The Role of Systems Biologic Approach in Cell Signaling and Drug Development Responses—A Mini Review
Abstract
1. Introduction
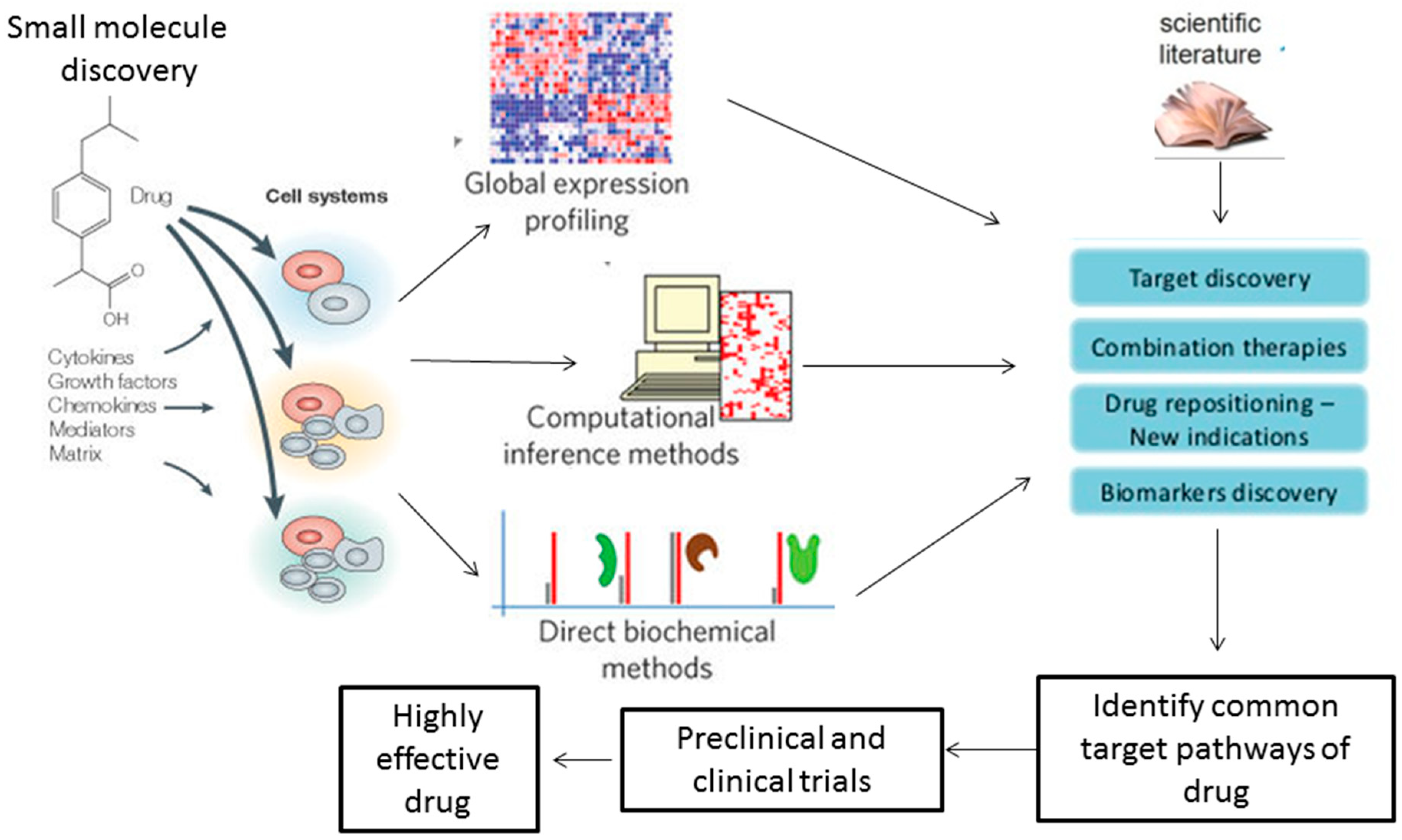
2. Role of Systems Biology in Drug Discovery and Development
3. Notch Signaling
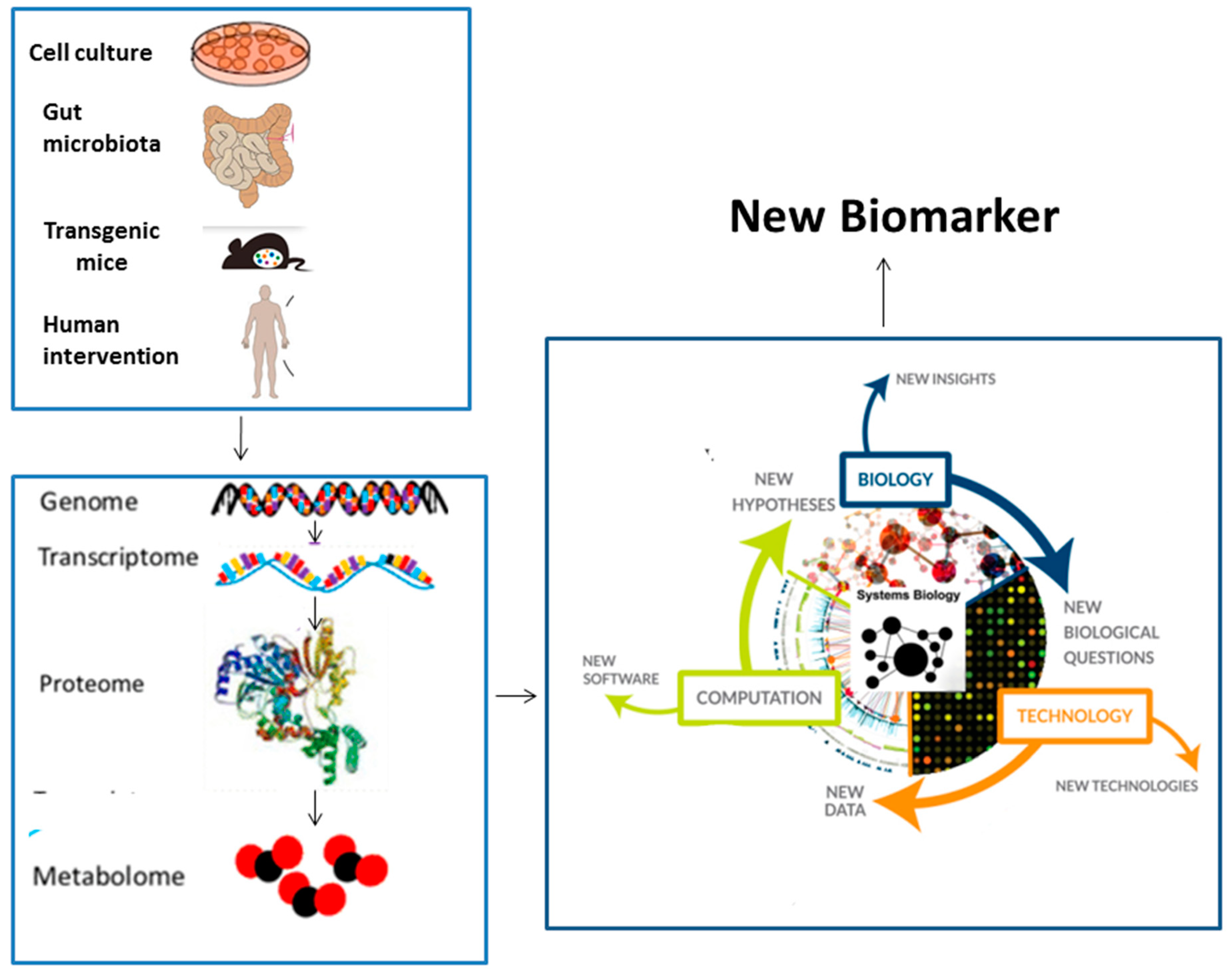
4. Systems Biology as a Biomarker
5. Summary
Acknowledgments
Conflicts of Interest
References
- Karaboga, I.; Demirtas, S.; Karaca, T. Investigation of the relationship between the Th17/IL-23 pathway and innate-adaptive immune system in TNBS-induced colitis in rats. Iran. J. Basic Med. Sci. 2017, 20, 870–879. [Google Scholar] [PubMed]
- Collins, D.; Chenard-Poirier, M.; Lopez, J. The PI3K pathway at the crossroads of cancer and the immune system: Strategies for next generation immunotherapy combinations. Curr. Cancer Drug Targets 2018, 18, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Sun, G.; Sun, X.; Tian, D.; Liu, K.; Liu, T.; Cong, M.; Xu, H.; Li, X.; Shi, W.; et al. A novel differentiation pathway from CD4+ T cells to CD4− T cells for maintaining immune system homeostasis. Cell Death Dis. 2016, 7, e2193. [Google Scholar] [CrossRef] [PubMed]
- Tomar, N.; De, R.K. A model of an integrated immune system pathway in Homo sapiens and its interaction with superantigen producing expression regulatory pathway in Staphylococcus aureus: Comparing behavior of pathogen perturbed and unperturbed pathway. PLoS ONE 2013, 8, e80918. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Takiuchi, M.; Iida, K.; Nagaki, K.; Inai, S. The activation mechanism of the alternative pathway of the human complement system by the immune precipitate formed with F(ab’)2 of rabbit IgG antibody: The generation of C3- and C5-cleaving enzymes on the immune precipitate. Immunochemistry 1977, 14, 25–30. [Google Scholar] [CrossRef]
- Basta, S.; Alatery, A. The cross-priming pathway: A portrait of an intricate immune system. Scand. J. Immunol. 2007, 65, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Pangburn, M.K.; Pangburn, K.L.; Koistinen, V.; Meri, S.; Sharma, A.K. Molecular mechanisms of target recognition in an innate immune system: Interactions among factor H, C3b, and target in the alternative pathway of human complement. J. Immunol. 2000, 164, 4742–4751. [Google Scholar] [CrossRef] [PubMed]
