Transcriptomic and Metagenomic Biomarkers in Peri-Implantitis: A Systematic Review, Diagnostic Meta-Analysis, and Functional Meta-Synthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. PICO Framework
- Population (P): Human-derived peri-implant tissue samples affected by peri-implantitis.
- Intervention (I): Application of transcriptomic, metagenomic, or machine learning methods for biomarker discovery.
- Comparator (C): Healthy peri-implant or periodontal tissues in matched or comparative analyses.
- Outcomes (O): Identification of key transcriptomic, microbial, or bioinformatic biomarkers as the primary outcome, and diagnostic performance metrics (e.g., ROC-AUC), methodological strategies, pathway classification, and immune cell deconvolution as secondary outcomes.
2.4. Information Sources and Search Strategy
2.5. Study Selection
2.6. Data Extraction
2.7. Outcome Measures
2.8. Risk of Bias and Evidence Certainty
2.9. Data Synthesis and Functional Meta-Synthesis
2.10. Diagnostic Meta-Analysis
3. Results
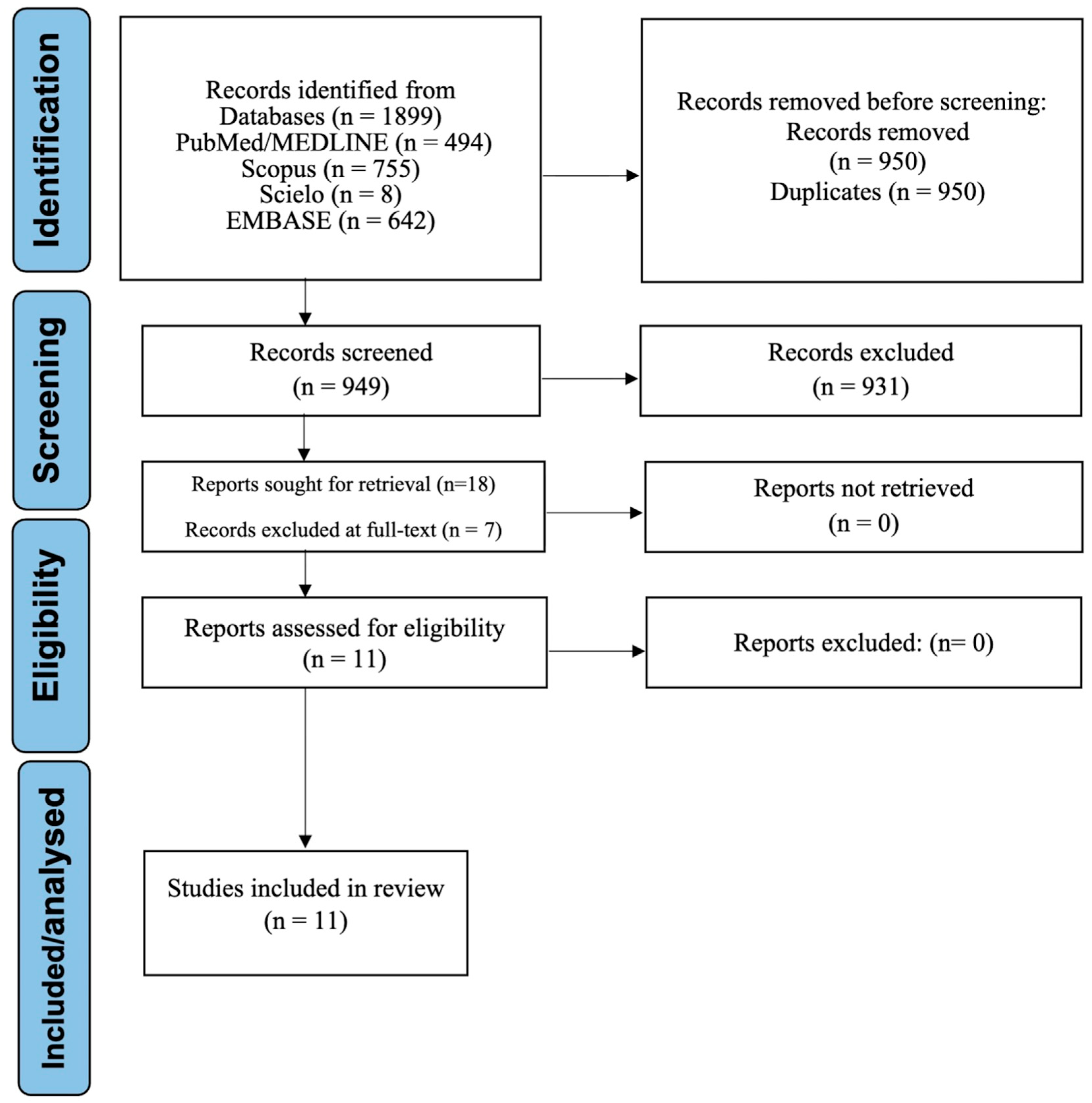
3.1. Study Selection
3.2. Overview of Included Studies
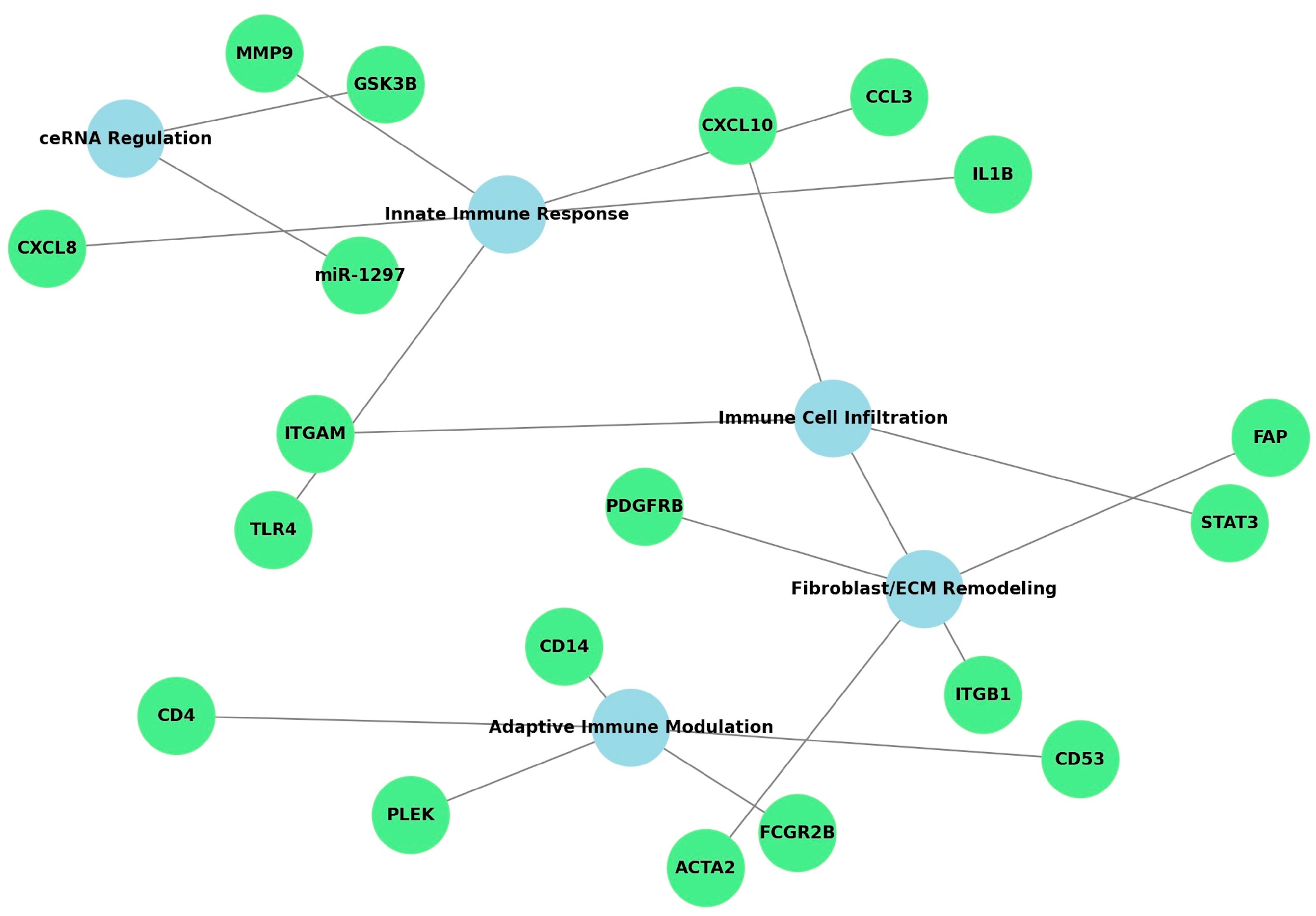
3.3. Functional Meta-Synthesis of Biomarkers and Pathways
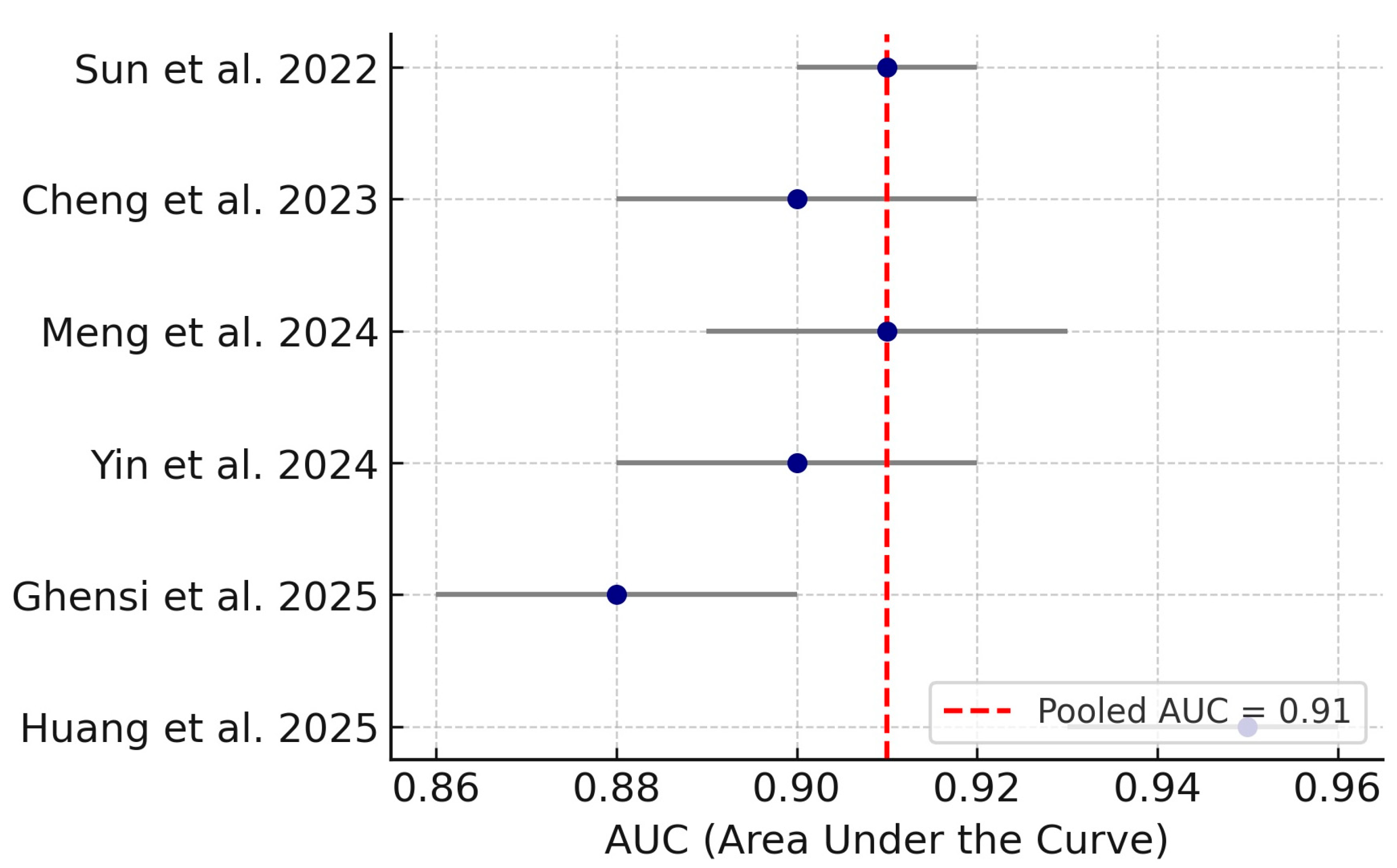
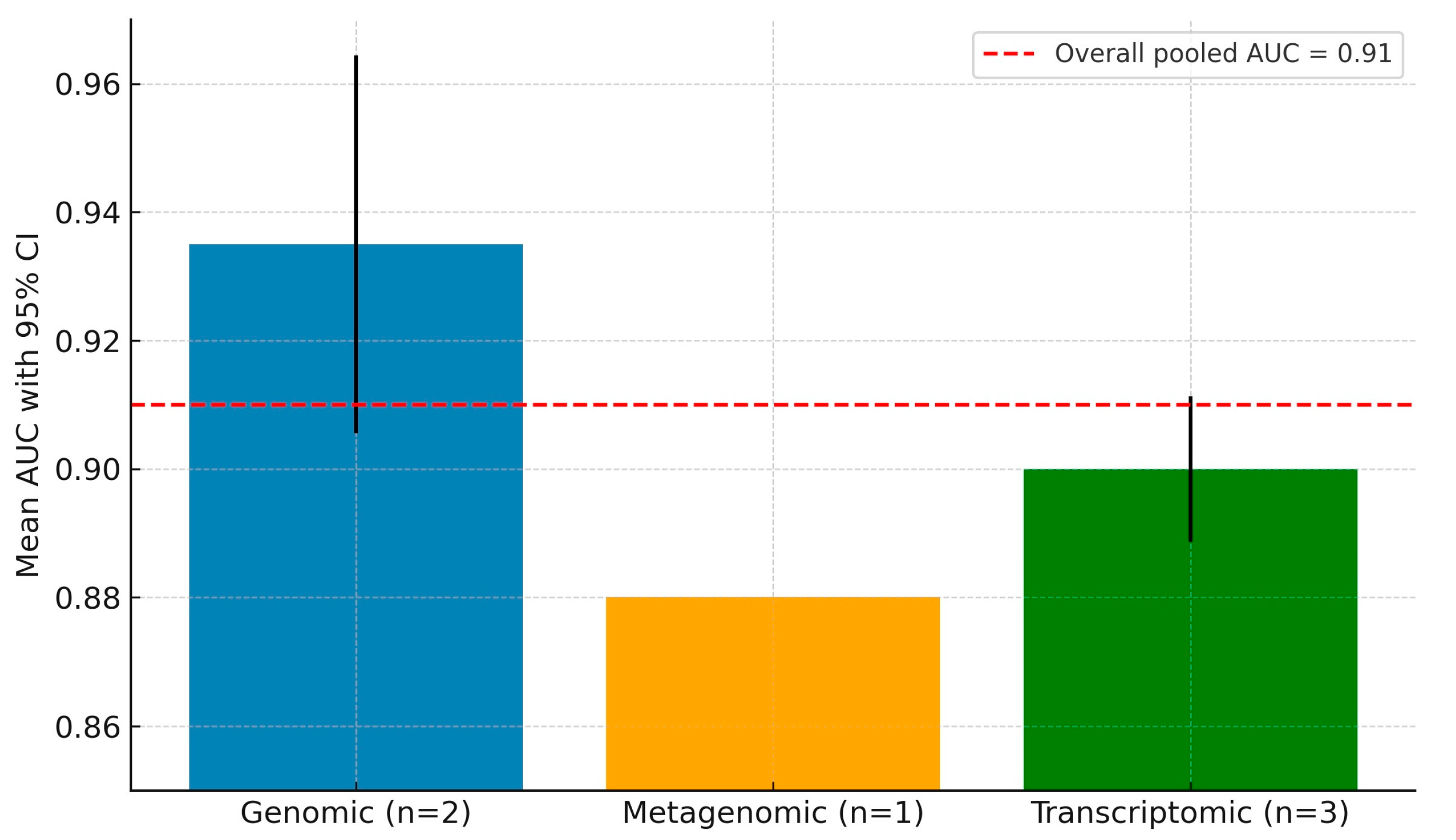
3.4. Diagnostic Meta-Analysis of Machine Learning Models
3.5. Additional Functional Insights and Secondary Outcomes
3.6. Risk of Bias and Evidence Certainty
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ardila, C.M.; Vivares-Builes, A.M. Antibiotic Resistance in Patients with Peri-Implantitis: A Systematic Scoping Review. Int. J. Environ. Res. Public Health 2022, 19, 15609. [Google Scholar] [CrossRef] [PubMed]
- Berglundh, T.; Armitage, G.; Araujo, M.G.; Avila-Ortiz, G.; Blanco, J.; Camargo, P.M.; Chen, S.; Cochran, D.; Derks, J.; Figuero, E.; et al. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S286–S291. [Google Scholar] [CrossRef]
