Computational Identification of RNF114 nsSNPs with Potential Roles in Psoriasis and Immune Dysregulation
Abstract
1. Introduction
2. Methods and Materials
2.1. Data Retrieval
2.2. Prediction of Deleterious Ns SNPs of RNF114 Gene
2.3. Prediction of Missense Variant Pathogenicity
2.4. Protein Stability Analysis of Predicted RNF114 nsSNPs
2.5. Three-Dimensional Structure Prediction and Visualization
2.6. Conservation Analysis and Surface Accessibility Prediction of RNF114
2.7. Identification of nsSNPs in RNF114 Protein Domains
2.8. Prediction of Protein-Protein Interactions
3. Results
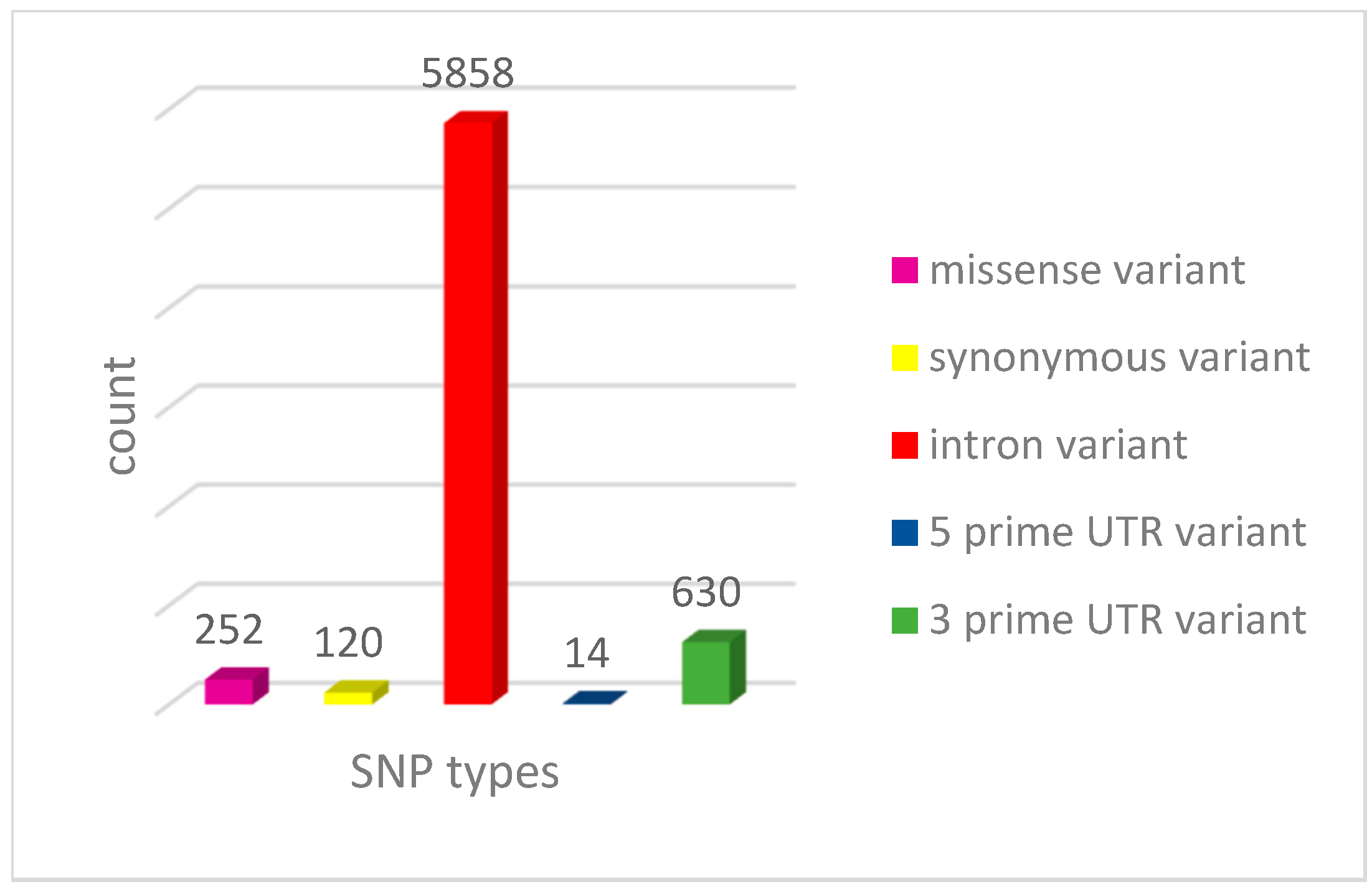
3.1. Distribution of RNF114 Gene SNPs
3.2. Detection of Functionally Damaging Missense Variants
3.3. Impact of nsSNPs on RNF114 Protein Stability
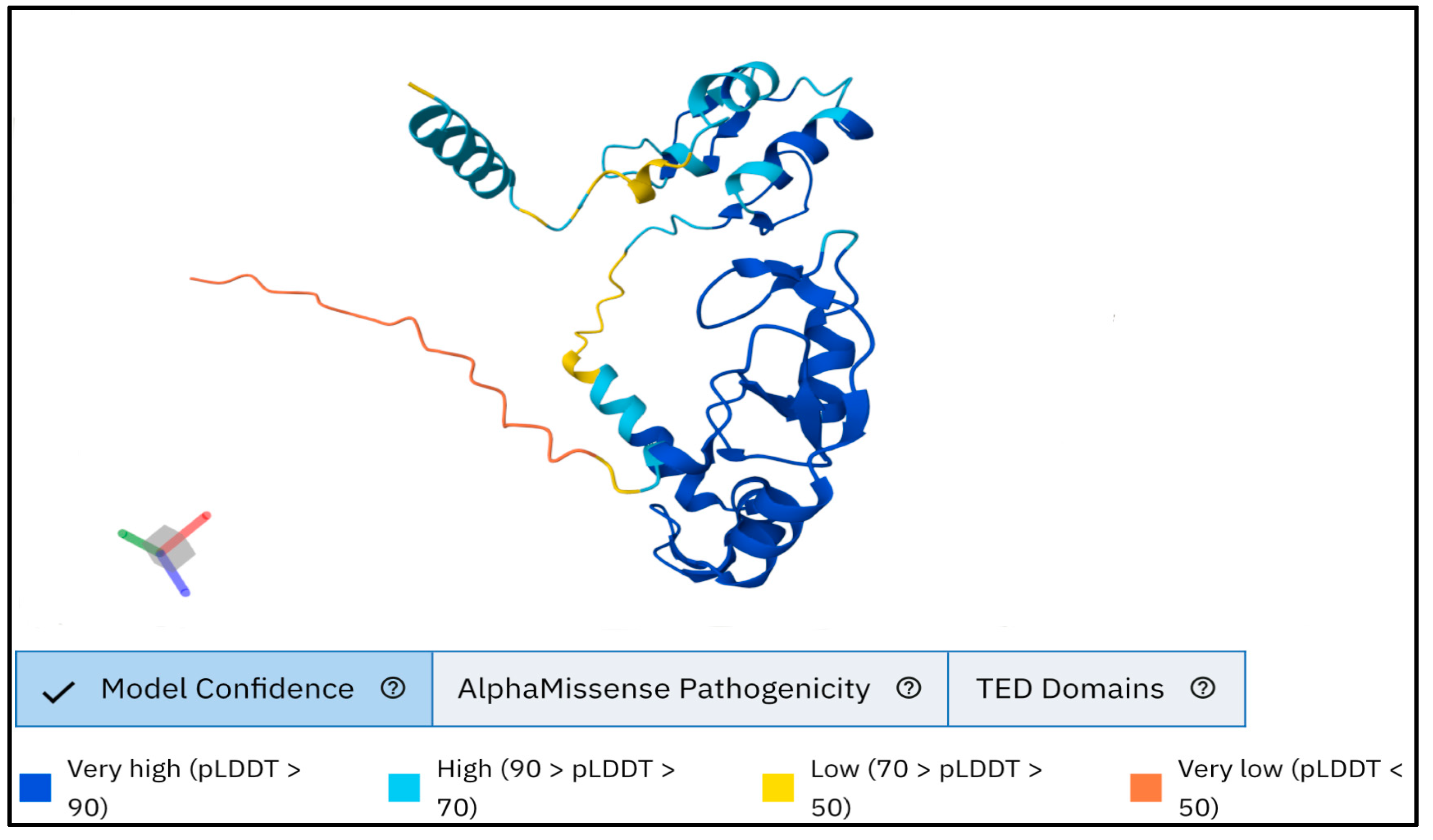
3.4. Three-Dimensional Structure Prediction by AlphaFold and Visualization of Wild and Mutant Type Proteins by ChimeraX
3.5. Evaluation of Conservation and Predicted Surface Accessibility for RNF114 Variants
3.6. Domain Identification of the RNF114 Protein by the InterPro Server
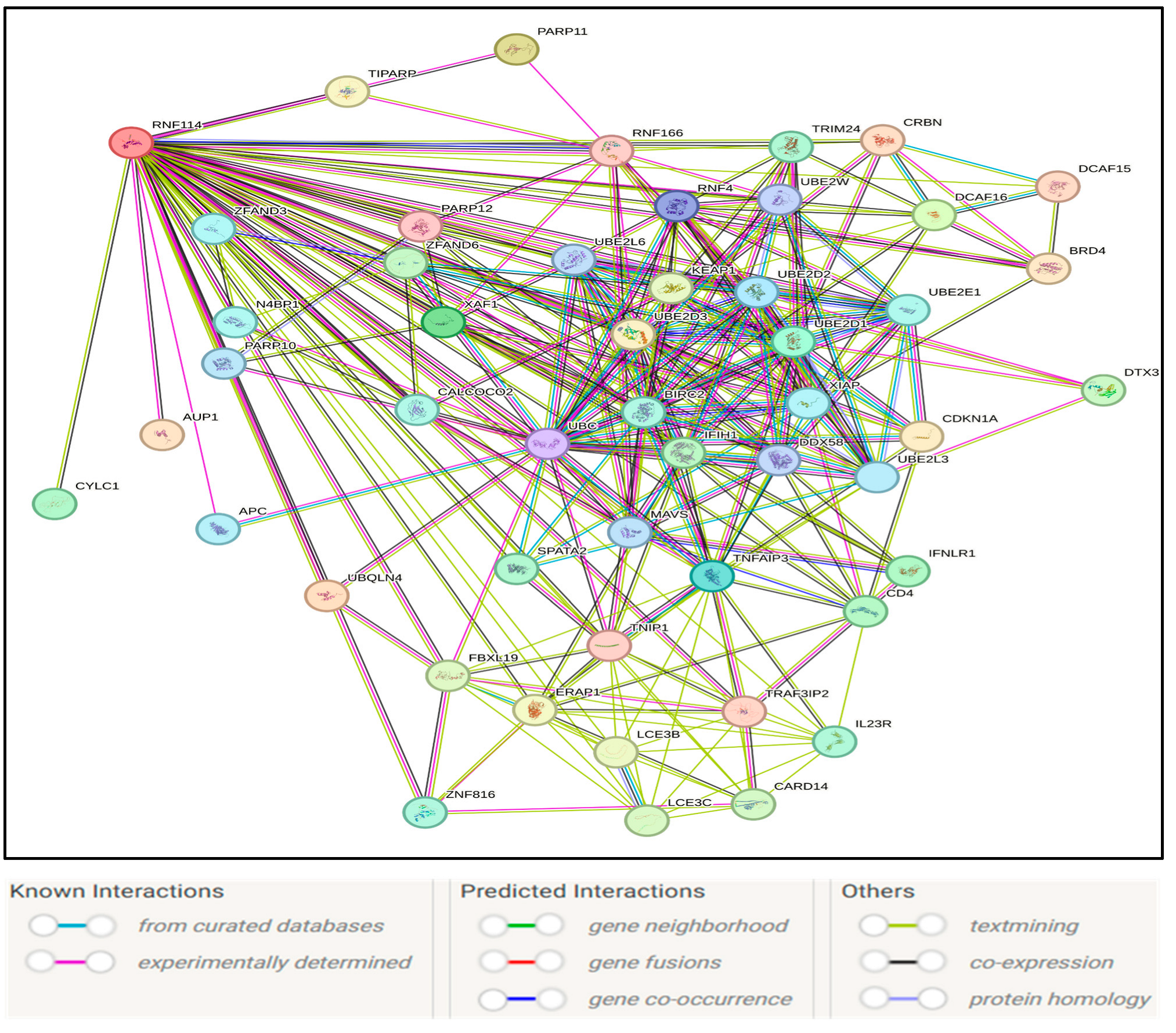
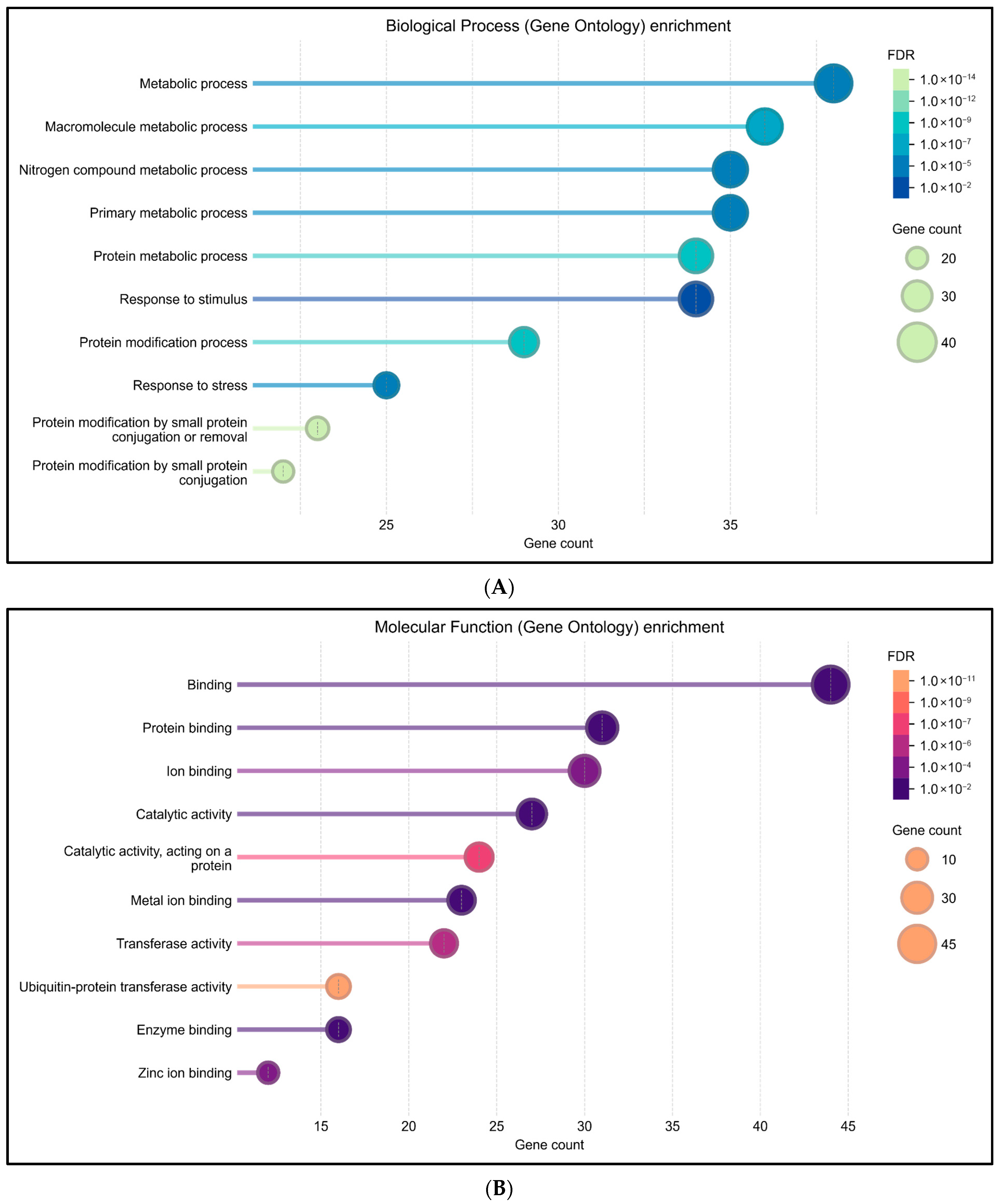
3.7. RNF114-Interacting Proteins and Gene Ontology Analysis
4. Discussion
5. Conclusions
6. Recommendation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ippagunta, S.K.; Gangwar, R.; Finkelstein, D.; Vogel, P.; Pelletier, S.; Gingras, S.; Redecke, V.; Häcker, H. Keratinocytes contribute intrinsically to psoriasis upon loss of Tnip1 function. Proc. Natl. Acad. Sci. USA 2016, 113, E6162–E6171. [Google Scholar] [CrossRef]
- Ni, X.; Lai, Y. Keratinocyte: A trigger or an executor of psoriasis? J. Leukoc. Biol. 2020, 108, 485–491. [Google Scholar] [CrossRef]
- Yang, L.; Fan, X.; Wang, G. 016 Hyperactivation of Nrf2 contributes to keratinocyte hyperplasia in psoriasis by promoting Keratin 6, 16 and 17 expressions. J. Investig. Dermatol. 2017, 137, S3. [Google Scholar] [CrossRef]