- Schon, K.; Koeppe, H.W. (Non-specific immune mechanisms. The alternate pathway in the complement system). Med. Klin. 1979, 74, 461–469. [Google Scholar] [PubMed]
- Fuchs, O. Important genes in the pathogenesis of 5q-syndrome and their connection with ribosomal stress and the innate immune system pathway. Leuk. Res. Treat. 2012, 2012, 179402. [Google Scholar] [CrossRef] [PubMed]
- Jalili, M. Network biology: Describing biological systems by complex networks: Comment on “Network science of biological systems at different scales: A review” by M. Gosak et al. Phys. Life Rev. 2018, 24, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, P.R.; Venkatesh, K.V. A conceptual review on systems biology in health and diseases: From biological networks to modern therapeutics. Syst. Synth. Biol. 2014, 8, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Lusis, A.J. A thematic review series: Systems biology approaches to metabolic and cardiovascular disorders. J. Lipid Res. 2006, 47, 1887–1890. [Google Scholar] [CrossRef] [PubMed]
- Demir, E.; Babur, O.; Rodchenkov, I.; Aksoy, B.A.; Fukuda, K.I.; Gross, B.; Sumer, O.S.; Bader, G.D.; Sander, C. Using biological pathway data with paxtools. PLoS Comput. Biol. 2013, 9, e1003194. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.I.; Yamagata, Y.; Takagi, T. FREX: A query interface for biological processes with hierarchical and recursive structures. Silico Biol. 2004, 4, 63–79. [Google Scholar]
- Peyvandipour, A.; Saberian, N.; Shafi, A.; Donato, M.; Draghici, S. A novel computational approach for drug repurposing using systems biology. Bioinformatics 2018. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhao, X.; Zhou, J.; Yang, J.; Zhang, Y.; Kuang, W.; Peng, J.; Chen, L.; Zeng, J. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nature Commun. 2017, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, P.P.; Orta, D.J.; Pena, I.; Flores, E.C.; Ramirez, J.U.; Beltran, H.I.; Alas, S.J. A Computational Approach to Studying Protein Folding Problems Considering the Crucial Role of the Intracellular Environment. J. Comput. Biol. 2017, 24, 995–1013. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Mourad, R.; Li, L.; Cuvier, O. Uncovering direct and indirect molecular determinants of chromatin loops using a computational integrative approach. PLoS Comput. Biol. 2017, 13, e1005538. [Google Scholar] [CrossRef] [PubMed]
