Automatic Classification of Morphologically Similar Fish Species Using Their Head Contours
Abstract
:1. Introduction
2. Materials and Methods
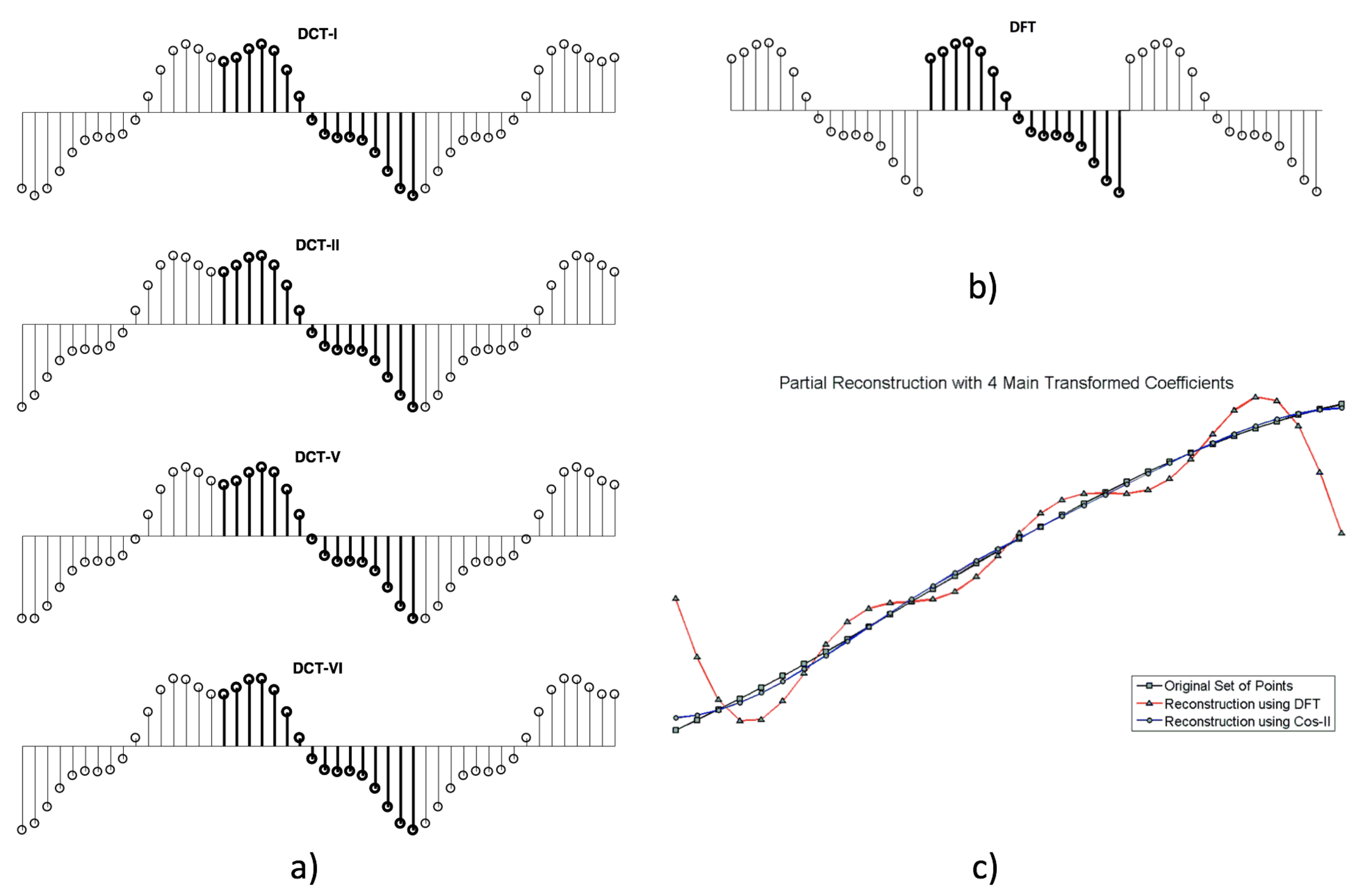
2.1. Discrete Transforms and Signal Reconstructions from a Reduced Set of Coefficients
2.1.1. Discrete Cosine Transform (Type-II)
2.1.2. Discrete Fourier Transform
2.1.3. A Note on the Implicit Extension of the Sequences Depending on the Discrete Transform Used and the Case of Open Contours
2.2. Data Used
2.3. Open Contour Extraction and Normalization
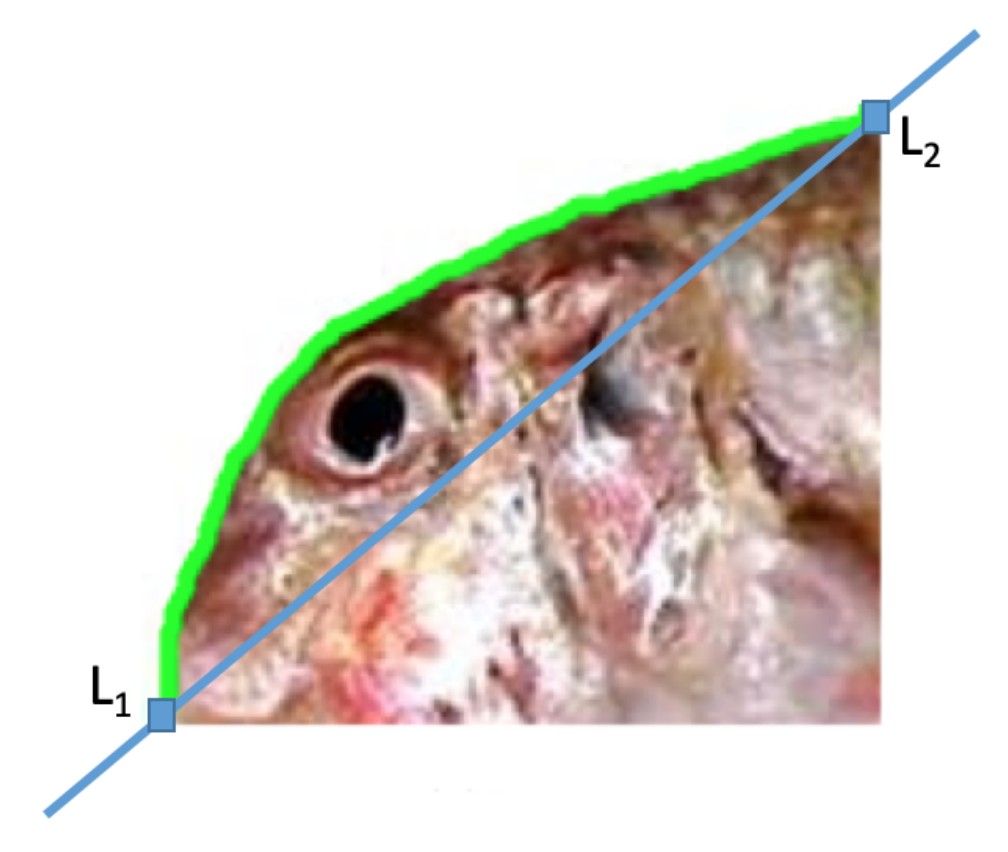
Open Contour Normalization
- First, for each open contour, we compute the Euclidean distance between () and (), and the angle that the segment defined by these two points forms with the horizontal. Seen graphically in Figure 7, the points () and () correspond to the landmarks and , respectively. As all the segments begin at , we have:Then, and are used to rotate and re-scale all the contours points according to:After this operation, without changing the aspect ratio, all profiles have been re-scaled and rotated so that they start at point (0,0) and end at point (1,0). Figure 8 separately shows both the scale and scale-and-rotation effects on the original Red Mullet contours. Note in Figure 8 that the relevant information to discriminate the contours is concentrated in (the vertical axis after the rotation) while (the horizontal axis after the rotation) will be very close for all contours and will always go from 0 to 1 in approximately constant increments.
- Second, in order to balance the number of points, the sequence of points is re-sampled so that they all have 256 values. The re-sampling is performed by cubic splines in order to have values at equispaced intervals in the range going from 0 to 1. We call the re-sampled sequence . Finally the contours are represented with a single sequence. In addition, the points and practically take the same value (0), which allows the DFT to be used as well.