- Niehues, H.; Rikken, G.; van Vlijmen-Willems, I.M.J.J.; Rodijk-Olthuis, D.; van Erp, P.E.J.; Zeeuwen, P.L.J.M.; Schalkwijk, J.; van den Bogaard, E.H. Identification of keratinocyte mitogens: Implications for hyperproliferation in psoriasis and atopic dermatitis. JID Innov. 2022, 2, 100066. [Google Scholar] [CrossRef]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of Psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Stuart, P.E.; Tian, C.; Gudjonsson, J.E.; Das, S.; Zawistowski, M.; Ellinghaus, E.; Barker, J.N.; Chandran, V.; Dand, N.; et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 2017, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bijlmakers, M.-J.; Kanneganti, S.K.; Barker, J.N.; Trembath, R.C.; Capon, F. Functional analysis of the RNF114 psoriasis susceptibility gene implicates innate immune responses to double-stranded RNA in disease pathogenesis. Hum. Mol. Genet. 2011, 20, 3129–3137. [Google Scholar] [CrossRef]
- Capon, F.; Bijlmakers, M.-J.; Wolf, N.; Quaranta, M.; Huffmeier, U.; Allen, M.; Timms, K.; Abkevich, V.; Gutin, A.; Smith, R.; et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum. Mol. Genet. 2008, 17, 1938–1945. [Google Scholar] [CrossRef]
- Kamal, E. In Silico Prioritization of STAT1 3′ UTR SNPs Identifies rs190542524 as a miRNA-Linked Variant with Potential Oncogenic Impact. Non-Coding RNA 2025, 11, 32. [Google Scholar] [CrossRef]
- Kamal, E.; Kaddam, L.A.; Ahmed, M.; Alabdulkarim, A. Integrating Artificial Intelligence and Bioinformatics Methods to Identify Disruptive STAT1 Variants Impacting Protein Stability and Function. Genes 2025, 16, 303. [Google Scholar] [CrossRef]
- Armstrong, D.L.; Zidovetzki, R.; E Alarcón-Riquelme, M.; Tsao, B.P.; A Criswell, L.; Kimberly, R.P.; Harley, J.B.; Sivils, K.L.; Vyse, T.J.; Gaffney, P.M.; et al. GWAS identifies novel SLE susceptibility genes and explains the association of the HLA region. Genes Immun. 2014, 15, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Onoufriadis, A.; Simpson, M.A.; Burden, A.D.; Barker, J.N.; Trembath, R.C.; Capon, F. Identification of Rare, Disease-Associated Variants in the Promoter Region of the RNF114 Psoriasis Susceptibility Gene. J. Investig. Dermatol. 2012, 132, 1297–1299. [Google Scholar] [CrossRef] [PubMed]