- Romandini, M.; Lima, C.; Pedrinaci, I.; Araoz, A.; Soldini, M.C.; Sanz, M. Prevalence and risk/protective indicators of peri-implant diseases: A university-representative cross-sectional study. Clin. Oral Implant. Res. 2021, 32, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Yadalam, P.K.; Ardila, C.M. Deep Neural Networks Based on Sp7 Protein Sequence Prediction in Peri-Implant Bone Formation. Int. J. Dent. 2025, 2025, 7583275. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, D.; Xie, G.; Li, J.; Tang, J.; Tang, L. LncRNA-mediated ceRNA network was identified as a crucial determinant of differential effects in periodontitis and periimplantitis by high-throughput sequencing. Clin. Implant. Dent. Relat. Res. 2020, 22, 424–450. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Li, Z.; Acharya, A.; Chen, D.; Chen, Z.; Mattheos, N.; Chen, Z.; Huang, B. Long non-coding RNA and mRNA expression profiles in peri-implantitis vs periodontitis. J. Periodontal Res. 2020, 55, 342–353. [Google Scholar] [CrossRef]
- Barbagallo, G.; Santagati, M.; Guni, A.; Torrisi, P.; Spitale, A.; Stefani, S.; Ferlito, S.; Nibali, L. Microbiome differences in periodontal, peri-implant, and healthy sites: A cross-sectional pilot study. Clin. Oral Investig. 2022, 26, 2771–2781. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.J.; Lai, Y.R.; Huang, Q.R.; Li, Y.R.; Zhang, Y.J.; Chen, R.Y.; Qian, S.J. Single-cell sequencing identifies inflammation-promoting fibroblast-neutrophil interaction in peri-implantitis. J. Clin. Periodontol. 2024, 51, 196–208. [Google Scholar] [CrossRef]
- Ghensi, P.; Manghi, P.; Zolfo, M.; Armanini, F.; Pasolli, E.; Bolzan, M.; Bertelle, A.; Dell’Acqua, F.; Dellasega, E.; Waldner, R.; et al. Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics. NPJ Biofilms Microbiomes 2020, 6, 47. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, G.; Wu, H.; Huang, X.; Xu, R.; Deng, F.; Li, Y. Ebselen restores peri-implantitis-induced osteogenic inhibition via suppressing BMSCs ferroptosis. Exp. Cell Res. 2023, 427, 113612. [Google Scholar] [CrossRef] [PubMed]
- Varoquaux, G.; Colliot, O. Evaluating Machine Learning Models and Their Diagnostic Value. In Machine Learning for Brain Disorders; Colliot, O., Ed.; Humana: New York, NY, USA, 2023; Chapter 20. [Google Scholar] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Critical Appraisal Skills Programme (CASP). CASP Qualitative Checklist [Internet]; CASP UK: Oxford, UK, 2018; Available online: https://casp-uk.net/casp-tools-checklists/ (accessed on 28 July 2025).