2.4. Features Development
2.5. Extreme Learning Machines, Training and Classification
3. Results
3.1. Features Based on the Dct and the Dft. a Comparative Study
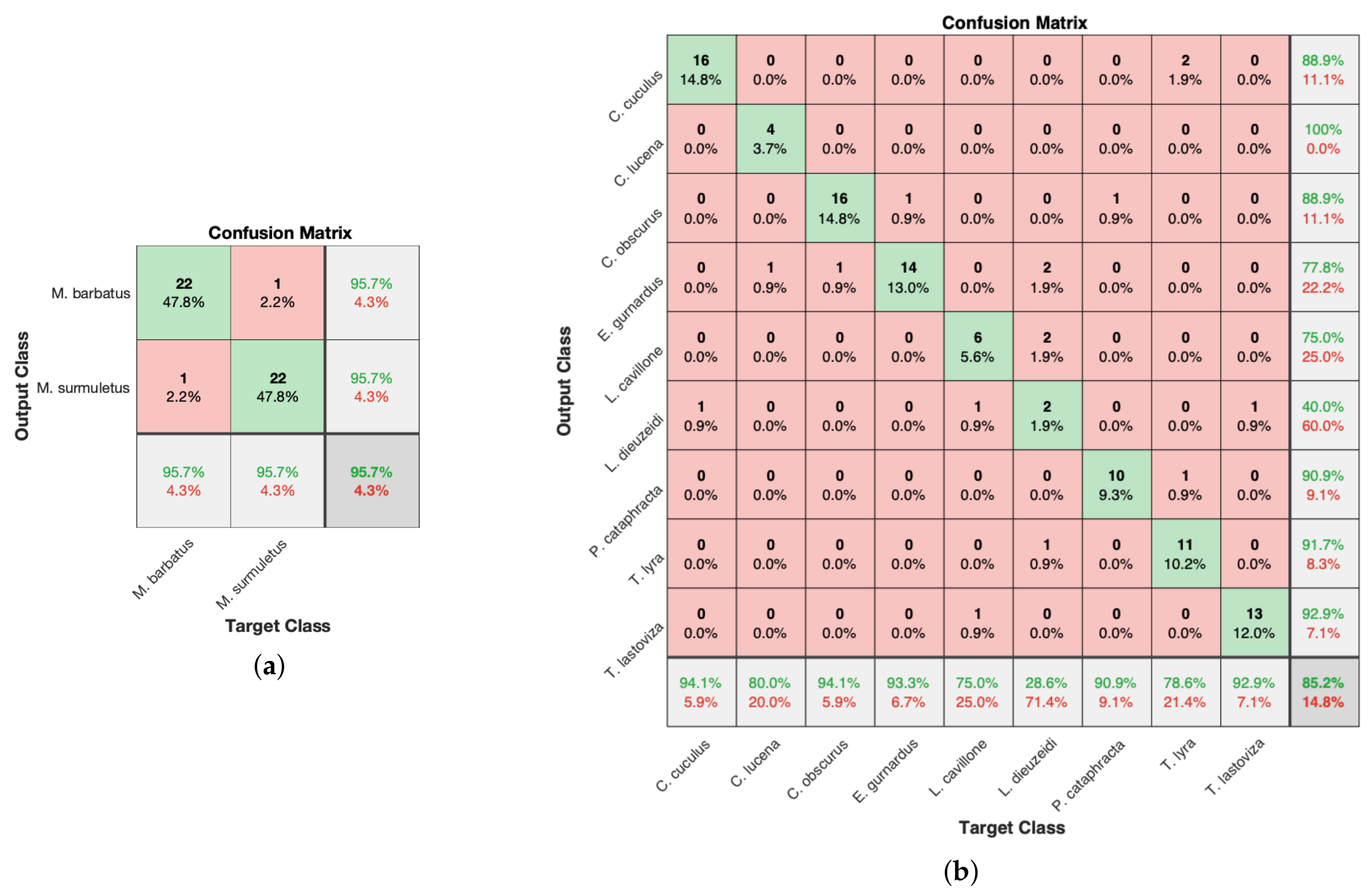
3.1.1. Leave-One-Out Cross-Validation
3.1.2. LOO-CV Tests
3.2. Regarding Automation of the Whole Process
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Azzurro, E.; Tuset, V.M.; Lombarte, A.; Maynou, F.; Simberloff, D.; Rodríguez-Pérez, A.; Solé, R.V. External morphology explains the success of biological invasions. Ecol. Lett. 2014, 17, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Stergiou, K.; Petrakis, G.; Papaconstantinou, C. The Mullidae (Mullus barbatus, M. surmuletus) fishery in Greek waters, 1964–1986. In FAO Fisheries Report (FAO); FAO: Rome, Italy, 1992. [Google Scholar]
- Renones, O.; Massuti, E.; Morales-Nin, B. Life history of the red mullet Mullus surmuletus from the bottom-trawl fishery off the Island of Majorca (north-west Mediterranean). Mar. Biol. 1995, 123, 411–419. [Google Scholar] [CrossRef]
- Bautista-Vega, A.; Letourneur, Y.; Harmelin-Vivien, M.; Salen-Picard, C. Difference in diet and size-related trophic level in two sympatric fish species, the red mullets Mullus barbatus and Mullus surmuletus, in the Gulf of Lions (north-west Mediterranean Sea). J. Fish Biol. 2008, 73, 2402–2420. [Google Scholar] [CrossRef]
- Cresson, P.; Bouchoucha, M.; Miralles, F.; Elleboode, R.; Mahe, K.; Marusczak, N.; Thebault, H.; Cossa, D. Are red mullet efficient as bio-indicators of mercury contamination? A case study from the French Mediterranean. Mar. Pollut. Bull. 2015, 91, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Golani, D.; Galil, B. Trophic relationships of colonizing and indigenous goatfishes (Mullidae) in the eastern Mediterranean with special emphasis on decapod crustaceans. Hydrobiologia 1991, 218, 27–33. [Google Scholar] [CrossRef]
- Labropoulou, M.; Eleftheriou, A. The foraging ecology of two pairs of congeneric demersal fish species: Importance of morphological characteristics in prey selection. J. Fish Biol. 1997, 50, 324–340. [Google Scholar] [CrossRef]
- Lombarte, A.; Recasens, L.; González, M.; de Sola, L.G. Spatial segregation of two species of Mullidae (Mullus surmuletus and M. barbatus) in relation to habitat. Mar. Ecol. Prog. Ser. 2000, 206, 239–249. [Google Scholar] [CrossRef]
- Maravelias, C.D.; Tsitsika, E.V.; Papaconstantinou, C. Environmental influences on the spatial distribution of European hake (Merluccius merluccius) and red mullet (Mullus barbatus) in the Mediterranean. Ecol. Res. 2007, 22, 678–685. [Google Scholar] [CrossRef]
- Bougis, P. Recherches Biométriques sur les Rougets,‘Mullus barbatus’ L. et‘Mullus surmuletus’ L...; Centre National de la Recherche Scientifique: Paris, France, 1952. [Google Scholar]
