Abstract
Crystalline bedrock has been chosen for deep geologic long-term storage of used nuclear fuel in Finland. The risks generated by the deep subsurface microbial communities in these disposal sites need to be well characterised in advance to ensure safety. Deep subsurface microbial communities in a steady state are unlikely to contribute to known risk factors, such as corrosion or gas production. However, the construction of the geological final-disposal facility, bedrock disturbances, and hydraulic gradients cause changes that affect the microbial steady-state. To study the induced metabolism of deep microbial communities in changing environmental conditions, the activating effect of different electron donors and acceptors were measured with redox sensing fluorescent dyes (5-Cyano-2,3-ditolyl tetrazolium chloride, CTC and RedoxSensor™ Green, RSG). Fluids originating from two different fracture zones of the Finnish disposal site in Olkiluoto were studied. These fracture fluids were very dissimilar both chemically and in terms of bacterial and archaeal diversity. However, the microbial communities of both fracture fluids were activated, especially with acetate, which indicates the important role of acetate as a preferred electron donor for Olkiluoto deep subsurface communities.
1. Introduction
Microbial communities inhabiting deep terrestrial groundwater systems utilise all available energy sources [1]. These energy sources and the biogeochemical cycles of elements, such as carbon, nitrogen, and sulphur, are connected to each other. However, at least part of the energy or nutrient sources are often severely limited, and microbial communities adapted accordingly [2]. Diverse communities may form in such conditions, although the core microbiome often consist of only a few species [3,4], or even of a single-species ecosystem [5]. Such microbial communities may achieve relatively stable community structures over the course of time. The deep subsurface life in undisturbed condition exists in a balance that is maintained in a steady state by one or a few limiting energetic or nutrient factors. However, the rare microbiome in the deep subsurface normally comprises significantly more taxa than the core microbiome, and offers a considerable amount of genomic functionality and potential to respond to changing environmental conditions [3,4].
The crystalline bedrock has been chosen for deep geologic long-term storage of used nuclear fuel in Finland. Thus, the evaluation of risks posed by microbial communities in these geological disposal sites is highly important. Deep subsurface microbial communities in a steady state are unlikely to contribute to known risk factors, such as corrosion or gas formation. However, the construction of a geological final-disposal facility, bedrock disturbances, and hydraulic gradients cause changes that affect the microbial steady-state. In Finland, the Olkiluoto final-disposal site contains different groundwater types including sulphate-rich, brackish chloride, and highly saline waters [6]. Through conductive fracture zones, blending of these water types with different redox states will occur when tunnels are excavated. In addition, gases such as methane can be found extensively in some water types. Mixing these groundwater types may cause metabolic activation of the resident microorganisms, and in some cases, may cause the accumulation of corrosive sulphide [6].
Distinguishing between microbial viability, activity, dormancy, and death is complicated [7,8,9]. This is especially challenging in the deep subsurface, where the physiological state of microbes is poorly explained by laboratory cultures [2]. There are several approaches to study the microbial state of viability and activity, and methods have their benefits and drawbacks in terms of different ecosystems, populations, and applications [7,8]. These techniques include targeting the morphological/membrane integrity, membrane potential, efflux pump activity, respiratory activity, enzymatic activity, substrate uptake, and cellular energy of microorganisms. In addition, fast developing sequencing technology, especially metatranscriptomics, provides new possibilities for studying microbial activation. A more simple option for activation research is the reverse transcribed quantitative PCR (RT-qPCR) of transcripts of specifically-chosen marker genes. An overall mixing of different approaches presents a wider image of the studied diverse microbial communities than applying only one technique.
CTC, 5-cyano-2,3-ditolyl tetrazolium chloride, is a redox-sensing fluorescent dye that is incorporated into cells by metabolically-active microbes, and is reduced by the electron transport system components and dehydrogenases to red fluorescent formazan [10]. It has been widely used to study the metabolic activity of aerobic bacteria. In addition, facultative and obligate anaerobic microorganisms, including fermentative bacteria and methanogens, have been shown to reduce CTC to intracellular CTC-formazan crystals [11]. RSG (RedoxSensor™ Green) is a fluorogenic redox indicator dye that turns into a green fluorescent form by, notably, the microbial reductases of electron transport systems [12]. RSG has been suggested as an alternative for CTC, as, unlike CTC, RSG does not suppress the cellular metabolism of some species [12].
Acetate is a key electron donor in the deep subsurface, and has been shown to be produced by e.g., autotrophic, acetogenic bacteria [13], but could also be produced by anaerobic methane-oxidising archaea [14] and through fermentation [15]. In addition, hydrogen from different deep subsurface sources has been identified as a key energy source, which can be oxidised in anaerobic environments with nitrate, ferric iron, sulphate, or carbon dioxide as electron acceptors e.g., [16,17]. Thus, we hypothesise that acetate and hydrogen are factors limiting the microbial metabolic activity in the deep subsurface groundwater of Olkiluoto. Together with suitable electron acceptors, these electron donors may considerably increase the low metabolic rates of the steady-state microbial communities. We aimed to simulate the groundwater mixing which occurs in the deep nuclear waste disposal site, and which introduces previously limited electron donors and acceptors for microbial communities, and generates community activation in bedrock. Two fracture fluids from the deep geologic spent nuclear fuel disposal site in Olkiluoto, Finland, from the drillholes OL-KR6/125–130 m and OL-KR15/446–460 m, were studied. The primary energy sources driving the OL-KR6 fracture fluid subsurface community in the original steady-state were, based on earlier geochemical and microbiological data [18], assessed to be sulphate reduction and methane oxidation. Other processes such as nitrate, ferric iron, and nitrite reduction, as well as formate and acetate synthesis and consumption were also active in the water [18]. No information about the energy sources driving the OL-KR15 fracture fluid community were known in advance. However, gases such as methane and carbon dioxide were probable energy sources, as well as the low amount of sulphate that was known to be present in the fluid. Fracture fluid samples were depleted of most of the prevalent energy sources before the beginning of the study to enable community activation with the addition of different electron acceptors and donors. All electron donor and acceptor amendments were selected for studying different sulphur reactions, because sulphate is abundant in the Olkiluoto deep subsurface. In addition, sulphide accumulation poses a risk for the long-term safety of the nuclear waste repository, as it is corrosive to metals. Acetate and hydrogen were selected to activate sulphate-reducing bacteria. Anaerobic methanotrophs from the group ANME-2d archaea have frequently been detected in different Olkiluoto groundwaters [19,20,21]. This group has recently been found to perform methane oxidation coupled to sulphate, nitrate, or ferric iron reduction [14,22,23], and may thus possibly produce sulphide also in the Olkiluoto subsurface. For this reason, different methane amendment experiments were conducted. In addition, we hoped to see activation arising from the sulphide oxidation, as, on some occasions, sulphur oxidisers have been abundantly detected in Olkiluoto fracture fluids [19,20,21].