- Emon, M.A.; Kodamullil, A.T.; Karki, R.; Younesi, E.; Hofmann-Apitius, M. Using Drugs as Molecular Probes: A Computational Chemical Biology Approach in Neurodegenerative Diseases. J. Alzheimer Dis. 2017, 56, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Kloft, C.; Trame, M.N.; Ritter, C.A. Systems pharmacology in drug development and therapeutic use—A forthcoming paradigm shift. Eur. J. Pharm. Sci. 2016, 94, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lotsch, J.; Ultsch, A. Process Pharmacology: A Pharmacological Data Science Approach to Drug Development and Therapy. CPT Pharmacomet. Syst. Pharmacol. 2016, 5, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, G.; Barone, A.W.; Dziak, R. The effect of ascorbic acid on bone cancer cells in vitro. Cogent Biol. 2017, 3, 1288335. [Google Scholar] [CrossRef]
- Fernandes, G.; Barone, A.W.; Dziak, R. Effects of Verapamil on Bone Cancer Cells in vitro. J. Cell Biol. Cell Metab. 2016, 3, 13. [Google Scholar] [CrossRef]
- Trame, M.N.; Biliouris, K.; Lesko, L.J.; Mettetal, J.T. Systems pharmacology to predict drug safety in drug development. Eur. J. Pharm. Sci. 2016, 94, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, I.O.; AlYahya, S.A.; Ebrahimie, E.; Mohammadi-Dehcheshmeh, M. Computational systems biology analysis of biomarkers in lung cancer; unravelling genomic regions which frequently encode biomarkers, enriched pathways, and new candidates. Gene 2018, 659, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.; Alford, B.; Soliman, H. Emerging Therapeutic Strategies in Breast Cancer. South. Med. J. 2017, 110, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Walsh, L.A.; Alvarez, M.J.; Sabio, E.Y.; Reyngold, M.; Makarov, V.; Mukherjee, S.; Lee, K.W.; Desrichard, A.; Turcan, S.; Dalin, M.G.; et al. An Integrated Systems Biology Approach Identifies TRIM25 as a Key Determinant of Breast Cancer Metastasis. Cell Rep. 2017, 20, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Todhunter, M.E.; LaBarge, M.A. Cell and Tissue Biology Paves a Path to Breast Cancer Prevention. Trends Cancer 2017, 3, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, E.J.; Plavec, I.; Nguyen, D.; Melrose, J.; Rosler, E.S.; Kao, L.T.; Wang, Y.; Hytopoulos, E.; Bishop, A.C.; Bateman, R.; et al. Rapid structure-activity and selectivity analysis of kinase inhibitors by BioMAP analysis in complex human primary cell-based models. Assay Drug Dev. Technol. 