- Tortonese, E.; di Entomologia, A.N.I.; Italiana, U.Z. Osteichthyes (Pesci Ossei): Parte Seconda; Edizioni Calderini Bologna: Bologna, France, 1975. [Google Scholar]
- Lombarte, A.; Aguirre, H. Quantitative differences in the chemoreceptor systems in the barbels of two species of Mullidae (Mullus surmuletus and M. barbatus) with different bottom habitats. Mar. Ecol. Prog. Ser. 1997, 150, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Aguirre, H. Presence of dentition in the premaxilla of juvenile Mullus barbatus and M. surmuletus. J. Fish Biol. 1997, 51, 1186–1191. [Google Scholar] [CrossRef]
- Aguirre, H.; Lombarte, A. Ecomorphological comparisons of sagittae in Mullus barbatus and M. surmuletus. J. Fish Biol. 1999, 55, 105–114. [Google Scholar] [CrossRef]
- Hureau, J.; Bauchot, M.; Nielsen, J.; Tortonese, E. Fishes of the North-Eastern Atlantic and the Mediterranean; Unesco: Paris, France, 1986; Volume 3. [Google Scholar]
- Kuhl, F.P.; Giardina, C.R. Elliptic Fourier features of a closed contour. Comput. Graph. Image Process. 1982, 18, 236–258. [Google Scholar] [CrossRef]
- El-ghazal, A.; Basir, O.; Belkasim, S. Farthest point distance: A new shape signature for Fourier descriptors. Signal Process. Image Commun. 2009, 24, 572–586. [Google Scholar] [CrossRef]
- Persoon, E.; Fu, K.S. Shape discrimination using Fourier descriptors. IEEE Trans. Syst. Man Cybern. 1977, 7, 170–179. [Google Scholar] [CrossRef]
- Tracey, S.R.; Lyle, J.M.; Duhamel, G. Application of elliptical Fourier analysis of otolith form as a tool for stock identification. Fish. Res. 2006, 77, 138–147. [Google Scholar] [CrossRef]
- Lestrel, P.E. Fourier Descriptors and Their Applications in Biology; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Mokhtarian, F.; Abbasi, S.; Kittler, J. Robust and E cient Shape Indexing through Curvature Scale Space. In Proceedings of the 1996 British Machine and Vision Conference BMVC, Edinburgh, UK, 9–12 September 1996; Volume 96. [Google Scholar]
- Dudek, G.; Tsotsos, J.K. Shape representation and recognition from multiscale curvature. Comput. Vis. Image Underst. 1997, 68, 170–189. [Google Scholar] [CrossRef] [Green Version]
- Mokhtarian, F.; Bober, M. Curvature Scale Space Representation: Theory, Applications, and MPEG-7 Standardization; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; Volume 25. [Google Scholar]
- Parisi-Baradad, V.; Lombarte, A.; García-Ladona, E.; Cabestany, J.; Piera, J.; Chic, O. Otolith shape contour analysis using affine transformation invariant wavelet transforms and curvature scale space representation. Mar. Freshw. Res. 2005, 56, 795–804. [Google Scholar] [CrossRef]
- Gibbs, J.W. Fourier’s series. Nature 1899, 59, 606. [Google Scholar] [CrossRef]
- Toubin, M.; Dumont, C.; Verrecchia, E.P.; Laligant, O.; Diou, A.; Truchetet, F.; Abidi, M. Multi-scale analysis of shell growth increments using wavelet transform. Comput. Geosci. 1999, 25, 877–885. [Google Scholar] [CrossRef] [Green Version]