- Momozawa, Y.; Mizukami, K. Unique roles of rare variants in the genetics of complex diseases in humans. J. Hum. Genet. 2021, 66, 11–23. [Google Scholar] [CrossRef]
- Folkersen, L.; Hooft, F.V.; Chernogubova, E.; Agardh, H.E.; Hansson, G.K.; Hedin, U.; Liska, J.; Syvänen, A.-C.; Paulsson-Berne, G.; Franco-Cereceda, A.; et al. Association of Genetic Risk Variants with Expression of Proximal Genes Identifies Novel Susceptibility Genes for Cardiovascular Disease. Circ. Cardiovasc. Genet. 2010, 3, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Clifford, R.J.; Edmonson, M.N.; Nguyen, C.; Scherpbier, T.; Hu, Y.; Buetow, K.H. Bioinformatics Tools for Single Nucleotide Polymorphism Discovery and Analysis. Ann. N. Y. Acad. Sci. 2004, 1020, 101–109. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Capriotti, E.; Altman, R.B.; Bromberg, Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genom. 2013, 14, S2. [Google Scholar] [CrossRef]
- Manfredi, M.; Savojardo, C.; Martelli, P.L.; Casadio, R. E-SNPs&GO: Embedding of protein sequence and function improves the annotation of human pathogenic variants. Bioinformatics 2022, 38, 5168–5174. [Google Scholar]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Calabrese, R.; Casadio, R. Predicting protein stability changes from sequences using support vector machines. Bioinformatics 2005, 21, ii54–ii58. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.; Baldi, P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct. Funct. Bioinform. 2006, 62, 1125–1132. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Biswas, M.; Bradley, P.; Bork, P.; Bucher, P.; et al. InterPro: An integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002, 3, 225–235. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Chen, X.; Nie, D.; Wang, J.; Wu, M. A RING finger protein 114 (RNF114) homolog from Chinese sturgeon (Acipenser sinensis) possesses immune-regulation properties via modulating RIG-I signaling pathway-mediated interferon expression. Fish Shellfish. Immunol. 2014, 41, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.S.; Egaña, I.; Lopitz-Otsoa, F.; Aillet, F.; Lopez-Mato, M.P.; Dorronsoro, A.; Lobato-Gil, S.; Sutherland, J.D.; Barrio, R.; Trigueros, C.; et al. The RING ubiquitin E3 RNF114 interacts with A20 and modulates NF-κB activity and T-cell activation. Cell Death Dis. 2014, 5, e1399. [Google Scholar] [CrossRef]
