Polymicrobial Infections in the Immunocompromised Host: The COVID-19 Realm and Beyond
Abstract
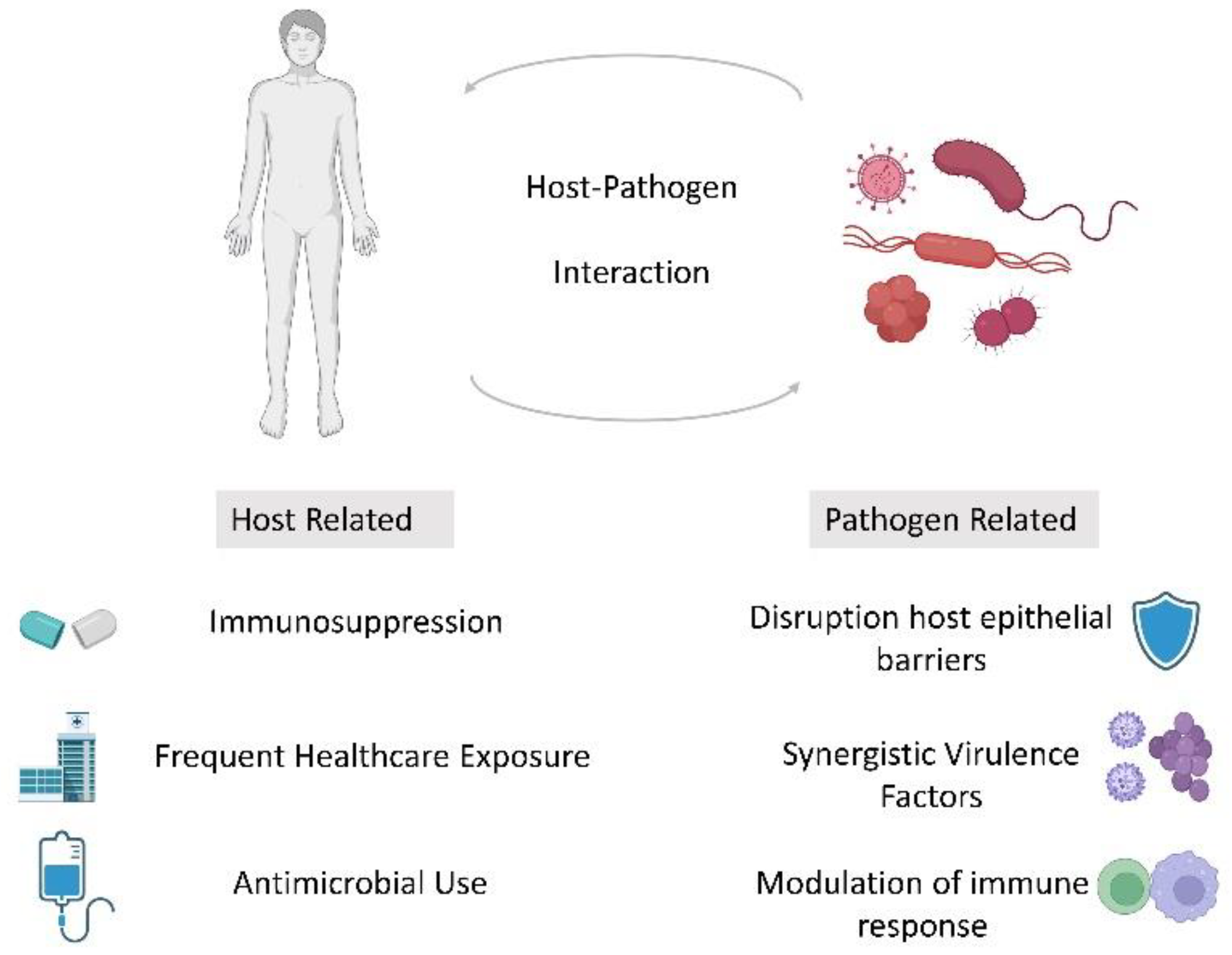
1. Introduction
2. Case Series
2.1. Case One
2.2. Case Two
2.3. Case Three
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, E.Y.; Monteforte, B.; Gupta, A.; Jiang, W.; May, L.; Hsieh, Y.H.; Dugas, A. The frequency of influenza and bacterial coinfection: A systematic review and meta-analysis. Influenza Other Respir. Viruses 2016, 10, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Rice, T.W.; Rubinson, L.; Uyeki, T.M.; Vaughn, F.L.; John, B.B.; Miller, R.R., III; Higgs, E.; Randolph, A.G.; Smoot, B.E.; Thompson, B.T. Critical illness from 2009 pandemic influenza A (H1N1) virus and bacterial co-infection in the United States. Crit. Care Med. 2012, 40, 1487. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Mishra, T.; Kumar, N.; Soubani, A.O. Influenza-associated aspergillosis: Nationwide trends, predictors and outcomes from 2005 to 2014. Chest 2020, 158, 1857–1866. [Google Scholar] [CrossRef] [PubMed]
- Koehler, P.; Bassetti, M.; Chakrabarti, A.; Chen, S.C.; Colombo, A.L.; Hoenigl, M.; Klimko, N.; Lass-Flörl, C.; Oladele, R.O.; Vinh, D.C. Defining and managing COVID-19-associated pulmonary aspergillosis: The 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect. Dis. 2021, 21, e149–e162. [Google Scholar] [CrossRef]
- Sia, I.G.; Patel, R. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant recipients. Clin. Microbiol. Rev. 2000, 13, 83–121. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balk, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, D.M.; et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef]
- Seo, S.; Waghmare, A.; Scott, E.M.; Xie, H.; Kuypers, J.M.; Hackman, R.C.; Campbell, A.P.; Choi, S.M.; Leisenring, W.M.; Jerome, K.R.; et al. Human rhinovirus detection in the lower respiratory tract of hematopoietic cell transplant recipients: Association with mortality. Haematologica 2017, 102, 1120–1130. [Google Scholar] [CrossRef]
- Khan, T.; Das, R.S.; Chaudhary, A.; Chatterjee, J.; Bhattacharya, S.D. Association of nasopharyngeal viruses and pathogenic bacteria in children and their parents with and without HIV. Pneumonia 2021, 13, 8. [Google Scholar] [CrossRef]
- Hofstra, J.J.; Matamoros, S.; van de Pol, M.A.; de Wever, B.; Tanck, M.W.; Wendt-Knol, H.; Deijs, M.; van der Hoek, L.; Wolthers, K.C.; Molenkamp, R.; et al. Changes in microbiota during experimental human Rhinovirus infection. BMC Infect. Dis. 2015, 15, 336. [Google Scholar] [CrossRef]
- Passariello, C.; Schippa, S.; Conti, C.; Russo, P.; Poggiali, F.; Garaci, E.; Palamara, A.T. Rhinoviruses promote internalisation of Staphylococcus aureus into non-fully permissive cultured pneumocytes. Microbes Infect. 2006, 8, 758–766. [Google Scholar] [CrossRef]