- Allen, E.G. New approaches to Fourier analysis of ammonoid sutures and other complex, open curves. Paleobiology 2006, 32, 299–315. [Google Scholar] [CrossRef]
- Yang, C.; Tiebe, O.; Shirahama, K.; Łukasik, E.; Grzegorzek, M. Evaluating contour segment descriptors. Mach. Vis. Appl. 2017, 28, 373–391. [Google Scholar] [CrossRef]
- Dommergues, C.H.; Dommergues, J.L.; Verrecchia, E.P. The discrete cosine transform, a Fourier-related method for morphometric analysis of open contours. Math. Geol. 2007, 39, 749–763. [Google Scholar] [CrossRef] [Green Version]
- Wilczek, J.; Monna, F.; Barral, P.; Burlet, L.; Chateau, C.; Navarro, N. Morphometrics of Second Iron Age ceramics–strengths, weaknesses, and comparison with traditional typology. J. Archaeol. Sci. 2014, 50, 39–50. [Google Scholar] [CrossRef]
- Stefanini, M.I.; Carmona, P.M.; Iglesias, P.P.; Soto, E.M.; Soto, I.M. Differential Rates of Male Genital Evolution in Sibling Species of Drosophila. Evol. Biol. 2018, 45, 211–222. [Google Scholar] [CrossRef]
- Ahmed, N.; Natarajan, T.; Rao, K.R. Discrete cosine transform. IEEE Trans. Comput. 1974, 100, 90–93. [Google Scholar] [CrossRef]
- Kou, W.; Mark, J.W. A new look at DCT-type transforms. IEEE Trans. Acoust. Speech Signal Process. 1989, 37, 1899–1908. [Google Scholar] [CrossRef]
- Martucci, S. Symmetric convolution and the discrete sine and cosine transforms. IEEE Trans. Signal Process. 1994, 42, 1038–1051. [Google Scholar] [CrossRef]
- Strang, G. The discrete cosine transform. SIAM Rev. 1999, 41, 135–147. [Google Scholar] [CrossRef]
- Suresh, K.; Sreenivas, T. Linear filtering in DCT IV/DST IV and MDCT/MDST domain. Signal Process. 2009, 89, 1081–1089. [Google Scholar] [CrossRef]
- Britanak, V.; Yip, P.C.; Rao, K.R. Discrete Cosine and Sine Transforms: General Properties, Fast Algorithms and Integer Approximations; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Tsitsas, N.L. On block matrices associated with discrete trigonometric transforms and their use in the theory of wave propagation. J. Comput. Math. 2010, 28, 864–878. [Google Scholar] [CrossRef]
- Ito, I.; Kiya, H. A computing method for linear convolution in the DCT domain. In Proceedings of the 2011 19th European Signal Processing Conference, Barcelona, Spain, 29 August–2 September 2011; pp. 323–327. [Google Scholar]
- Rao, K.R.; Yip, P. Discrete Cosine Transform: Algorithms, Advantages, Applications; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Wang, Z. Fast algorithms for the discrete W transform and for the discrete Fourier transform. IEEE Trans. Acoust. Speech Signal Process. 1984, 32, 803–816. [Google Scholar] [CrossRef]
- Roma, N.; Sousa, L. A tutorial overview on the properties of the discrete cosine transform for encoded image and video processing. Signal Process. 2011, 91, 2443–2464. [Google Scholar] [CrossRef]