Consequently, the study focused especially on the role of sulphate. We aimed to clarify the roles of acetate and hydrogen as electron donors for sulphate reduction, as well as the importance of the anaerobic sulphate-driven methane oxidation in Olkiluoto deep subsurface groundwater communities. The two studied groundwaters were chosen based on differences in chemical compositions. Microbial activations were studied with respiratory activity measurements with CTC and RSG, and cellular energy measurement with ATP.
2. Materials and Methods
Sampling of fracture fluids. The Olkiluoto site has been extensively described [6] previously. For this study, two fracture fluids from Olkiluoto drillholes OL-KR6/125–130 m and OL-KR15/446–460 m were sampled in spring 2017. In order to obtain indigenous fracture fluids, the packer-isolated fracture zones were purged by removing stagnant drillhole water by pumping for a minimum of four weeks before the sample water was collected. The fracture fluid samples were collected into sterile, anaerobic glass bottles through a sterile silicon tube with an injection needle through the butyl rubber stopper. The gas from the bottles was led out through another injection needle equipped with a sterile filter and silicon tube with a water lock to prevent air from entering the sampling bottle. Samples were kept chilled and protected from light during transportation to the laboratory the next day.
Fracture fluid chemistry. The OL-KR6/125–130 m fracture fluid was a sulphate-rich water with low organic, but higher inorganic, carbon content than in OL-KR15/446–460 m fracture fluid, which had a low sulphate content, traces of sulphide, and it originated from a greater depth with more saline conditions than the OL-KR6 fluid (Table 1). Chemistry results were provided by Posiva Oy, and the methods applied are listed in Table 1. In general, at depths greater than 300 m, nitrogen is the most dominant dissolved gas, and the amount of methane increases after 300 m depth until 800 m (from a few to over 800 mL L−1 water) in Olkiluoto site [6]. The hydrogen content has generally been low but measurable (in the range of µL:s L−1) in fracture fluids, and carbon dioxide is present mainly at concentrations lower than 1 mL L−1.

Table 1.
OL-KR6/125–130 m and OL-KR15/446–460 m drillhole fracture fluid hydrogeochemical characteristics from Olkiluoto, Finland. na: not analysed, m b.s.l.; meters below sea level.
Estimation of microbial community sizes. The size of the microbial community in the original fracture fluids was determined by epifluorescence microscopy of 4′,6 diamidino-2-phenylindole (DAPI|Sigma-Aldrich, St. Louis, MO, USA) stained cells, as described in [24] from 5 mL subsamples. The microscopy slides were examined using an Axio Imager M2 epifluorescence microscope (Carl Zeiss Microscopy) equipped with a digital camera (AxioCam MRm, Carl Zeiss). The images were acquired and analysed using the Zen 2.3 (blue edition) software (Carl Zeiss), and the number of cells was counted from 30 random microscopy fields. The microbial cell number was determined from an average of two replicate filters of each sample.
Nucleic acid isolation. Community biomass for DNA extraction was collected by filtration of the fracture fluids on nitrocellulose acetate (CA) 0.2 µm filters (Corning Inc., Corning, NY, USA) from duplicate 1 L of each fracture fluid by vacuum suction. The filters were cut from the filter funnels with sterile scalpels and directly frozen at −80 °C in sterile 50 mL plastic test tubes. The frozen CA filters were thawed on ice and cut into two pieces each. DNA was extracted using the NucleoSpin Soil DNA extraction kit (Macherey-Nagel). Each membrane half was inserted into a separate bead tube for microbial cell lysis. The DNA was extracted using lysis buffer SL1 with Enhancer solution SX according to the manufacturer’s instructions. After the lysis step, the supernatant of the filter pair halves were combined for the remainder of the extraction protocol. The DNA was eluted into 100 µL elution buffer SE. Negative DNA isolation controls were included in the extraction procedure. The DNA concentration of each sample was determined using the Qubit 2.0 spectrophotometer (Life Technologies, Carlsbad, CA, USA).
Quantitative PCR (qPCR) was applied to estimate the number of bacteria and archaea (16S rRNA gene), sulphate reducers (dsrB), nitrate reducers (narG), and methanogens (mcrA) in the fracture fluid samples. The abundance of bacterial 16S rRNA genes was determined by qPCR, as described in [25] targeting bacterial 16S rRNA gene with primers P1 and P2 [26], which specifically target the V1–V3 region of the bacterial 16S rDNA gene. The size of the archaeal population in the fracture groundwaters was determined using primers ARC344f [27] and Ar744r (reverse complement from [28]) flanking the V4–V6 region of the archaeal 16S rRNA gene, as described in [3]. Sulphate reducers (dsrB) were quantified, as described in [24], and bacterial nitrate reducers (narG) and archaeal methanogens (mcrA) were determined according to [29] and [20] respectively.
