Biomed. Eng. 2008, 56, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Pfaeffli, M.; Vock, P.; Dirnhofer, R.; Braun, M.; Bolliger, S.A.; Thali, M.J. Post-mortem radiological CT identification based on classical ante-mortem X-ray examinations. Forensic Sci. Int. 2007, 171, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Shinkawa, N.; Hirai, T.; Nishii, R.; Yukawa, N. Usefulness of 2D fusion of postmortem CT and antemortem chest radiography studies for human identification. Jpn. J. Radiol. 2017, 35, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Niespodziewanski, E.; Stephan, C.N.; Guyomarc’h, P.; Fenton, T.W. Human Identification via Lateral Patella Radiographs: A Validation Study. J. Forensic Sci. 2016, 61, 134–140. [Google Scholar] [CrossRef] [PubMed]
- D’Alonzo, S.S.; Guyomarc’h, P.; Byrd, J.E.; Stephan, C.N. A Large-Sample Test of a Semi-Automated Clavicle Search Engine to Assist Skeletal Identification by Radiograph Comparison. J. Forensic Sci. 2017, 62, 181–186. [Google Scholar] [CrossRef]
- Gómez, O.; Ibáñez, O.; Valsecchi, A.; Cordón, O.; Kahana, T. 3D-2D silhouette-based image registration for comparative radiography-based forensic identification. Pattern Recognit. 2018, 83, 469–480. [Google Scholar] [CrossRef]
- Thali, M.J.; Brogdon, B.; Viner, M.D. Forensic Radiology; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Gómez, O.; Mesejo, P.; Ibáñez, O.; Valsecchi, A.; Cordón, O. Deep architectures for high-resolution multi-organ chest X-ray image segmentation. Neural Comput. Appl. 2019, 1–15. [Google Scholar] [CrossRef]
- Gómez, Ó.; Mesejo, P.; Ibáñez, Ó.; Cordón, Ó. Deep architectures for the segmentation of frontal sinuses in XRay images: Towards an automatic forensic identification system in comparative radiography. Neurcomputing 2020, in press. [Google Scholar]
- Gómez, Ó.; Mesejo, P.; Ibáñez, Ó.; Valsecchi, A.; Cordón, Ó. A real-coded evolutionary algorithm-based registration approach for forensic identification using the radiographic comparison of frontal sinuses. In Proceedings of the 22nd IEEE Congress on Evolutionary Computation (IEEE CEC), Glasgow, UK, 19–24 July 2020. [Google Scholar]
- Iino, M.; Fujimoto, H.; Yoshida, M.; Matsumoto, H.; Fujita, M.Q. Identification of a jawless skull by superimposing post-mortem and ante-mortem CT. J. Forensic Radiol. Imaging 2016, 6, 31–37. [Google Scholar] [CrossRef]
- Ruder, T.D.; Brun, C.; Christensen, A.M.; Thali, M.J.; Gascho, D.; Schweitzer, W.; Hatch, G.M. Comparative radiologic identification with CT images of paranasal sinuses—Development of a standardized approach. J. Forensic Radiol. Imaging 2016, 7, 1–9. [Google Scholar] [CrossRef]
- Hacl, A.; Costa, A.L.F.; Oliveira, J.M.; Tucunduva, M.J.; Girondi, J.R.; Nahás-Scocate, A.C.R. Three-dimensional volumetric analysis of frontal sinus using medical software. J. Forensic Radiol. Imaging 2017, 11, 1–5. [Google Scholar] [CrossRef]
- Deloire, L.; Diallo, I.; Cadieu, R.; Auffret, M.; Alavi, Z.; Ognard, J.; Ben Salem, D. Post-mortem X-ray computed tomography (PMCT) identification using ante-mortem CT-scan of the sphenoid sinus. J. Neuroradiol. 2019, 46, 248–255. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, D.; Foong, K.W.; Sim, T.; San Wong, Y.; Cheng, H.L. Towards automated pose invariant 3D dental biometrics. In Proceedings of the 2011 International Joint Conference on Biometrics (IJCB), Washington, DC, USA, 11–13 October 2011; pp. 1–7. [Google Scholar]