2004, 2, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.G.; Gross, B.E.; Demir, E.; Rodchenkov, I.; Babur, O.; Anwar, N.; Schultz, N.; Bader, G.D.; Sander, C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011, 39, D685–D690. [Google Scholar] [CrossRef] [PubMed]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. InnateDB: Systems biology of innate immunity and beyond—Recent updates and continuing curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Kankel, M.W.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Gene 2012, 13, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Hori, K.; Obar, R.A.; Artavanis-Tsakonas, S. Proteomic analysis of the Notch interactome. Methods Mol. Biol. 2014, 1187, 181–192. [Google Scholar] [PubMed]
- Guruharsha, K.G.; Obar, R.A.; Mintseris, J.; Aishwarya, K.; Krishnan, R.T.; Vijayraghavan, K.; Artavanis-Tsakonas, S. Drosophila protein interaction map (DPiM): A paradigm for metazoan protein complex interactions. Fly 2012, 6, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lin, X.; Yu, H. Identifying genes related with rheumatoid arthritis via system biology analysis. Gene 2015, 571, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Polytarchou, C.; Koukos, G.; Iliopoulos, D. Systems biology in inflammatory bowel diseases: Ready for prime time. Curr. Opin. Gastroenterol. 2014, 30, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Balbas-Martinez, V.; Ruiz-Cerda, L.; Irurzun-Arana, I.; Gonzalez-Garcia, I.; Vermeulen, A.; Gomez-Mantilla, J.D.; Troconiz, I.F. A systems pharmacology model for inflammatory bowel disease. PLoS ONE 2018, 13, e0192949. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Zhao, B.; Deng, Y.; Shangguan, S.; Zhou, F.; Zhou, W.; Li, X.; Li, Y.; Chen, G. Notch signaling in cerebrovascular diseases (Review). Mol. Med. Rep. 2016, 14, 2883–2898. [Google Scholar] [CrossRef] [PubMed]