- Lewin, S.; Booth, A.; Glenton, C.; Munthe-Kaas, H.; Rashidian, A.; Wainwright, M.; Bohren, M.A.; Tunçalp, Ö.; Colvin, C.J.; Garside, R.; et al. Applying GRADE-CERQual to qualitative evidence synthesis findings: Introduction to the series. Implement. Sci. 2018, 13 (Suppl. 1), 2. [Google Scholar] [CrossRef]
- Huang, J.; Zou, Y.; Deng, H.; Zha, J.; Pathak, J.L.; Chen, Y.; Ge, Q.; Wang, L. Integration of bioinformatics and machine learning strategies identifies ferroptosis and immune infiltration signatures in peri-implantitis. Int. J. Mol. Sci. 2025, 26, 4306. [Google Scholar] [CrossRef]
- Ghensi, P.; Heidrich, V.; Bazzani, D.; Asnicar, F.; Armanini, F.; Bertelle, A.; Dell’ACqua, F.; Dellasega, E.; Waldner, R.; Vicentini, D.; et al. Shotgun metagenomics identifies in a cross-sectional setting improved plaque microbiome biomarkers for peri-implant diseases. J. Clin. Periodontol. 2025, 52, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.M.; Kim, Y.; Lee, H.S.; Son, H.; Heo, H.J.; Baek, S.E.; Kim, E.K.; Lee, J.-Y.; Lee, K.E.; Kim, Y.H.; et al. Paired transcriptional analysis of periodontitis and peri-implantitis within same host: A pilot study. J. Dent. 2024, 151, 105366. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Fang, Y.; Liao, Y.; Chen, Z.; Liu, S.; Zhu, H.; Song, K.; Shi, B. Bioinformatics investigation of adaptive immune-related genes in peri-implantitis and periodontitis: Characteristics and diagnostic values. Immun. Inflamm. Dis. 2024, 12, e1272. [Google Scholar] [CrossRef]
- Meng, Q.; Han, J.; Zhang, X.; Su, W.; Liu, B.; Liu, T. Comprehensive analysis of immune infiltration and key genes in peri-implantitis using bioinformatics and molecular biology approaches. Med. Sci. Monit. 2024, 30, e941870. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yan, Q.; Huang, S.; Li, Y. Identification of key immune biomarkers in peri-implantitis through transcriptome analysis. Oral Dis. 2024, 30, 3982–3992. [Google Scholar] [CrossRef]