- Massutí, E.; Reñones, O. Demersal resource assemblages in the trawl fishing grounds off the Balearic Islands (western Mediterranean). Sci. Mar. 2005, 69, 167–181. [Google Scholar] [CrossRef] [Green Version]
- Balcells, M.; Fernández-Arcaya, U.; Lombarte, A.; Ramon, M.; Abelló, P.; Mecho, A.; Company, J.; Recasens, L. Effect of a small-scale fishing closure area on the demersal community in the NW Mediterranean Sea. In Rapports et Procès-Verbaux des Réunions de la Commission Internationale pour l’Exploration Scientifique de la Mer Méditerranée; Mediterranean Science Commission: Kiel, Germany, 2016; pp. 41–517. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.B.; Chen, L.; Siew, C.K. Universal approximation using incremental constructive feedforward networks with random hidden nodes. IEEE Trans. Neural Netw. 2006, 17, 879–892. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 91–99. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Álvarez-Ellacuría, A.; Palmer, M.; Catalán, I.A.; Lisani, J.L. Image-based, unsupervised estimation of fish size from commercial landings using deep learning. ICES J. Mar. Sci. 2019. [Google Scholar] [CrossRef]











| Binary-Class Classification Accuracy (LOO-CV 1000 Iterations) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Hidden | DCT-5 | DFT-3 | ||||||
| Nodes | mean | std | max | min | mean | std | max | min |
| 7 | 88.44 | 0.031 | 95.65 | 78.26 | 88.46 | 0.032 | 95.65 | 76.09 |
| 8 | 89.09 | 0.031 | 95.65 | 78.26 | 89.09 | 0.030 | 95.65 | 78.26 |
| 9 | 89.37 | 0.030 | 97.83 | 80.44 | 89.29 | 0.031 | 95.65 | 76.09 |
| 10 | 89.30 | 0.031 | 95.65 | 78.26 | 89.36 | 0.028 | 95.65 | 80.44 |
| 11 | 89.34 | 0.029 | 95.65 | 80.44 | 89.19 | 0.030 | 95.65 | 78.26 |
| 12 | 89.33 | 0.031 | 95.65 | 80.44 | 89.20 | 0.030 | 95.65 | 78.26 |
| 13 | 89.04 | 0.031 | 95.65 | 78.26 | 89.10 | 0.030 | 95.65 | 80.44 |
| 14 | 88.76 | 0.032 | 95.65 | 78.26 | 88.64 | 0.032 | 95.65 | 78.26 |
| 15 | 88.80 | 0.031 | 95.65 | 78.26 | 88.57 | 0.031 | 95.65 | 76.09 |
| 16 | 88.40 | 0.032 | 95.65 | 78.26 | 88.14 | 0.033 | 95.65 | 73.91 |
| 17 | 88.05 | 0.033 | 95.65 | 78.26 | 87.92 | 0.033 | 95.65 | 76.09 |
| Multi-Class Classification Accuracy (LOO-CV 1000 Iterations) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Hidden | DCT-5 | DFT-5 | ||||||
| Nodes | mean | std | max | min | mean | std | max | min |
| 5 | 72.66 | 0.024 | 79.63 | 64.82 | 56.78 | 0.035 | 66.67 | 44.44 |
| 10 | 72.66 | 0.024 | 79.63 | 64.82 | 72.29 | 0.029 | 81.48 | 63.89 |
| 15 | 76.66 | 0.023 | 84.26 | 68.52 | 77.78 | 0.024 | 85.19 | 69.44 |
| 20 | 78.62 | 0.022 | 85.19 | 72.22 | 80.52 | 0.024 | 88.89 | 72.22 |
| 25 | 79.84 | 0.023 | 87.04 | 72.22 | 82.04 | 0.021 | 87.96 | 74.07 |
| 30 | 80.42 | 0.023 | 87.04 | 72.22 | 82.59 | 0.021 | 88.89 | 75.00 |