The qPCR reactions were performed in 10 µL reaction volumes using the KAPA2 × Syrb® FAST qPCR kit on a LightCycler480 qPCR machine (Roche Applied Science, Penzberg, Germany) on white 96-well plates (Roche Applied Science) sealed with transparent adhesive seals (4titude®, Ockley, UK). Each reaction contained 2.5 µM of the relevant forward and reverse primer and 1 µL DNA extract. Each reaction was run in triplicate from duplicate samples. No-template control reactions were used to determine background fluorescence in the reactions. The number of amplified genes were determined by comparing the amplification result (Cp) to that of a standard dilution series (101–107 copies µL−1) of Esherichia coli (ATCC 31608) 16S rRNA genes in plasmid, genomic DNA of Halobacterium salinarum (DSM 3754) for archaea, Desulfobacterium autotrophicum (E-001658) dsrB genes in plasmid for sulphate reducers, Pseudomonas aeruginosa (VTT E-84219) narG genes in plasmid for nitrate reducers, and mcrA genes of Methanothermobacter thermoautotrophicus (DSM 1053) in plasmid for methanogens. The melting curve analysis and inhibition of the PCR was tested by comparing the amplification of the standard to the amplification in reactions containing the sample DNA spiked with 1 µL standard DNA. Inhibition of the qPCR assay by all template DNAs was found to be low. Nucleic acid extraction and reagent controls were run in all qPCR assays in parallel with the samples.
Amplicon library preparation. Amplicon libraries for high throughput sequencing with the Ion Torrent PGM platform were prepared by PCR from the DNA samples. Bacterial 16S rRNA genes were amplified with primers S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 [30], targeting the variable region V3-V4 region of the 16S rDNA gene and archaeal 16S genes with primers S-D-Arch-0349-a-S-17/S-D-Arch-0787-a-A-20 [31], targeting the V4 region of the gene. PCR amplification was performed in parallel 25 μL reactions for every sample containing 1x MyTaqTM Red Mix (Bioline, London, UK), 20 pmol of each primer, up to 25 μL molecular-biology-grade water (Sigma) and 2 μL of template DNA. The PCR program consisted of an initial denaturation step at 95 °C for 3 min, 35 cycles for bacteria, and 40 cycles for archaea of 15 s at 95 °C, 15 s at 50 °C, and 15 s at 72 °C. A final elongation step of 30 s was performed at 72 °C. The PCR products were verified with agarose gel electrophoresis. Amplicons were sent for sequencing to Bioser (Oulu, Finland), and amplicons were purified and size selected before sequencing by the staff at Bioser.
Amplicon sequence analysis. The Ion Torrent sequence data was analysed with mothur v. 1.39.5 [32]. Sequence reads were first subjected to quality control where raw sequence reads were subjected to quality trimming. Minimum sequence lengths were set to 250 nucleotides. A maximum of 8 homopolymers and 1 primer difference were allowed. Default settings were used with the exception of a lower qwindowaverage set to 25 to compensate for the Iontorrent sequencing platform. Sequences were aligned to the Silva seed alignment v.132 [33] for clustering and distance matrix calculation. Chimeric sequences were identified using the chimera.slayer in mothur, and were subsequently removed. Pairwise distances between unique sequences were calculated with a cutoff of 0.03. Classification of bacterial and archaeal sequences and OTUs was done against the Silva version 132 taxonomy [33]. Chao1 richness estimates and diversity indices were calculated for the bacterial and archaeal communities with 97% sequence similarity from the normalised data. Sequence data was subsampled to 5608 reads for bacteria and 13,513 reads for archaea.
The sequences were deposited in the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/) under accession number PRJEB28560.
Activation of fracture fluid microbial communities with electron donors and acceptors. The deep fracture waters of OL-KR6/125–130 m and OL-KR15/446–460 m were incubated without amendment for six-weeks protected from light at 14 °C before the activation experiments in order to exhaust the electron acceptor and donor sources of the original fracture fluids and record the microbial baseline activity. Altogether, 10 different electron acceptor or donor amendment and one control treatment without added substrates were prepared (Table 2). Additions were chosen based on the needed electron donors/acceptors in sulphur related reactions especially in sulphate reduction and anaerobic methane oxidation. The substrate concentrations were low in order to not inhibit oligotrophic microorganisms.

Table 2.
Activation experiments and electron acceptors and donors added to OL-KR6/125–130 m and OL-KR15/446–460 m fracture fluid samples.
All experiments were performed anaerobically with all reagents sterilised and rendered anoxic with nitrogen gas purging, with the exception of the sulphide. The used redox sensing fluorescent dyes CTC (5-Cyano-2,3-ditolyl tetrazolium chloride, Polysciences) and RSG (RedoxSensor™ Green, Molecular Probes) were diluted in distilled sterile water according to the manufacturers’ instructions, with final concentrations of CTC and RSG at 4.5 mM and 0.1 µM in the fracture fluids, respectively. Experiments were performed in 20 mL sterile glass microcosms in an anaerobic glove box containing nitrogen gas. The redox sensing dye was added to the sample water, mixed well, and divided into duplicate microcosms of 3.3 mL. Activating compounds (Table 2) were then added into the microcosms. Methane addition was performed by first removing 8 mL of microcosm headspace nitrogen gas with a sterile injection needle and syringe through the butyl rubber stopper, and then adding 13 mL of sterile filtered methane gas (100%). As an exception, hydrogen-containing microcosms were at first purged with a gas mixture consisting of hydrogen and nitrogen (10:90%), after which the microcosms were sealed with a gas tight butyl rubber stopper. Sample water containing the redox-sensing dyes was subsequently injected into the microcosms through the butyl rubber stopper using a hypodermic needle and syringe. In addition, 5 mL of sterile filtered hydrogen-nitrogen gas mixture was injected into microcosms to induce sufficient hydrogen over-pressure and dissolution in to the water phase. Microcosms were incubated for three hours at 14 °C in the dark, to ensure proper microbial activation, but no longer than that in order to minimise the formation of metabolites and further activation, as well as to diminish the possible toxic effect of the CTC stain.
After 3 h of incubation at 14 °C in the dark, 0.3 mL of fluid was taken from each microcosm for duplicate measurements with the ATP Biomass Kit HS (BioThema, Handen, Sweden) according to the manufacturer’s recommendations. The remaining sample in the microcosms was stained with DAPI by adding 10 µL mL−1 stain (0.5 mg mL−1) to the microcosms in the anaerobic chamber. Then, the microcosms were incubated for an additional 15 min. Each microcosm water was frozen into triplicate cryo-tubes containing 5% glycerol in 1×TE buffer (final concentrations, pH 8) at −80 °C.