- Cheng, L.; Jin, S.; Lu, Q.; Zhou, J.; Liu, J.; Guan, X.; Xia, H.; He, H. Identification of immunological bioprocesses involved in peri-implantitis using weighted gene co-expression network analysis. J. Periodontol. 2023, 94, 392–403. [Google Scholar] [CrossRef]
- Sun, B.; Zhang, W.; Song, X.; Wu, X. Gene Correlation Network Analysis to Identify Biomarkers of Peri-Implantitis. Medicina 2022, 58, 1124. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Hu, L.; Shen, X.; Liu, C. Identification of Potential Genetic Biomarkers and Target Genes of Peri-Implantitis Using Bioinformatics Tools. BioMed Res. Int. 2021, 2021, 1759214. [Google Scholar] [CrossRef]
- Li, S.; Zhou, C.; Xu, Y.; Wang, Y.; Li, L.; Pelekos, G.; Ziebolz, D.; Schmalz, G.; Qin, Z. Similarity and Potential Relation Between Periimplantitis and Rheumatoid Arthritis on Transcriptomic Level: Results of a Bioinformatics Study. Front Immunol. 2021, 12, 702661. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, J.; Gong, C.; Lan, K.; Shen, Y.; Ding, X. Development of an immunogenomic landscape for the competing endogenous RNAs network of peri-implantitis. BMC Med. Genet. 2020, 21, 208. [Google Scholar] [CrossRef]
- Li, J.; Wei, J.J.; Wu, C.H.; Zou, T.; Zhao, H.; Huo, T.Q.; Wei, C.J.; Yang, T. Epimedin A inhibits the PI3K/AKT/NF-κB signalling axis and osteoclast differentiation by negatively regulating TRAF6 expression. Mol. Med. 2024, 30, 125. [Google Scholar] [CrossRef]
- Shuid, A.N.; Abdul Nasir, N.A.; Ab Azis, N.; Shuid, A.N.; Razali, N.; Ahmad Hairi, H.; Mohd Miswan, M.F.; Naina Mohamed, I. A Systematic Review on the Molecular Mechanisms of Resveratrol in Protecting Against Osteoporosis. Int. J. Mol. Sci. 2025, 26, 2893. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Zhao, Y.; Chen, R.; Li, W. Diagnostic performance of transcriptomic biomarkers in peri-implantitis: A machine learning approach. BMC Oral Health 2021, 21, 585. [Google Scholar]
- Bunte, K.; Beikler, T. Th17 Cells and the IL-23/IL-17 Axis in the Pathogenesis of Periodontitis and Immune-Mediated Inflammatory Diseases. Int. J. Mol. Sci. 2019, 20, 3394. [Google Scholar] [CrossRef]