- Shembade, N.; Ma, A.; Harhaj, E.W. Inhibition of NF-κB Signaling by A20 Through Disruption of Ubiquitin Enzyme Complexes. Science 2010, 327, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Maroof, A.; Patel, D.D. TNF-α-induced protein 3 (A20): The immunological rheostat. J. Allergy Clin. Immunol. 2018, 142, 401–402. [Google Scholar] [CrossRef] [PubMed]


| Bioinformatics Tool/Server | Function | Input | Cut-Off Value |
|---|---|---|---|
| SIFT | Predicts whether an amino acid substitution affects protein function based on sequence homology. | SNP ID | Tolerance Index (TI) < 0.05 indicates deleterious. ≥0.05 indicates tolerated |
| PolyPhen-2 | Assesses the impact of amino acid substitutions on protein structure and function using structural and evolutionary data. | Amino acid substitution | score ≥ 0.85 indicates probably damaging score > 0.5 < 0.85 possibly damaging <0.5 benign |
| PROVEAN | Evaluates the effect of amino acid substitutions on protein function by comparing sequences | Amino acid substitution | Score ≤ −2.5 indicates deleterious >−2.5 indicates benign |
| META-SNP | Predicts the pathogenicity of SNPs by integrating multiple algorithms (SIFT, SNAP, and PhD-SNP) for consensus. | Amino acid substitution | A consensus score >0.5 indicates pathogenic |
| E-SNPs&GO | Integrates support vector machine with functional annotations derived from Gene Ontology terms. | Protein sequence in the FASTA format and the list of variations | RI closer to 0 indicating benign variants, RI closer to 10 indicating pathogenic variants |
| PANTHER | Classifies nsSNPs based on evolutionary relationships and functional annotations. | Protein Sequence, and amino acid acid substitution | probably damaging (PSEP time > 450 my, possibly damaging(PSEP 450 my > time > 200 my, and probably benign (PSEP time < 200 my). |