- Zhong, X.; Yu, D.; Wong, Y.S.; Sim, T.; Lu, W.F.; Foong, K.W.C.; Cheng, H.L. 3D dental biometrics: Alignment and matching of dental casts for human identification. Comput. Ind. 2013, 64, 1355–1370. [Google Scholar] [CrossRef]
- Zhang, Z.; Ong, S.H.; Zhong, X.; Foong, K.W.C. Efficient 3D dental identification via signed feature histogram and learning keypoint detection. Pattern Recognit. 2016, 60, 189–204. [Google Scholar] [CrossRef]
- Gibelli, D.; Cellina, M.; Cappella, A.; Gibelli, S.; Panzeri, M.M.; Oliva, A.G.; Termine, G.; De Angelis, D.; Cattaneo, C.; Sforza, C. An innovative 3D-3D superimposition for assessing anatomical uniqueness of frontal sinuses through segmentation on CT scans. Int. J. Leg. Med. 2019, 133, 1159–1165. [Google Scholar] [CrossRef]
- Decker, S.J.; Ford, J.M. Forensic personal identification utilizing part-to-part comparison of CT-derived 3D lumbar models. Forensic Sci. Int. 2019, 294, 21–26. [Google Scholar] [CrossRef]
- Dirnhofer, R.; Jackowski, C.; Vock, P.; Potter, K.; Thali, M.J. VIRTOPSY: Minimally invasive, imaging-guided virtual autopsy. Radiographics 2006, 26, 1305–1333. [Google Scholar] [CrossRef]
- O’Sullivan, S.; Holzinger, A.; Zatloukal, K.; Saldiva, P.; Sajid, M.I.; Wichmann, D. Machine learning enhanced virtual autopsy. Autopsy Case Rep. 2017, 7, 3. [Google Scholar] [CrossRef]
- O’Sullivan, S.; Holzinger, A.; Wichmann, D.; Saldiva, P.H.N.; Sajid, M.I.; Zatloukal, K. Virtual autopsy: Machine learning and ai provide new opportunities for investigating minimal tumor burden and therapy resistance by cancer patients. Autopsy Case Rep. 2018, 8, e2018003. [Google Scholar] [CrossRef]
- O’Sullivan, S.; Heinsen, H.; Grinberg, L.T.; Chimelli, L.; Amaro, E.; do Nascimento Saldiva, P.H.; Jeanquartier, F.; Jean-Quartier, C.; Martin, M.d.G.M.; Sajid, M.I.; et al. The role of artificial intelligence and machine learning in harmonization of high-resolution post-mortem MRI (virtopsy) with respect to brain microstructure. Brain Inform. 2019, 6, 3. [Google Scholar] [CrossRef][Green Version]
- Holzinger, A. Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Inform. 2016, 3, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, T.; Leue, B.; Lorensen, B.; Lauric, A. Computer-Aided Forensics: Metal Object Detection. Stud. Health Technol. Inform. 2006, 119, 249. [Google Scholar] [PubMed]
- Ebert, L.C.; Heimer, J.; Schweitzer, W.; Sieberth, T.; Leipner, A.; Thali, M.; Ampanozi, G. Automatic detection of hemorrhagic pericardial effusion on PMCT using deep learning—A feasibility study. Forensic Sci. Med. Pathol. 2017, 13, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Peña-Solórzano, C.; Albrecht, D.; Harris, P.; Bassed, R.; Gillam, J.; Dimmock, M. Semi-supervised labelling of the femur in a whole-body post-mortem CT database using deep learning. Comput. Biol. Med. 2020, 122, 103797. [Google Scholar] [CrossRef]
- Nickerson, B.A.; Fitzhorn, P.A.; Koch, S.K.; Charney, M. A methodology for near-optimal computational superimposition of two-dimensional digital facial photographs and three-dimensional cranial surface meshes. J. Forensic Sci. 1991, 36, 480–500. [Google Scholar] [CrossRef]
- Yoshino, M.; Imaizumi, K.; Miyasaka, S.; Seta, S. Evaluation of anatomical consistency in craniofacial superimposition images. Forensic Sci. Int. 1995, 74, 125–134. [Google Scholar] [CrossRef]
- Yoshino, M.; Matsuda, H.; Kubota, S.; Imaizumi, K.; Miyasaka, S.; Seta, S. Computer assisted skull identification system using video superimposition. Forensic Sci. Int. 1997, 90, 231–244. [Google Scholar] [CrossRef]