- Ardila, C.M.; González-Arroyave, D.; Tobón, S. Machine learning for predicting antimicrobial resistance in critical and high-priority pathogens: A systematic review considering antimicrobial susceptibility tests in real-world healthcare settings. PLoS ONE 2025, 20, e0319460. [Google Scholar] [CrossRef] [PubMed]
- Yadalam, P.K.; Natarajan, P.M.; Ardila, C.M. Variational graph autoencoder for reconstructed transcriptomic data associated with NLRP3 mediated pyroptosis in periodontitis. Sci. Rep. 2025, 15, 1962. [Google Scholar] [CrossRef] [PubMed]
- Kadkhodazadeh, M.; Tabari, Z.A.; Pourseyediyan, T.; Najafi, K.; Amid, R. Relationship between Genetic Polymorphisms with Periodontitis and Peri-Implantitis in the Iranian Population: A Literature Review. J. Long-Term Eff. Med. Implant. 2016, 26, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Cai, Y.; He, L.; Jiang, H. Identification of a CircRNA-miRNA-mRNA Network and Integrated Analysis of Immune Infiltration in Oral Squamous Cell Carcinoma. J. Cancer 2023, 14, 250–261. [Google Scholar] [CrossRef]
- Wu, T.; Duan, Y.; Zhang, T.; Tian, W.; Liu, H.; Deng, Y. Research Trends in the Application of Artificial Intelligence in Oncology: A Bibliometric and Network Visualization Study. Front. Biosci. (Landmark Ed.) 2022, 27, 254. [Google Scholar] [CrossRef] [PubMed]
- Koçak, M.; Akçalı, Z. The published role of artificial intelligence in drug discovery and development: A bibliometric and social network analysis from 1990 to 2023. J. Cheminformatics 2025, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.A.; Vallania, F.; Liu, C.; Bongen, E.; Tomczak, A.; Andres-Terrè, M.; Lofgren, S.; Tam, A.; Deisseroth, C.A.; Li, M.D.; et al. Empowering multi-cohort gene expression analysis to increase reproducibility. Pac. Symp. Biocomput. 2017, 22, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.D.; Ensor, J.; Snell, K.I.E.; Harrell FEJr Martin, G.P.; Reitsma, J.B.; Moons, K.G.M.; Collins, G.; van Smeden, M. Calculating the sample size required for developing a clinical prediction model. BMJ 2020, 368, m441. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wang, C.; Jiang, W.; Wei, Y.; Li, Q.; Song, Z.; Li, S.; Sun, F.; Liu, Z.; Wang, Y.; et al. Comparative Transcriptome Analysis of Gingival Immune-Mediated Inflammation in Peri-Implantitis and Periodontitis Within the Same Host Environment. J. Inflamm. Res. 2022, 15, 3119–3133. [Google Scholar] [CrossRef]
- Delucchi, F.; Canepa, C.; Canullo, L.; Pesce, P.; Isola, G.; Menini, M. Biomarkers from Peri-Implant Crevicular Fluid (PICF) as Predictors of Peri-Implant Bone Loss: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 3202. [Google Scholar] [CrossRef]
- Grosse, S.D.; Gudgeon, J.M. Cost or price of sequencing? Implications for economic evaluations in genomic medicine. Genet. Med. 2021, 23, 1833–1835. [Google Scholar] [CrossRef] [PubMed]
- Condor, A.M.; Kui, A.; Condor, D.C.; Negucioiu, M.; Buduru, S.D.; Lucaciu, P.O. Metabolomics Applications for Diagnosing Peri-Implantitis: A Systematic Review of In Vivo Studies. Diagnostics 2025, 15, 990. [Google Scholar] [CrossRef]
- Bai, Y.; Min, R.; Chen, P.; Mei, S.; Deng, F.; Zheng, Z.; Jiang, C.; Miao, R.; Wu, Z.; Zhang, P.; et al. Disulfiram blocks inflammatory TLR4 signaling by targeting MD-2. Proc. Natl. Acad. Sci. USA 2023, 120, e2306399120. [Google Scholar] [CrossRef]
- Patra, M.C.; Achek, A.; Kim, G.Y.; Panneerselvam, S.; Shin, H.J.; Baek, W.Y.; Lee, W.H.; Sung, J.; Jeong, U.; Cho, E.Y.; et al. A Novel Small-Molecule Inhibitor of Endosomal TLRs Reduces Inflammation and Alleviates Autoimmune Disease Symptoms in Murine Models. Cells 2020, 9, 1648. [Google Scholar] [CrossRef] [PubMed]
- Akwata, D.; Kempen, A.L.; Dayal, N.; Brauer, N.R.; Sintim, H.O. Identification of a Selective FLT3 Inhibitor with Low Activity against VEGFR, FGFR, PDGFR, c-KIT, and RET Anti-Targets. ChemMedChem. 2024, 19, e202300442. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Fei, Z.; Haghani, A.; Robeck, T.R.; Zoller, J.A.; Li, C.Z.; Lowe, R.; Yan, Q.; Zhang, J.; Vu, H.; et al. Universal DNA methylation age across mammalian tissues. Nat. Aging 2023, 3, 1144–1166. [Google Scholar] [CrossRef] [PubMed]
| Author (Year) | Country | Design | Omics Type | Sample Type | Biomarker Type | ML/Bioinfo Method | Diagnostic Metric |