| Alpha Missense | Predicts the impact of missense mutations on protein function using structural modelling. | UniProt ID, And official gene symbol | Pathogenicity score: 0–0.34: likely benign, 0.34–0.564: ambiguous, 0.564–0.78: likely pathogenic 0.78–1.0: likely pathogenic |
| I-Mutant2.0 | Predicts changes in protein stability upon mutation by calculating ΔΔG values. | Amino acid sequence and Amino acid substitution | ΔΔG > 0 indicates decreased stability |
| MUpro | Predicts the effect of mutations on protein stability using machine-learning algorithms. | Amino acid sequence and Amino acid substitution | Score < 0.0 indicates destabilizing effect Score > 0.0 indicates stabilizing effect. |
| AlphaFold | Provides highly accurate protein structure predictions based on amino acid sequences | UniProt ID, official gene symbol | pLDDT > 90 Very high confidence 90 > pLDDT > 70 High confidence 70 > pLDDT > 50 Low confidence pLDDT < 50 Very low confidence |
| ChimeraX | Visualizes molecular structures and interactions, allowing for detailed analysis of protein conformations and mutations. | Protein structure data | N/A |
| ConSurf | Analyzes evolutionary conservation of amino acids to determine their functional importance. | protein sequence | Conservation score 9 indicates the highest conservation |
| InterPro | Integrates diverse protein family, domain, and functional site information to provide comprehensive functional annotations. | Protein sequence | N/A |
| STRING | Predicts protein-protein interactions to provide biological context for nsSNPs. | Protein name | Interaction score > 0.4 indicates significant interaction |
| nsSNP ID | Amino Acid Changes | SIFT | PolyPhen2.0 | Provean | META-SNP | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | T1 | Effect | Score | Effect | Score | Prediction | Score | |||
| 1 | rs2090286290 | F27S | deleterious | 0 | possibly damaging | 0.843 | Deleterious | −4.344 | Disease | 0.71 |
| 2 | rs1386412786 | C29R | deleterious | 0 | probably damaging | 1 | Deleterious | −9.438 | Disease | 0.858 |
| 3 | rs773986175 | V40A | deleterious | 0.03 | possibly damaging | 0.765 | Deleterious | −2.68 | Disease | 0.675 |
| 4 | rs1462212625 | H46Y | deleterious | 0 | probably damaging | 0.958 | Deleterious | −4.792 | Disease | 0.791 |