In addition to the activation microcosms, unamended sample waters were stained with CTC, RSG, or DAPI, incubated for 3 h, and frozen in 5% GlyTE buffer at −80 °C. The number of microbial cells (DAPI), activated microbial cells (CTC, RSG), and uncoloured particles was determined from the pre-stained thawed microcosm waters using flow cytometry (BD FACSAria IIu, Becton Dickinson, NJ, USA). Undyed samples were analysed first, in order to determine the background fluorescence, followed by the samples dyed with DAPI alone to determine the cell fraction of the samples. Finally, the double-stained samples (DAPI + CTC or DAPI + RSG) were run to detect the metabolically-active cell population. For each sample, 10,000 events were acquired. All experiments were run in duplicate. The activation of the microbial communities in the groundwater was measured by comparing the number of fluorescent CTC or RSG dyed cells to the total number of microbial cells stained with DAPI.
3. Results
3.1. Microbial Community Analyses
The cell numbers in OL-KR6/125–130 m and OL-KR15/446–460 m fracture fluids were 3.8 × 104 and 4.0 × 104 cells mL−1, respectively, based on DAPI staining before the six-week incubation. The number of bacterial and archaeal 16S rRNA genes were higher in OL-KR15 fracture fluid (0.9–1.7 × 105 copies mL−1) than in OL-KR6 water (2.2–2.4 × 104 copies mL−1) (Figure 1). Sulphate reducer (dsrB) and especially nitrate reducer (narG) gene copy amounts were also higher in OL-KR15 than in OL-KR6 water. In contrast, methanogen (mcrA) genes were detected in the OL-KR6 fracture fluid, but not in the OL-KR15 water.
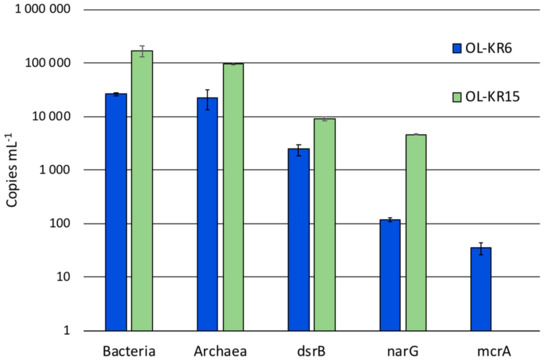
Figure 1.
The average number of bacterial and archaeal 16S rRNA, dsrB, narG and mcrA gene copies determined by qPCR from OL-KR6/125–130 m (blue) and OL-KR15/446–460 m (orange) fracture fluids (mL−1) from duplicate samples with triplicate reactions.
3.2. Sequence Statistics, Diversity Estimates and Microbial Communities in the Original Water
The number of sequence reads obtained from the two fracture fluid samples varied between 5608 and 16,958 for bacteria, and from 13,513 to 16,827 for archaea (Table 3). Compared to the actual numbers of observed operational taxonomic units (OTUs), on average, 55.7% and 61.2% of the Chao1 estimated numbers of bacterial OTUs were detected from OL-KR6 and from OL-KR15 fracture fluids normalised data, respectively. For archaea, the average OTU coverage was 69.4% and 72.9% from OL-KR6 and from OL-KR15 fracture fluids, respectively, and thus, higher for archaea than for bacteria in both samples. The Shannon diversity index H′ was higher in OL-KR6 water for both the bacterial and archaeal communities, compared to OL-KR15 (Table 3). Furthermore, H’ for the bacterial communities was higher than the archaeal H′ in both fracture fluids. From the bacterial sequences, altogether 35 different phyla and candidate phyla were detected from OL-KR6 and OL-KR15 combined, while the archaeal communities contained a total of nine phyla and candidate phyla. Of these phyla, only six bacterial and four archaeal ones were represented by more than 1% of the sequences in either sample (Figure 2 and Figure 3). The share of unclassified bacterial and archaeal sequences was 4.3% and 16.8% in the OL-KR6 sample, and less than 0.3% for both bacteria and archaea in OL-KR15 fracture fluid.

Table 3.
Total number of sequence reads and observed and estimated (Chao1) numbers of OTUs, number of singleton and doubleton OTUs, and the Shannon diversity index H′ of bacterial and archaeal 16S rRNA gene data from duplicate (A/B) samples of OL-KR6/125–130 m and OL-KR15/446–460 m fracture fluids normalised to 5608 (bacteria) and 13,513 (archaea) sequences.
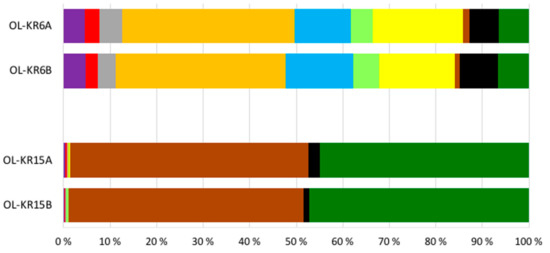
Figure 2.
Relative abundance of the bacterial classes belonging to the dominant (>1% of the sequence reads) bacterial phyla in OL-KR6/125–130 m and OL-KR15/446–460 m fracture fluids obtained by Ion Torrent sequencing. A and B indicate replicates of the same sample.
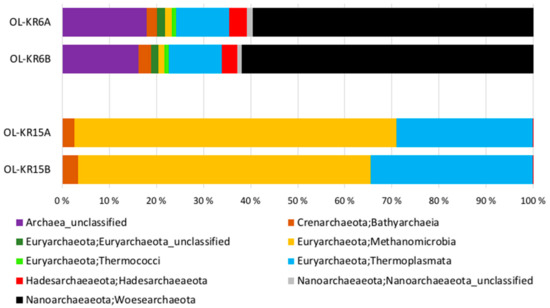
Figure 3.
Relative abundance of the archaeal classes belonging to the dominant (>1% of the sequence reads) archaeal phyla in OL-KR6/125–130 m and OL-KR15/446–460 m fracture fluids obtained by Ion Torrent sequencing. A and B indicate replicates of the same sample.