|---|---|---|---|---|---|---|---|
| Huang et al. (2025) [15] | China | Transcriptomic ML | Microarray | Peri-implant tissue | Ferroptosis, immune | LASSO, SVM-RFE, Boruta | AUC: 0.95 |
| Ghensi et al. (2025) [16] | Italy | Metagenomic | Shotgun metagenomics | Submucosal plaque | Microbial | Taxonomic/functional + ML | AUC: 0.88 |
| Oh et al. (2024) [17] | South Korea | Transcriptomic | RNA-seq | PI vs. Periodontitis tissue | Fibroblast markers | DEG analysis | Not reported |
| Yin et al. (2024) [18] | China | Transcriptomic | Microarray | Peri-implant tissue | Adaptive immune genes | Enrichment, ROC, qPCR | AUC: 0.90 |
| Meng et al. (2024) [19] | China | Transcriptomic ML | Microarray | Gingival tissue | Immune genes | LASSO, SVM-RFE, qPCR | AUC: 0.92 |
| Chen et al. (2024) [20] | China | Transcriptomic | Microarray | PI vs. healthy gingiva | Immune + DEG | PPI, enrichment, IHC | Not reported |
| Cheng et al. (2023) [21] | China | Transcriptomic | RNA-seq | Peri-implant tissues | Immune cell profiles | WGCNA, ssGSEA | AUC: 0.89 |
| Sun et al. (2022) [22] | China | Transcriptomic ML | Microarray | Peri-implant tissue | DEG + function | ML modeling, enrichment | AUC: 0.91 |
| Zhang et al. (2021) [23] | China | Transcriptomic | RNA-seq | Peri-implant tissue | Immune hub genes | WGCNA, clustering | Not reported |
| Li J et al. (2021) [24] | China | Transcriptomic ML | Microarray | Peri-implant tissues | Diagnostic DEGs | Feature selection, ML | AUC: 0.89 |
| Li M et al. (2020) [25] | China | Transcriptomic | Microarray | Peri-implant gingiva | Immune + ceRNA | ceRNA, DEG analysis | Not reported |
| Biological Theme | Contributing Studies | Key Genes | Associated Pathways |
|---|---|---|---|
| Innate Immune Response | Chen et al. [20], Cheng et al. [21], Zhang et al. [23] | IL1B, TLR4, CXCL8, CCL3, MMP9 | Cytokine-cytokine receptor interaction, TLR signaling |
| Adaptive Immune Modulation | Yin et al. [18], Li et al. [24], Chen et al. [20] | CD4, CD14, FCGR2B, CD53, PLEK | T cell receptor, NF-κB, Osteoclast differentiation |