- Dubberke, E.; Burdette, S.; AST Infectious Diseases Community of Practice. Clostridium difficile infections in solid organ transplantation. Am. J. Transplant. 2013, 13, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Theel, E.S.; Ramanan, P. Clinical significance of low-positive histoplasma urine antigen results. J. Clin. Microbiol. 2014, 52, 3444–3446. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.P.; Shah, P.K.; Azzi, J.M.; Chemaly, R.F. Parainfluenza virus infections in hematopoietic cell transplant recipients and hematologic malignancy patients: A systematic review. Cancer Lett. 2016, 370, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Erard, V.; Chien, J.W.; Kim, H.W.; Nichols, W.G.; Flowers, M.E.; Martin, P.J.; Corey, L.; Boeckh, M. Airflow decline after myeloablative allogeneic hematopoietic cell transplantation: The role of community respiratory viruses. J. Infect. Dis. 2006, 193, 1619–1625. [Google Scholar] [CrossRef]
- Hirsch, H.; Martino, R.; Ward, K.; Boeckh, M.; Einsele, H.; Ljungman, P. Guidelines for diagnosis and treatment of human respiratory syncytial virus parainfluenza virus, metapneumovirus, rhinovirus and coronavirus. Clin. Infect. Dis. 2013, 56, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Feldman, C.; Anderson, R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia 2021, 13, 5. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M.; et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 2021, 27, 83–88. [Google Scholar] [CrossRef]
- Krumbein, H.; Kümmel, L.S.; Fragkou, P.C.; Thölken, C.; Hünerbein, B.L.; Reiter, R.; Papathanasiou, K.A.; Renz, H.; Skevaki, C. Respiratory viral co-infections in patients with COVID-19 and associated outcomes: A systematic review and meta-analysis. Rev. Med. Virol. 2022, e2365. [Google Scholar] [CrossRef]
- Crotty, M.P.; Akins, R.; Nguyen, A.; Slika, R.; Rahmanzadeh, K.; Wilson, M.H.; Dominguez, E.A. Investigation of subsequent and co-infections associated with SARS-CoV-2 (COVID-19) in hospitalized patients. medRxiv 2020. [Google Scholar]
- Zhang, G.; Hu, C.; Luo, L.; Fang, F.; Chen, Y.; Li, J.; Peng, Z.; Pan, H. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J. Clin. Virol. 2020, 127, 104364. [Google Scholar] [CrossRef]
- Lv, Z.; Cheng, S.; Le, J.; Huang, J.; Feng, L.; Zhang, B.; Li, Y. Clinical characteristics and co-infections of 354 hospitalized patients with COVID-19 in Wuhan, China: A retrospective cohort study. Microbes Infect. 2020, 22, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Amin-Chowdhury, Z.; Aiano, F.; Mensah, A.; Sheppard, C.L.; Litt, D.; Fry, N.K.; Andrews, N.; Ramsay, M.E.; Ladhani, S.N. Impact of the Coronavirus Disease 2019 (COVID-19) Pandemic on Invasive Pneumococcal Disease and Risk of Pneumococcal Coinfection With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Prospective National Cohort Study, England. Clin. Infect. Dis. 2021, 72, e65–e75. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.M.; Ashar, H.K.; Chow, V.T.; Teluguakula, N. Lethal synergism between influenza and Streptococcus pneumoniae. Infect. Pulm. Dis. 2016, 2. [Google Scholar] [CrossRef]
- Kim, E.-H.; Nguyen, T.-Q.; Casel, M.A.B.; Rollon, R.; Kim, S.-M.; Kim, Y.-I.; Yu, K.-M.; Jang, S.-G.; Yang, J.; Poo, H.; et al. Coinfection with SARS-CoV-2 and Influenza A Virus Increases Disease Severity and Impairs Neutralizing Antibody and CD4+ T Cell Responses. J. Virol. 2022, 96, e01873-21. [Google Scholar] [CrossRef]