The bacterial and archaeal communities differed markedly from each other in the two studied fracture fluids. The bacterial community of OL-KR6 fracture fluid consisted of classes of Epsilonproteobacteria (35%), Parcubacteria (17%), Omnitrohicaeota (13%), Deltaproteobacteria (7%), and Gammaproteobacteria (6%), whereas the OL-KR15 fracture fluid contained mainly Alphaproteobacteria (49%) and Gammaproteobacteria (45%) (Figure 2). The archaeal community of OL-KR6 was dominated by Woesearchaeota (61%), together with an unclassified archaeal group (17%) and Thermoplasmata (11%). In OL-KR15, the archaeal microbiome consisted mainly of Methanomicrobia (65%) and Thermoplasmata (32%) (Figure 3).
3.3. Activation of Microbial Communities in the Fracture Fluids
Activation of the microbial communities in the two slightly alkaline fracture fluids was successful. Both CTC and RSG were actively transported into cells and reduced by the microbial activity, becoming fluorescent in the studied fracture fluids. After the six-week incubation period, the percent of fluorescent cells in the fracture fluids ranged from 36% to 54%, and from 75% to 90% after staining with CTC or RSG, respectively (Figure 4 and Figure 5). However, this high activity hampered the detection of further activation with added electron donors and acceptors. Based on the lower number of active microbial cells detected with CTC, further activation experiments were performed only with CTC. ATP amounts measured from the six-week incubated fracture fluids showed ATP amounts in OL-KR15 (0.28 pmol mL−1) that were more than six times higher than those in the OL-KR6 (0.04 pmol mL−1) fracture fluid. This was in agreement with the redox dyes, and indicated that OL-KR15 fracture fluid microorganisms were relatively active despite the six-week incubation period. However, as the ATP amounts were quite small and the deviations considerable, comparisons of the activated fracture fluids with electron donors and acceptors could not produce reliable results (data not shown).
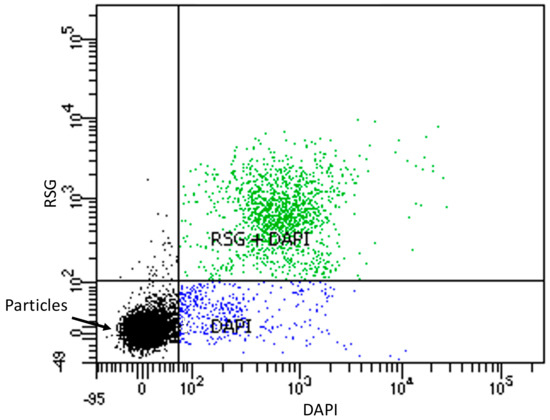
Figure 4.
Scatter plot of RSG and DAPI stained starved OL-KR6/125–130 m fracture fluid. 10,000 events were measured with flow cytometry: black dots are particles, blue dots are cells stained with DAPI and green dots are cells stained with both DAPI and RSG.
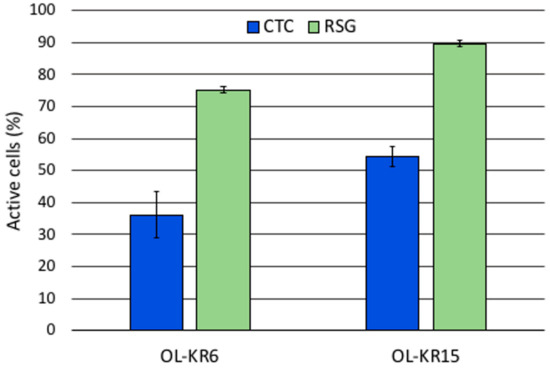
Figure 5.
Share of fluorescent cells in the six-weeks incubated groundwaters of OL-KR6/125–130 m and OL-KR15/446–460 m with CTC (5-Cyano-2,3-ditolyl tetrazolium chloride) and RSG (RedoxSensor Green) redox sensing fluorescent dyes from the DAPI (4′,6 diamidino-2-phenylindole) stained cells.
The microbial community of the OL-KR6 fracture fluid was activated with all tested electron donor and acceptor additions (Figure 6). Acetate and nitrate had the highest activation effect on the microbial cells. It is likely that acetate notably activated sulphate reducers (SRBs), that probably suffered from electron donor shortage due to the preceding six-week incubation. SRBs were abundantly detected from the original groundwater in the qPCR assay (Figure 1). Nitrate seemed to be quite effectively used as an electron acceptor, and nitrate reductase (narG) genes were also detected in the original groundwater with qPCR. Methane was an average activator, and possibly activated methanotrophs. Nitrate addition, together with methane, increased the amounts of activated cells, although methane addition may have inhibited the activation level of microbes that might have been activated by nitrate addition alone. However, the difference in the activation capacity between nitrate and nitrate together with methane remained within the standard deviations. Sulphide and hydrogen were the least effective activators. OL-KR6 fracture water contained a lot of putative sulphur oxidisers, as an abundant part of the bacterial sequences belonged to Epsilonproteobacteria, and more specifically, to Sulfurimonas spp. (33%). However, sulphide activated only a minor part of the community, and a shortage of suitable electron acceptors was possibly a limiting factor. For the low activation effect by hydrogen, the reason may be that there was no carbon dioxide available, and thus, hydrogenotrophic methanogens and autotrophic SRB could not be activated. It is also likely that hydrogen was not the first choice as an electron donor for sulphate reductions by the SRB, and at least some detected groups are known to utilise acetate, especially in the Desulfobacteraceae [34] and Desulfarculaceae [35] families. Overall, it should also be noted that one third of the community was already active before the addition of substrates, and these electron donor and acceptor additions only targeted cells that were not already actively taking up the CTC stain.
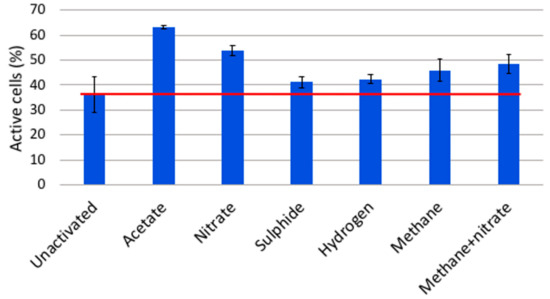
Figure 6.