| 5 | rs1600868749 | C49R | deleterious | 0 | probably damaging | 1 | Deleterious | −11.048 | Disease | 0.827 |
| 6 | rs1305658652 | C52R | deleterious | 0 | probably damaging | 1 | Deleterious | −10.453 | Disease | 0.772 |
| 7 | rs745318334 | R68C | deleterious | 0 | probably damaging | 0.916 | Deleterious | −5.546 | Disease | 0.793 |
| 8 | rs758000156 | R68H | deleterious | 0 | probably damaging | 0.993 | Deleterious | −3.848 | Disease | 0.773 |
| 9 | rs772338253 | S101C | deleterious | 0.04 | possibly damaging | 0.881 | Deleterious | −2.605 | Disease | 0.648 |
| 10 | rs2090326008 | S101P | deleterious | 0.02 | possibly damaging | 0.799 | Deleterious | −3.274 | Disease | 0.715 |
| 11 | rs775686162 | R104Q | deleterious | 0.03 | possibly damaging | 0.67 | Deleterious | −2.649 | Disease | 0.734 |
| 12 | rs1017816166 | R104W | deleterious | 0.02 | probably damaging | 0.997 | Deleterious | −6.345 | Disease | 0.787 |
| 13 | rs2090326107 | H106N | deleterious | 0.02 | probably damaging | 0.965 | Deleterious | −6.284 | Disease | 0.785 |
| 14 | rs751133548 | C110S | deleterious | 0 | probably damaging | 0.998 | Deleterious | −8.968 | Disease | 0.792 |
| 15 | rs759089769 | K112T | deleterious | 0.02 | possibly damaging | 0.782 | Deleterious | −4.729 | Disease | 0.569 |
| 16 | rs775076155 | R138C | deleterious | 0 | probably damaging | 0.992 | Deleterious | −5.423 | Disease | 0.736 |
| 17 | rs2090330323 | C143R | deleterious | 0 | probably damaging | 0.999 | Deleterious | −10.948 | Disease | 0.766 |
| 18 | rs2090330344 | P144S | deleterious | 0 | probably damaging | 0.999 | Deleterious | −6.444 | Disease | 0.721 |
| 19 | rs1568920426 | N150K | deleterious | 0 | probably damaging | 0.992 | Deleterious | −4.835 | Disease | 0.505 |
| 20 | rs866758335 | P174L | deleterious | 0.02 | probably damaging | 0.999 | Deleterious | −8.527 | Disease | 0.577 |
| 21 | rs1443484485 | P174T | deleterious | 0 | probably damaging | 0.999 | Deleterious | −6.91 | Disease | 0.576 |
| 22 | rs766819838 | C176R | deleterious | 0 | probably damaging | 1 | Deleterious | −11.035 | Disease | 0.755 |