| 35 | 80.64 | 0.022 | 87.04 | 73.19 | 82.49 | 0.022 | 87.96 | 75.00 |
| 40 | 80.54 | 0.023 | 87.96 | 73.19 | 81.94 | 0.022 | 88.89 | 74.07 |
| 45 | 80.00 | 0.024 | 87.04 | 72.22 | 81.18 | 0.029 | 87.04 | 74.07 |
| 50 | 79.38 | 0.025 | 87.04 | 71.30 | 79.79 | 0.024 | 86.11 | 72.22 |
| 55 | 78.18 | 0.025 | 87.04 | 69.44 | 78.15 | 0.025 | 85.19 | 68.52 |
| 60 | 76.71 | 0.026 | 85.18 | 68.52 | 76.26 | 0.027 | 85.19 | 68.52 |
| Multi-Class Classification Accuracy (LOO-CV 1000 Iterations) | ||||
|---|---|---|---|---|
| Hidden | DCT-5 | DFT-5 | ||
| nodes | mean | std | max | min |
| 20 | 80.70 | 0.024 | 87.96 | 73.15 |
| 21 | 81.50 | 0.023 | 87.96 | 75.00 |
| 24 | 82.08 | 0.022 | 88.89 | 75.00 |
| 26 | 82.48 | 0.022 | 88.89 | 75.93 |
| 28 | 82.89 | 0.022 | 89.82 | 75.00 |
| 30 | 83.15 | 0.022 | 90.74 | 75.93 |
| 32 | 83.41 | 0.023 | 91.67 | 75.00 |
| 34 | 83.53 | 0.022 | 89.82 | 76.85 |
| 36 | 83.40 | 0.023 | 89.82 | 75.00 |
| 38 | 83.34 | 0.023 | 89.82 | 75.00 |
| 40 | 83.48 | 0.023 | 89.82 | 75.93 |
| 42 | 83.20 | 0.022 | 89.82 | 76.85 |
| 44 | 83.20 | 0.024 | 89.82 | 75.00 |
| 46 | 82.82 | 0.023 | 89.82 | 74.07 |
| 48 | 82.57 | 0.023 | 89.82 | 75.00 |
| 50 | 82.11 | 0.024 | 88.89 | 75.00 |
| Feature Extraction Network | ||
|---|---|---|
| Layer | Type | Main Characteristics |
| 1 | Image Input | 32 × 32 × 3 images with ‘zerocenter’ normalization |
| 2 | Convolution | 32 5 × 5 × 3 convolutions with stride [1 1] and padding [2 2 2 2] |
| 3 | Max Pooling | 3 × 3 max pooling with stride [2 2] and padding [0 0 0 0] |
| 4 | ReLU | ReLU |
| 5 | Convolution | 32 5 × 5 × 32 convolutions with stride [1 1] and padding [2 2 2 2] |
| 6 | ReLU | ReLU |
| 7 | Average Pooling | 3 × 3 average pooling with stride [2 2] and padding [0 0 0 0] |
| 8 | Convolution | 64 5 × 5 × 32 convolutions with stride [1 1] and padding [2 2 2 2] |
| 9 | ReLU | ReLU |
| 10 | Average Pooling | 3 × 3 average pooling with stride [2 2] and padding [0 0 0 0] |
| 11 | Fully Connected | 64 fully connected layer |
| 12 | ReLU | ReLU |
| 13 | Fully Connected | 2 fully connected layer |
| 14 | Softmax | softmax |
| 15 | Classification Output | crossentropyex |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marti-Puig, P.; Manjabacas, A.; Lombarte, A. Automatic Classification of Morphologically Similar Fish Species Using Their Head Contours. Appl. Sci. 2020, 10, 3408. https://doi.org/10.3390/app10103408
Marti-Puig P, Manjabacas A, Lombarte A. Automatic Classification of Morphologically Similar Fish Species Using Their Head Contours. Applied Sciences. 2020; 10(10):3408. https://doi.org/10.3390/app10103408
Chicago/Turabian StyleMarti-Puig, Pere, Amalia Manjabacas, and Antoni Lombarte. 2020. "Automatic Classification of Morphologically Similar Fish Species Using Their Head Contours" Applied Sciences 10, no. 10: 3408. https://doi.org/10.3390/app10103408