Comparison of the relative amounts of active cells in the microbial community detected with the fluorescent redox dye CTC in OL-KR6/124–130 m groundwater. The baseline sample did not receive any electron donor or acceptor additions, and indicates the baseline level of active microbial cells. The added compounds are shown on the x-axis, and the columns represent the mean per cent of active microbial cells after the addition and 3 h incubation of different electron donors and acceptors.
The microbial community in OL-KR15 fracture fluid behaved very differently from that of the OL-KR6 fracture fluid. The active share of the community in the baseline water was over 50% (Figure 7), which made it more difficult to study the effect and activating ability of the added electron donors and acceptors. Only acetate and acetate with sulphate activated the OL-KR15 microbial community, with an approximately similar effect in either case. As with OL-KR6, it is likely that acetate was a limiting electron donor for SRB and aceticlastic methanogens, even though the SRB abundance was low based on the sequencing of the bacterial communities of the original fracture fluid.
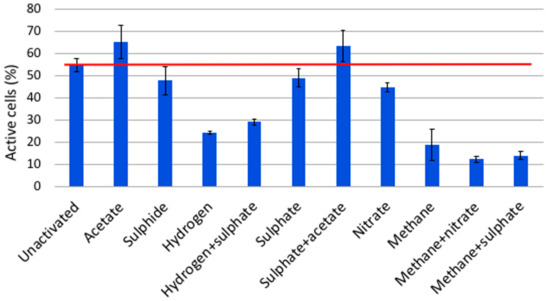
Figure 7.
Comparison of the relative amounts of active cells in the microbial community detected with fluorescent redox dye CTC in OL-KR15/446–460 m fracture fluid. The unactivated sample did not receive any electron donor or acceptor additions, indicating the baseline level of active microbial cells. The added compounds are shown on the x-axis, and the columns represent the mean per cent of active microbial cells after substrate addition and 3 h incubation.
The qPCR results showed overall higher copy numbers for bacterial and archaeal 16S rRNA, dsrB and narG genes in OL-KR15, compared to those in the OL-KR6 fracture fluid. Only the copy numbers of mcrA genes were lower and below detection in the OL-KR15 sample (Figure 1). Sulphate and sulphide additions had little effect on the community activity, which may indicate that they were not limiting factors for sulphate reduction or sulphide oxidation. Nitrate addition had, surprisingly, a slightly inhibiting effect on the community activity when compared to the narG gene qPCR of the original sample, as there was a relatively high number of narG gene copies detected in the water, but no activation occurred. Hydrogen addition with or without sulphate also had an inhibitory effect on community activity. Again, hydrogen with carbon dioxide might have activated methanogens in the archaeal community, and the SRBs were probably not activated with hydrogen. The major part (60–66%) of the archaeal community was found to consist of anaerobic methanotrophs (ANME), more specifically of group ANME-2d archaea (Figure 3) in OL-KR15 fracture fluid, based on sequence results. However, no mcrA genes were detected with qPCR, as the used primers may not have detected mcrA genes of ANME archaea. In addition, all methane additions with or without sulphate or nitrate clearly inhibited the community activity. This indicates that the ANME-2d archaea were not oxidising methane coupled to nitrate or sulphate reduction. It is also possible that they were dormant or inactivated during the preceding incubation period, or possible suffered from the CTC stain toxicity.
4. Discussions
The microbial communities inhabiting OL-KR6 and OL-KR15 fracture fluids behaved differently in activation tests with several electron donors and acceptors, as expected. The main reason for this was in the chemical composition of the fracture fluids, as well as the original bedrock environment, which both affected the fracture fluid microbiomes. Overall, the OL-KR6 fracture fluid, retrieved from 94 m below sea level (m b.s.l.), contained abundant amounts of sulphate and inorganic carbon, and had a more diverse microbial community, compared to that of the OL-KR15 sample. However, this community was less active in terms of measured ATP amounts per volume, and respiratory activity measured with redox dyes compared to the OL-KR15 fracture fluid community. The OL-KR15 fracture fluid originated from 434 mbsl, and had higher salinity compared to the OL-KR6 fracture fluid. Nevertheless, the OL-KR15 fracture fluid had higher organic carbon content than that of OL-KR6, which could have warranted for a higher level of microbial activity, in comparison to that detected in OL-KR6. The six-week incubation period before the activations was needed to decrease the share of active cells in the sample fluids. However, this relatively long incubation probably favoured heterotrophic microorganisms over autotrophs, especially in the OL-KR15 fracture fluid, where the HCO3 amount was only one tenth of that found in the OL-KR6 fracture fluid. The community composition in both fracture fluids probably changed due to the incubation period before the activations were started.
The OL-KR6 fracture fluid community was activated, especially with acetate and nitrate (Figure 6). In addition to SRB being detected with qPCR and sequencing, other bacterial groups able to utilise acetate, such as Sulfurimonas spp. (33%) [36] from Epsilonproteobacteria and Dehalococcoidia (4%) [37], were detected with sequencing. Based on the archaeal sequencing, few sequences of known aceticlastic methanogens, such as Methanosaeta spp. and Methanosarcina spp., were detected. However, Hadesarchaea and Bathyarchaeota, that were found in the archaeal sequences (2–3% each), have been found to contain genes encoding for acetyl-CoA synthetase for conversion of acetate into acetyl-CoA [38,39], and may have been activated by acetate addition. The concentration of acetate in the Olkiluoto fracture fluids is often below the detection limit [40], as it was also in both studied fracture fluids (Table 1). However, it seems that detectable and even relatively high amounts of acetate can be found sporadically in the Olkiluoto fracture water [41] Nevertheless, acetate production is likely in these subsurface fluids, as both autotrophic and heterotrophic acetogens have been cultivated from many Olkiluoto fracture fluids [42], and an active pathway for acetogenesis has been detected in the studied OL-KR6/125–130 m fracture fluid by metagenomic and metatranscriptomic sequencing [18]. In addition to utilising acetate, some of the SRB also produce acetate as a by-product of sulphate reduction when organic compounds are incompletely degraded to acetate, such as Desulfovibrionaceae and Desulfovibrio species [43].