| 23 | rs754977174 | M179T | deleterious | 0 | possibly damaging | 0.834 | Deleterious | −4.604 | Disease | 0.599 |
| 24 | rs2090346648 | P184S | deleterious | 0.02 | probably damaging | 0.985 | Deleterious | −5.709 | Disease | 0.501 |
| 25 | rs772186536 | H194R | deleterious | 0 | probably damaging | 0.967 | Deleterious | −7.013 | Disease | 0.67 |
| 26 | rs866406478 | R198Q | deleterious | 0 | probably damaging | 0.978 | Deleterious | −2.902 | Disease | 0.671 |
| 27 | rs776148672 | R198W | deleterious | 0.04 | probably damaging | 0.998 | Deleterious | −5.858 | Disease | 0.722 |
| 28 | rs761458275 | R200W | deleterious | 0 | possibly damaging | 0.631 | Deleterious | −3.729 | Disease | 0.696 |
| 29 | rs2090346937 | T205A | deleterious | 0 | probably damaging | 0.998 | Deleterious | −3.802 | Disease | 0.524 |
| 30 | rs1262555907 | T205I | deleterious | 0 | probably damaging | 0.999 | Deleterious | 4.809 | Disease | 0.673 |
| 31 | rs1160221496 | F206S | deleterious | 0 | probably damaging | 0.978 | Deleterious | −5.9 | Disease | 0.755 |
| 32 | rs2090357290 | D212G | deleterious | 0.01 | probably damaging | 0.998 | Deleterious | −5.718 | Disease | 0.686 |
| NsSNP | Amino Acid Change | ESNP & GO | Panther | |||
|---|---|---|---|---|---|---|
| Prediction | RI | Effect | PSEP | |||
| 1 | rs146221262 | H46Y | Pathogenic | 7 | Probably damaging | 1628 |
| 2 | rs1600868749 | C49R | Pathogenic | 8 | Probably damaging | 1628 |
| 3 | rs745318334 | R68C | Pathogenic | 7 | Probably damaging | 910 |
| 4 | rs758000156 | R68H | Pathogenic | 4 | Probably damaging | 910 |
| NsSNP | Amino Acid Change | Alpha-Missense Pathogenicity | Alpha-Missense Prediction | |
|---|---|---|---|---|
| 1 | rs1600868749 | C49R | 0.998 | Likely Pathogenic |
| 2 | rs745318334 | R68C | 0.947 | Likely Pathogenic |
| 3 | rs758000156 | R68H | 0.887 | Likely Pathogenic |
| NsSNP | Amino Acid Change | I-Mutant 2.0 | Mupro | ||||
|---|---|---|---|---|---|---|---|
| Stability | RI | DDG (kcal/mol) | Stability | DDG (kcal/mol) | |||
| 1 | rs1600868749 | C49R | Decrease | 5 | −0.84 | Decrease | −0.20914 |
| 2 | rs745318334 | R68C | Decrease | 5 | −0.27 | Decrease | −0.22222442 |