| Immune Cell Infiltration | Chen et al. [20], Li et al. [25], Cheng et al. [21] | ITGAM, STAT3, CXCL10 | Chemokine signaling, Leukocyte migration |
| Fibroblast Activation and ECM | Oh et al. [17] | ACTA2, FAP, PDGFRB | PI3K-Akt, ECM-receptor interaction |
| ceRNA Networks | Li et al. [25] | GSK3B, miR-1297 | ceRNA regulation, Wnt signaling |
| Study | Clear Aim and Design | Data Appropriateness | Analytic Transparency | Validation Robustness |
|---|---|---|---|---|
| Huang et al. [15] | Yes | Yes | Yes | Yes |
| Ghensi et al. [16] | Yes | Yes | Yes | Moderate |
| Oh et al. [17] | Yes | Yes | Moderate | No |
| Yin et al. [18] | Yes | Yes | Yes | Yes |
| Meng et al. [19] | Yes | Yes | Yes | Yes |
| Chen et al. [20] | Yes | Yes | Moderate | Moderate |
| Cheng et al. [21] | Yes | Yes | Yes | Moderate |
| Sun et al. [22] | Yes | Yes | Yes | Yes |
| Zhang et al. [23] | Yes | Moderate | Moderate | No |
| Li J et al. [24] | Yes | Yes | Yes | Moderate |
| Li M et al. [25] | Yes | Moderate | Moderate | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ardila, C.M.; Pineda-Vélez, E.; Vivares-Builes, A.M. Transcriptomic and Metagenomic Biomarkers in Peri-Implantitis: A Systematic Review, Diagnostic Meta-Analysis, and Functional Meta-Synthesis. Med. Sci. 2025, 13, 187. https://doi.org/10.3390/medsci13030187
Ardila CM, Pineda-Vélez E, Vivares-Builes AM. Transcriptomic and Metagenomic Biomarkers in Peri-Implantitis: A Systematic Review, Diagnostic Meta-Analysis, and Functional Meta-Synthesis. Medical Sciences. 2025; 13(3):187. https://doi.org/10.3390/medsci13030187
Chicago/Turabian StyleArdila, Carlos M., Eliana Pineda-Vélez, and Anny M. Vivares-Builes. 2025. "Transcriptomic and Metagenomic Biomarkers in Peri-Implantitis: A Systematic Review, Diagnostic Meta-Analysis, and Functional Meta-Synthesis" Medical Sciences 13, no. 3: 187. https://doi.org/10.3390/medsci13030187
APA StyleArdila, C. M., Pineda-Vélez, E., & Vivares-Builes, A. M. (2025). Transcriptomic and Metagenomic Biomarkers in Peri-Implantitis: A Systematic Review, Diagnostic Meta-Analysis, and Functional Meta-Synthesis. Medical Sciences, 13(3), 187. https://doi.org/10.3390/medsci13030187