- Price, N.D.; Magis, A.T.; Earls, J.C.; Glusman, G.; Levy, R.; Lausted, C.; McDonald, D.T.; Kusebauch, U.; Moss, C.L.; Zhou, Y.; et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 2017, 35, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Heerspink, H.J.; Aschauer, C.; Heinzel, A.; Heinze, G.; Kainz, A.; Sunzenauer, J.; Perco, P.; de Zeeuw, D.; Rossing, P.; et al. Systems Biology-Derived Biomarkers to Predict Progression of Renal Function Decline in Type 2 Diabetes. Diabetes Care 2017, 40, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Schett, G.; Elewaut, D.; McInnes, I.B.; Dayer, J.M.; Neurath, M.F. How cytokine networks fuel inflammation: Toward a cytokine-based disease taxonomy. Nat. Med. 2013, 19, 822–824. [Google Scholar] [CrossRef] [PubMed]
- MacIsaac, K.D.; Wang, I.M.; Menetski, J.; Roberts, C. Genomic and systems approaches to translational biomarker discovery in immunological diseases. Drug Discov. Today 2014, 19, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Saberi Anvar, M.; Minuchehr, Z.; Shahlaei, M.; Kheitan, S. Gastric cancer biomarkers; A systems biology approach. Biochem. Biophys. Rep. 2018, 13, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Piening, B.D.; Zhou, W.; Contrepois, K.; Rost, H.; Gu Urban, G.J.; Mishra, T.; Hanson, B.M.; Bautista, E.J.; Leopold, S.; Yeh, C.Y.; et al. Integrative Personal Omics Profiles during Periods of Weight Gain and Loss. Cell Syst. 2018, 6, 157–170. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abhyankar, V.; Bland, P.; Fernandes, G. The Role of Systems Biologic Approach in Cell Signaling and Drug Development Responses—A Mini Review. Med. Sci. 2018, 6, 43. https://doi.org/10.3390/medsci6020043
Abhyankar V, Bland P, Fernandes G. The Role of Systems Biologic Approach in Cell Signaling and Drug Development Responses—A Mini Review. Medical Sciences. 2018; 6(2):43. https://doi.org/10.3390/medsci6020043
Chicago/Turabian StyleAbhyankar, Vrushali, Paul Bland, and Gabriela Fernandes. 2018. "The Role of Systems Biologic Approach in Cell Signaling and Drug Development Responses—A Mini Review" Medical Sciences 6, no. 2: 43. https://doi.org/10.3390/medsci6020043
APA StyleAbhyankar, V., Bland, P., & Fernandes, G. (2018). The Role of Systems Biologic Approach in Cell Signaling and Drug Development Responses—A Mini Review. Medical Sciences, 6(2), 43. https://doi.org/10.3390/medsci6020043