- Ghosh, A.; Sinha, P. An economised craniofacial identification system. Forensic Sci. Int. 2001, 117, 109–119. [Google Scholar] [CrossRef]
- Santamaría, J.; Cordón, O.; Damas, S. Evolutionary approaches for automatic 3D modeling of skulls in forensic identification. In Workshops on Applications of Evolutionary Computation; Springer: Berlin/Heidelberg, Germany, 2007; pp. 415–422. [Google Scholar]
- Santamaría, J.; Cordón, O.; Damas, S.; García-Torres, J.M.; Quirin, A. Performance evaluation of memetic approaches in 3D reconstruction of forensic objects. Soft Comput. 2009, 13, 883–904. [Google Scholar] [CrossRef]
- Ballerini, L.; Cordón, O.; Damas, S.; Santamaría, J. Automatic 3D modeling of skulls by scatter search and heuristic features. In Applications of Soft Computing. Updating the State of the Art; Avineri, E., Koepen, M., Dahal, K., Sunitiyoso, Y., Roy, R., Eds.; Springer: Berlin, Germany, 2009; pp. 149–158. [Google Scholar]
- Ibáñez, O.; Ballerini, L.; Cordón, O.; Damas, S.; Santamaría, J. An experimental study on the applicability of evolutionary algorithms to craniofacial superimposition in forensic identification. Inf. Sci. 2009, 179, 3998–4028. [Google Scholar] [CrossRef]
- Ibánez, O.; Cordon, Ó.; Damas, S.; Santamaria, J. Modeling the skull–face overlay uncertainty using fuzzy sets. IEEE Trans. Fuzzy Syst. 2011, 19, 946–959. [Google Scholar]
- Campomanes-Álvarez, B.R.; Cordón, O.; Damas, S. Evolutionary multiobjective optimization for mesh simplification of 3d open models. Integr. Comput. Aided Eng. 2013, 20, 375–390. [Google Scholar] [CrossRef]
- Duan, F.; Yang, Y.; Li, Y.; Tian, Y.; Lu, K.; Wu, Z.; Zhou, M. Skull identification via correlation measure between skull and face shape. IEEE Trans. Inf. Forensics Secur. 2014, 9, 1322–1332. [Google Scholar] [CrossRef]
- Campomanes-Álvarez, B.R.; Ibánez, O.; Campomanes-Álvarez, C.; Damas, S.; Cordón, O. Modeling facial soft tissue thickness for automatic skull-face overlay. IEEE Trans. Inf. Forensics Secur. 2015, 10, 2057–2070. [Google Scholar] [CrossRef]
- Campomanes-Alvarez, C.; Ibáñez, O.; Cordón, O. Design of criteria to assess craniofacial correspondence in forensic identification based on computer vision and fuzzy integrals. Appl. Soft Comput. 2016, 46, 596–612. [Google Scholar] [CrossRef]
- Bermejo, E.; Campomanes-Álvarez, C.; Valsecchi, A.; Ibáñez, O.; Damas, S.; Cordón, O. Genetic algorithms for skull-face overlay including mandible articulation. Inf. Sci. 2017, 420, 200–217. [Google Scholar] [CrossRef]
- Nagpal, S.; Singh, M.; Jain, A.; Singh, R.; Vatsa, M.; Noore, A. On matching skulls to digital face images: A preliminary approach. In Proceedings of the 2017 IEEE International Joint Conference on Biometrics (IJCB), Denver, CO, USA, 1–4 October 2017; pp. 813–819. [Google Scholar]
- Singh, M.; Nagpal, S.; Singh, R.; Vatsa, M.; Noore, A. Learning a shared transform model for skull to digital face image matching. In Proceedings of the 2018 IEEE 9th International Conference on Biometrics Theory, Applications and Systems (BTAS), Los Angeles, CA, USA, 22–25 October 2018; pp. 1–7. [Google Scholar]
- Campomanes-Alvarez, C.; Ibáñez, O.; Cordón, O.; Wilkinson, C. Hierarchical information fusion for decision making in craniofacial superimposition. Inf. Fusion 2018, 39, 25–40. [Google Scholar] [CrossRef]
- Campomanes-Alvarez, C.; Martos-Fernández, R.; Wilkinson, C.; Ibánez, O.; Cordón, O. Modeling skull-face anatomical/morphological correspondence for craniofacial superimposition-based identification. IEEE Trans. Inf. Forensics Secur. 2018, 13, 1481–1494. [Google Scholar] [CrossRef]