| 3 | rs758000156 | R68H | Decrease | 8 | −0.44 | Decrease | −0.72417906 |
| NsSNP | Amino Acid Change | Conservation Score | Prediction | |
|---|---|---|---|---|
| 1 | rs1600868749 | C49R | 9 | structural residue (highly conserved and buried) |
| 2 | rs745318334 | R68C | 9 | functional residue (highly conserved and exposed) |
| 3 | rs758000156 | R68H | 9 | functional residue (highly conserved and exposed) |
| RNF114 Protein Domains | Positions | nsSNPs |
|---|---|---|
| Zinc/RING finger domain, C3HC4 | 17–118 | C49R, R68C, R68H |
| Drought induced 19 protein (Di19), zinc-binding at position | 140–200 | |
| RING/U-box | 23–70 | C49R, R68C, R68H |
| RING-HC_RNF114 | 26–71 | C49R, R68C |
| Zinc finger RING-type profile | 29–68 | C49R, R68C, R68H |
| ring_2 | 29–67 | C49R |
| RING-type zinc-finger | 29–64 | C49R |
| Zinc finger C2HC RNF-type profile at position | 91–110 | |
| C2HC Zing finger domain | 85–117 | |
| Drought induced 19 protein (Di19), zinc-binding | 140–200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldoseri, G.M.; Alwabran, A.I.; Aldoseri, G.M.; Aldoseri, M.M.; Kamal, E. Computational Identification of RNF114 nsSNPs with Potential Roles in Psoriasis and Immune Dysregulation. Med. Sci. 2025, 13, 194. https://doi.org/10.3390/medsci13030194
Aldoseri GM, Alwabran AI, Aldoseri GM, Aldoseri MM, Kamal E. Computational Identification of RNF114 nsSNPs with Potential Roles in Psoriasis and Immune Dysregulation. Medical Sciences. 2025; 13(3):194. https://doi.org/10.3390/medsci13030194
Chicago/Turabian StyleAldoseri, Ghalia Mahfod, Arwa Ibrahim Alwabran, Ghanem Mahfod Aldoseri, Mobarak Mahfod Aldoseri, and Ebtihal Kamal. 2025. "Computational Identification of RNF114 nsSNPs with Potential Roles in Psoriasis and Immune Dysregulation" Medical Sciences 13, no. 3: 194. https://doi.org/10.3390/medsci13030194
APA StyleAldoseri, G. M., Alwabran, A. I., Aldoseri, G. M., Aldoseri, M. M., & Kamal, E. (2025). Computational Identification of RNF114 nsSNPs with Potential Roles in Psoriasis and Immune Dysregulation. Medical Sciences, 13(3), 194. https://doi.org/10.3390/medsci13030194