In the case of the OL-KR15 fracture fluid, acetate was also the most efficient activator, indicating electron donor limitation for sulphate reduction. It also appears that other acetate oxidisers or microorganisms using acetate as a carbon source were activated by acetate. Alphaproteobacterial Brevundimonas spp. sequences were abundantly found (38–42%) with sequencing. This bacterial group has previously been found from the deep subsurface environment, and has been shown to especially benefit from the acetate amendment [44]. Brevundimonas spp. bacteria have also been detected growing in activated sludge with sodium acetate as the sole carbon source [45]. It is likely that acetate, when available, is efficiently used by the fracture fluid communities in Olkiluoto by several different metabolic pathways and microbial taxa.
As the aim of this experiment was primarily to study sulphur reactions, sulphate and sulphate, together with electron donors, were tested in OL-KR15 fracture fluid, as the sulphate concentration was initially low, and after the six-week incubation, was shown to be even lower. However, sulphate had only a minor effect on the activation of the microbial community; it slightly increased the activating effects of hydrogen and methane, but had no effect together with acetate. This indicates that sulphate was not yet totally depleted in the fracture fluid, or its role as an electron acceptor in this fracture fluid was only minor.
Methane has been detected in minor amounts (1.81 ppm) in OL-KR6 fracture fluid [18], but in OL-KR15 fracture fluid that originates from the depth of 434 m b.s.l., dissolved methane is abundant [6]. However, it is likely that during sampling and fluid decompression, as well as during experiment initiation in the anaerobic chamber, the methane and other gases initially present in the fracture fluids mostly escaped. The addition of methane probably activated some methanotrophs in OL-KR6 fracture fluid. The amount of putative methanotrophs was relatively low based on archaeal sequence data. However, Bathyarchaeota have been found to have genes for methane metabolism [39], and they may also have been activated by methane addition in OL-KR6.
Unexpectedly, in OL-KR15 fracture fluid, methane alone or together with nitrate or sulphate had a strong inhibitory effect on the activity. This is very contradictory, as the archaea in the OL-KR15 fracture fluid were mostly (60–66%) identified to belong to the ANME-2d group, which is close to the genus Candidatus Methanoperedens that is described to perform nitrate [14] or sulphate [22,46] dependent AOM. However, AOM coupled with soluble ferric complexes [47] has also been observed, and AOM with solid iron or manganese oxide reduction has been suggested [23,48,49] for ANME archaea. In this study, it remains unclear if the ANME-2d archaea detected were unable to perform AOM coupled to nitrate or sulphate reduction, or if they preferred iron or manganese reduction coupled to AOM. However, it is more likely that these ANME-2d archaea were already active (over 50% of the community), or they were dormant, inactivated during the six-week incubation, or were sensitive to the toxicity of the CTC stain. CTC has some drawbacks, like toxicity to some species, causing inhibition of their metabolism and respiration, and some strains poorly incorporate it into the cell [50]. CTC toxicity could explain the detected inhibition effect, since higher cell activity could lead to higher amount of toxic CTC formazan in the cells, and cell death and lysis. In fact, in all activation experiments, but especially in methane, hydrogen, and sulphide activations of the OL-KR15 fracture fluid, there were particles that were stained only with CTC, and not with DAPI, detected by the flow cytometry. This may indicate cell lysis due to the CTC formazan effect. For this reason, all tests showing inhibition may be affected by these properties of the CTC stain. The other tested activity stain RSG could be more suitable in terms of lower toxicity, but it demands groundwaters with less active microbial communities, or a longer initial incubation period for depleting the electron acceptor and donor sources, which may change the community composition and present a higher risk of oxygen contamination and water chemistry change. Earlier experiments with deep subsurface fracture fluids from the Fennoscandian bedrock have shown a wide range of baseline activity for microbial communities from less than one percent to almost 90% with the CTC stain after a four week depletion incubation [51,52].
The efficiency and attractiveness of nitrate as an activator and electron acceptor is not surprising, as it offers higher oxidising potential than sulphate. In the OL-KR6 fracture fluid community, nitrate was quite effectively used as an electron acceptor, and the microbes had the potential for its utilisation, as nitrate reductase (narG) genes were also detected in the original fracture fluid with qPCR. Furthermore, an abundant part of the bacterial sequences belonged to Epsilonproteobacteria, mainly to Sulfurimonas spp., which are known to oxidise reduced sulphur compounds with oxygen, nitrate, or nitrite [53]. In addition, gammaproteobacterial Pseudomonas spp. that were detected in the bacterial sequences (6%) are known for nitrate reduction [54,55]. The concentration of nitrate is normally below the limit of detection in the deep fracture fluids in Olkiluoto [6]. However, it is possible that nitrate is used immediately after it is formed in the deep subsurface, and thus, evades measurement. Some nitrate may form e.g., by radiolysis of NH3 in anaerobic water [56]. In a groundwater community from 95 m b.s.l. depth (VLJ-KR9) in Olkiluoto, the amount of ammonia oxidising and nitrate reducing bacteria was shown to increase when ammonia and nitrate were available [57].
Interestingly, sulphide and hydrogen were not effective activators, even though the OL-KR6 fracture water contained a lot of known sulphur oxidising taxa (Sulfurimonas spp.) and methanogens and sulphate reducers that should be able to utilise hydrogen. The six-week incubation period may have depleted the needed electron acceptors for sulphide oxidation and methanogenesis to occur. More activation with hydrogen could probably have been detected if carbon dioxide had also been added in order to target methanogens in both fracture fluids. Experiments with hydrogen with carbon dioxide, and sulphide with nitrate, would shed light on the communities functioning in future experiments. In addition, activation on a bigger scale would offer the possibility of studying activated transcriptomics. Especially for samples with low cell counts and/or small available sample sizes, the fluorescent redox dye method for finding the most efficiently-activating substrates would help in choosing the most interesting activations for larger scale experiments.