- Valsecchi, A.; Damas, S.; Cordón, O. A Robust and Efficient Method for Skull-Face Overlay in Computerized Craniofacial Superimposition. IEEE Trans. Inf. Forensics Secur. 2018, 13, 1960–1974. [Google Scholar] [CrossRef]
- Faria-Porto, L.; Correia-Lima, L.; Flores, M.; Valsecchi, A.; Ibáñez, O.; Machado-Palharese, C.; de Barros-Vidala, F. Automatic cephalometric landmarks detection on frontal faces: An approach based on supervised learning techniques. Digit. Investig. 2019, 30, 108–116. [Google Scholar] [CrossRef]
- San Tan, J.; Liao, I.Y.; Venkat, I.; Belaton, B.; Jayaprakash, P. Computer-aided superimposition via reconstructing and matching 3D faces to 3D skulls for forensic craniofacial identifications. Vis. Comput. 2019, 1–15. [Google Scholar] [CrossRef]
- Park, H.K.; Chung, J.W.; Kho, H.S. Use of hand-held laser scanning in the assessment of craniometry. Forensic Sci. Int. 2006, 160, 200–206. [Google Scholar] [CrossRef]
- Cummaudo, M.; Guerzoni, M.; Marasciuolo, L.; Gibelli, D.; Cigada, A.; Obertovà, Z.; Ratnayake, M.; Poppa, P.; Gabriel, P.; Ritz-Timme, S.; et al. Pitfalls at the root of facial assessment on photographs: A quantitative study of accuracy in positioning facial landmarks. Int. J. Leg. Med. 2013, 127, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Austin-Smith, D.; Maples, W.R. The reliability of skull/photograph superimposition in individual identification. J. Forensic Sci. 1994, 39, 446–455. [Google Scholar] [CrossRef]
- Jayaprakash, P.T.; Srinivasan, G.J.; Amravaneswaran, M.G. Cranio-facial morphoanalysis: A new method for enhancing reliability while identifying skulls by photo superimposition. Forensic Sci. Int. 2001, 117, 121–143. [Google Scholar] [CrossRef]
- Pesce Delfino, V.; Vacca, E.; Potente, F.; Lettini, T.; Colonna, M. Shape analytical morphometry in computer-aided skull identification via video superimposition. In Forensic Analysis of the Skull: Craniofacial Analysis, Reconstruction and Identification; Wiley: New York, NY, USA, 1993; pp. 131–159. [Google Scholar]
- Ricci, A.; Marella, G.L.; Apostol, M.A. A new experimental approach to computer-aided face/skull identification in forensic anthropology. Am. J. Forensic Med. Pathol. 2006, 27, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, O.; Vicente, R.; Navega, D.; Wilkinson, C.; Jayaprakash, P.; Huete, M.; Briers, T.; Hardiman, R.; Navarro, F.; Ruiz, E.; et al. Study on the performance of different craniofacial superimposition approaches (I). Forensic Sci. Int. 2015, 257, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Vandermeulen, D.; Claes, P.; Loeckx, D.; De Greef, S.; Willems, G.; Suetens, P. Computerized craniofacial reconstruction using CT-derived implicit surface representations. Forensic Sci. Int. 2006, 159, S164–S174. [Google Scholar] [CrossRef]
- Vandermeulen, D.; Claes, P.; De Greef, S.; Willems, G.; Clement, J.; Suetens, P. Automated facial reconstruction. Craniofacial Identif. 2012, 203. [Google Scholar] [CrossRef]
- Tu, P.; Book, R.; Liu, X.; Krahnstoever, N.; Adrian, C.; Williams, P. Automatic face recognition from skeletal remains. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–7. [Google Scholar]
- Liu, C.; Li, X. Superimposition-guided facial reconstruction from skull. arXiv 2018, arXiv:1810.00107. [Google Scholar]