- Gupta, A.; Karyakarte, R.; Joshi, S.; Das, R.; Jani, K.; Shouche, Y.; Sharma, A. Nasopharyngeal microbiome reveals the prevalence of opportunistic pathogens in SARS-CoV-2 infected individuals and their association with host types. Microbes Infect. 2022, 24, 104880. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Shafiekhani, M.; Shekari, Z.; Boorboor, A.; Zare, Z.; Arabsheybani, S.; Azadeh, N. Bacterial and fungal co-infections with SARS-CoV-2 in solid organ recipients: A retrospective study. Virol. J. 2022, 19, 35. [Google Scholar] [CrossRef]
- Baghdadi, J.D.; Coffey, K.C.; Adediran, T.; Goodman, K.E.; Pineles, L.; Magder, L.S.; O’Hara, L.M.; Pineles, B.L.; Nadimpalli, G.; Morgan, D.J.; et al. Antibiotic Use and Bacterial Infection among Inpatients in the First Wave of COVID-19: A Retrospective Cohort Study of 64,691 Patients. Antimicrob. Agents Chemother. 2021, 65, e0134121. [Google Scholar] [CrossRef]
- McCullers, J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014, 12, 252–262. [Google Scholar] [CrossRef]
- Salazar, F.; Bignell, E.; Brown, G.D.; Cook, P.C.; Warris, A. Pathogenesis of Respiratory Viral and Fungal Coinfections. Clin. Microbiol. Rev. 2022, 35, e00094-21. [Google Scholar] [CrossRef]
- Croft, C.A.; Culibrk, L.; Moore, M.M.; Tebbutt, S.J. Interactions of Aspergillus fumigatus conidia with airway epithelial cells: A critical review. Front. Microbiol. 2016, 7, 472. [Google Scholar] [CrossRef] [PubMed]
- Singh, N. Interactions between viruses in transplant recipients. Clin. Infect. Dis. 2005, 40, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.E.; Caliendo, A.M.; Arias, C.A.; Englund, J.A.; Lee, M.J.; Loeb, M.; Patel, R.; El Alayli, A.; Kalot, M.A.; Falck-Ytter, Y. Infectious Diseases Society of America guidelines on the diagnosis of coronavirus disease 2019. Clin. Infect. Dis. 2020, ciaa760. [Google Scholar] [CrossRef] [PubMed]
- Trifilio, S.; Zhou, Z.; Fong, J.L.; Zomas, A.; Liu, D.; Zhao, C.; Zhang, J.; Mehta, J. Polymicrobial bacterial or fungal infections: Incidence, spectrum of infection, risk factors, and clinical outcomes from a large hematopoietic stem cell transplant center. Transpl. Infect. Dis. 2015, 17, 267–274. [Google Scholar] [CrossRef]
| Bacterial Coinfections in Respiratory Viral Disease [25,29] |
|
| Fungal Coinfections in Respiratory Viral Disease [30,31] |
|
| Viral Coinfections [32] |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Higgins, E.; Gupta, A.; Cummins, N.W. Polymicrobial Infections in the Immunocompromised Host: The COVID-19 Realm and Beyond. Med. Sci. 2022, 10, 60. https://doi.org/10.3390/medsci10040060
Higgins E, Gupta A, Cummins NW. Polymicrobial Infections in the Immunocompromised Host: The COVID-19 Realm and Beyond. Medical Sciences. 2022; 10(4):60. https://doi.org/10.3390/medsci10040060
Chicago/Turabian StyleHiggins, Eibhlin, Aanchal Gupta, and Nathan W. Cummins. 2022. "Polymicrobial Infections in the Immunocompromised Host: The COVID-19 Realm and Beyond" Medical Sciences 10, no. 4: 60. https://doi.org/10.3390/medsci10040060
APA StyleHiggins, E., Gupta, A., & Cummins, N. W. (2022). Polymicrobial Infections in the Immunocompromised Host: The COVID-19 Realm and Beyond. Medical Sciences, 10(4), 60. https://doi.org/10.3390/medsci10040060