- Imaizumi, K.; Taniguchi, K.; Ogawa, Y.; Matsuzaki, K.; Maekawa, H.; Nagata, T.; Moriyama, T.; Okuda, I.; Hayakawa, H.; Shiotani, S. Development of three-dimensional facial approximation system using head CT scans of Japanese living individuals. J. Forensic Radiol. Imaging 2019, 17, 36–45. [Google Scholar] [CrossRef]
- Mesejo, P.; Ibáñez, O.; Cordón, O.; Cagnoni, S. A survey on image segmentation using metaheuristic-based deformable models: State of the art and critical analysis. Appl. Soft Comput. 2016, 44, 1–29. [Google Scholar] [CrossRef]
- Claes, P.; Liberton, D.K.; Daniels, K.; Rosana, K.M.; Quillen, E.E.; Pearson, L.N.; McEvoy, B.; Bauchet, M.; Zaidi, A.A.; Yao, W.; et al. Modeling 3D facial shape from DNA. PLoS Genet. 2014, 10, e1004224. [Google Scholar] [CrossRef]
- Claes, P.; Roosenboom, J.; White, J.D.; Swigut, T.; Sero, D.; Li, J.; Lee, M.K.; Zaidi, A.; Mattern, B.C.; Liebowitz, C.; et al. Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat. Genet. 2018, 50, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Frudakis, T. Molecular Photofitting: Predicting Ancestry and Phenotype Using DNA; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Kayser, M.; Schneider, P.M. DNA-based prediction of human externally visible characteristics in forensics: Motivations, scientific challenges, and ethical considerations. Forensic Sci. Int. Genet. 2009, 3, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Sulem, P.; Gudbjartsson, D.F.; Stacey, S.N.; Helgason, A.; Rafnar, T.; Magnusson, K.P.; Manolescu, A.; Karason, A.; Palsson, A.; Thorleifsson, G.; et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007, 39, 1443. [Google Scholar] [CrossRef]
- Walsh, S.; Liu, F.; Ballantyne, K.N.; van Oven, M.; Lao, O.; Kayser, M. IrisPlex: A sensitive DNA tool for accurate prediction of blue and brown eye colour in the absence of ancestry information. Forensic Sci. Int. Genet. 2011, 5, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Rollo, R.; Ovenden, J.; Dudgeon, C.; Bennett, M.; Tucker, K.; Stephan, C. The utility of the IrisPlex system for estimating iris colour of Australians from their DNA. Forensic Sci. Int. 2018, 7, 98–115. [Google Scholar]
- Pneuman, A.; Budimlija, Z.M.; Caragine, T.; Prinz, M.; Wurmbach, E. Verification of eye and skin color predictors in various populations. Leg. Med. 2012, 14, 78–83. [Google Scholar] [CrossRef]
- Walsh, S.; Wollstein, A.; Liu, F.; Chakravarthy, U.; Rahu, M.; Seland, J.H.; Soubrane, G.; Tomazzoli, L.; Topouzis, F.; Vingerling, J.R.; et al. DNA-based eye colour prediction across Europe with the IrisPlex system. Forensic Sci. Int. Genet. 2012, 6, 330–340. [Google Scholar] [CrossRef]
- Kastelic, V.; Pośpiech, E.; Draus-Barini, J.; Branicki, W.; Drobnič, K. Prediction of eye color in the Slovenian population using the IrisPlex SNPs. Croat. Med. J. 2013, 54, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Spichenok, O.; Budimlija, Z.M.; Mitchell, A.A.; Jenny, A.; Kovacevic, L.; Marjanovic, D.; Caragine, T.; Prinz, M.; Wurmbach, E. Prediction of eye and skin color in diverse populations using seven SNPs. Forensic Sci. Int. Genet. 2011, 5, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Maroñas, O.; Phillips, C.; Söchtig, J.; Gomez-Tato, A.; Cruz, R.; Alvarez-Dios, J.; de Cal, M.C.; Ruiz, Y.; Fondevila, M.; Carracedo, Á.; et al. Development of a forensic skin colour predictive test. Forensic Sci. Int. Genet. 2014, 13, 34–44. [Google Scholar] [CrossRef]
- Ibáñez, Ó.; Corbal, I.; Gómez, I.; Gómez, Ó.; González, A.; Macías, M.; Prada, K.; Valsecchi, A.; Mesejo, P. Skeleton-ID: Artificial Intelligence at the service of Forensic Anthropology. In Proceedings of the 11th International Scientific Meeting of the Spanish Association of Forensic Anthropology and Odontology (AEAOF), Pastrana, Spain, 8–10 November 2019. [Google Scholar]
- Edgar, H.; Daneshvari Berry, S.; Moes, E.; Adolphi, N.; Bridges, P.; Nolte, K. New Mexico Decedent Image Database; Office of the Medical Investigator, University of New Mexico: Albuquerque, NM, USA, 2020. [Google Scholar]
- Halabi, S.S.; Prevedello, L.M.; Kalpathy-Cramer, J.; Mamonov, A.B.; Bilbily, A.; Cicero, M.; Pan, I.; Pereira, L.A.; Sousa, R.T.; Abdala, N.; et al. The RSNA pediatric bone age machine learning challenge. Radiology 2019, 290, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Li-wei, H.L.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Jack Jr, C.R.; Bernstein, M.A.; Fox, N.C.; Thompson, P.; Alexander, G.; Harvey, D.; Borowski, B.; Britson, P.J.L.; Whitwell, J.; Ward, C.; et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 2008, 27, 685–691. [Google Scholar] [CrossRef]
- Poldrack, R.A.; Barch, D.M.; Mitchell, J.; Wager, T.; Wagner, A.D.; Devlin, J.T.; Cumba, C.; Koyejo, O.; Milham, M. Toward open sharing of task-based fMRI data: The OpenfMRI project. Front. Neuroinform. 2013, 7, 12. [Google Scholar] [CrossRef]
- Dressel, J.; Farid, H. The accuracy, fairness, and limits of predicting recidivism. Sci. Adv. 2018, 4, eaao5580. [Google Scholar] [CrossRef]
- Valsecchi, A.; Irurita-Olivares, J.; Mesejo, P. Age estimation in forensic anthropology: Methodological considerations about the validation studies of prediction models. Int. J. Leg. Med. 2019, 133, 1915–1924. [Google Scholar] [CrossRef] [PubMed]
- Snell, J.; Swersky, K.; Zemel, R. Prototypical networks for few-shot learning. In Proceedings of the Annual Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4077–4087. [Google Scholar]
- Goodfellow, I. NIPS 2016 tutorial: Generative adversarial networks. arXiv 2016, arXiv:1701.00160. [Google Scholar]
- Hoshen, Y.; Li, K.; Malik, J. Non-adversarial image synthesis with generative latent nearest neighbors. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 5811–5819. [Google Scholar]
- Bertinetto, L.; Valmadre, J.; Henriques, J.F.; Vedaldi, A.; Torr, P.H. Fully-convolutional siamese networks for object tracking. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 850–865. [Google Scholar]
- Ji, S.; Xu, W.; Yang, M.; Yu, K. 3D convolutional neural networks for human action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Karpathy, A.; Fei-Fei, L. Densecap: Fully convolutional localization networks for dense captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 4565–4574. [Google Scholar]
- Lathuilière, S.; Mesejo, P.; Alameda-Pineda, X.; Horaud, R. DeepGUM: Learning deep robust regression with a Gaussian-Uniform mixture model. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 202–217. [Google Scholar]
- Lathuilière, S.; Juge, R.; Mesejo, P.; Munoz-Salinas, R.; Horaud, R. Deep mixture of linear inverse regressions applied to head-pose estimation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4817–4825. [Google Scholar]
- Samek, W.; Wiegand, T.; Müller, K.R. Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv 2017, arXiv:1708.08296. [Google Scholar]
- Castelvecchi, D. Can we open the black box of AI? Nat. News 2016, 538, 20. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Todd, T.W. Age changes in the pubic bone. I. The male white pubis. Am. J. Phys. Anthropol. 1920, 3, 285–334. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).