A large portion of the archaeal (60%) and bacterial (34%) communities in OL-KR6 groundwater consisted of microorganisms with potential parasitic or ectosymbiotic lifestyles. Woesearchaeota have small genome sizes and are predicted to live in symbiotic relationships with other microorganisms and/or by fermentation [58]. Patescibacteria also have small genomes [59,60], and Parcubacteria, a patescibacterial class, are found to lack biosynthetic capabilities, but to have potential attachment and adhesion proteins, which suggests ectosymbiotic or parasitic behaviour [61]. Recently it was suggested that a member of Omnitrophica has two physiological stages, i.e., free-living cells with low ribosome content, and cells attaching either to bacteria or archaea and having higher metabolic activity, which indicates the use of their surface polysaccharides and parasitic life style [62]. These small-sized microorganisms were detected regardless of the filter pore-size (0.2 µm) used for the collection of microorganisms. This further indicates their attachment to their symbionts or host cells as they were collected. The relevance of this high proportion of probable parasitic/symbiotic microorganisms detected by sequencing in this deep subsurface fracture fluid is not known. However, their activation is not as straight forward if they lack the pathways needed for the activation reactions or for the independent reduction of CTC. In addition, the activation of parasitic or symbiotic microorganisms attached to other active cells would not show in the detection of fluorescent particles in flow cytometry or in microscopy. The high share of parasitic/symbiotic microorganisms may also affect the results of the fluorescence microscopy compared to the 16S gene copy numbers measured by qPCR. The microscopy results indicated populations equivalent in the two waters, whereas qPCR showed one log higher copy numbers for OL-KR15 fracture fluid, where a large share of both the bacterial and the archaeal communities were possibly also symbiotic/parasitic. In this case, the fluorescence microscopy underestimates the size of the microbial community.
The activation of deep subsurface microbial populations detected with fluorescent redox dyes was an efficient screening method to study induced microbial metabolism. However, the used redox dyes (CTC and RSG) stained a large part of the studied fracture fluid communities without substrate additions, and thus, this part was not detectable in the activation experiments. The activation experiments showed that in the OL-KR6 fracture fluid, all tested amendments raised the community activity. Increased amounts of any of these tested compounds could lead to a shift in the community steady-state, as well as to changes of the water chemistry. In OL-KR15 fracture fluid, the activation was not as systematic, which was likely due to drawbacks in the used methodology, and possibly a shortage of the required electron acceptors for the methanogenesis or sulphide oxidation. Despite the relatively large differences in the chemistry and microbial diversity of the studied fracture fluids, both communities were activated, especially with acetate. Activation with acetate potentially leads to sulphate reduction and corrosive sulphide formation, which is a risk for the long-term safety of nuclear waste disposal.
Author Contributions
Conceptualisation, H.M., M.B. and M.V.; Methodology, H.M. and M.B.; Formal Analysis, H.M.; Investigation, H.M.; Writing—Original Draft Preparation, H.M.; Supervision, M.V.; Project Administration, M.V.; Writing—Review & Editing, H.M., M.B. and M.V.
Funding
This research was funded by the Horizon 2020 project MIND through funding from the Euratom research and training programme 2014–2018 under Grant Agreement no. 661880. The research was also funded by VTT.
Acknowledgments
We thank Posiva Ltd. for fracture fluid samples from the repository environment and for the chemical analysis results. Tiina Lamminmäki is acknowledged for the critical review of the manuscript. Mirva Pyrhönen is thanked for excellent assistance in the laboratory and Sirpa Jylhä for performing the FACS analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lovley, D.R.; Chapelle, F.H. Deep subsurface microbial processes. Rev. Geophys. 1995, 33, 365–381. [Google Scholar] [CrossRef]
- Hoehler, T.M.; Jørgensen, B.B. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 2013, 11, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Bomberg, M.; Lamminmäki, T.; Itävaara, M. Microbial communities and their predicted metabolic characteristics in deep fracture groundwaters of the crystalline bedrock at Olkiluoto, Finland. Biogeosciences 2016, 13, 6031–6047. [Google Scholar] [CrossRef]
- Purkamoa, L.; Bomberg, M.; Kietäväinen, R.; Salavirta, H.; Nyyssönen, M.; Nuppunen-Puputti, M.; Ahonen, L.; Kukkonen, I.; Itävaara, M. Microbial co-occurrence patterns in deep Precambrian bedrock fracture fluids. Biogeosciences 2016, 13, 3091–3108. [Google Scholar] [CrossRef]
- Chivian, D.; Brodie, E.L.; Alm, E.J.; Culley, D.E.; Dehal, P.S.; DeSantis, T.Z.; Gihring, T.M.; Lapidus, A.; Lin, L.-H.; Lowry, S.R.; et al. Environmental genomics reveals a single-species ecosystem deep within earth. Science 2008, 322, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Posiva Ltd. Olkiluoto Site Description 2011, 1st ed.; Posiva Ltd.: Eurajoki, Finland, 2012; p. 1028. ISBN 978-951-652-179-7. Available online: www.posiva.fi/tietopankki/olkiluoto_site_description_2011.1793.xhtml#.Wwz2Uqm-l0s (accessed on 30 October 2018).
- Hammes, F.; Berney, M.; Egli, T. Cultivation-independent assessment of bacterial viability. In High Resolution Microbial Single Cell Analytics, 1st ed.; Müller, D., Bley, T., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2011; Volume 124, pp. 123–150. ISBN 978-3-642-16886-4. [Google Scholar]
- Del Giorgio, P.A.; Gasol, J.M. Physiological structure and single-cell activity in marine bacterioplankton. In Microbial Ecology of the Oceans, 2nd ed.; Kirshman, D.L., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 244–298. ISBN 978-0-470-04344-8. [Google Scholar]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Créach, V.; Baudoux, A.-C.; Bertru, G.; Le Rouzic, B. Direct estimate of active bacteria: CTC use and limitations. J. Microbiol. Methods 2003, 52, 19–28. [Google Scholar] [CrossRef]
- Bhupathiraju, V.; Hernandez, M.; Landfear, D.; Alvarez-Cohen, L. Application of a tetrazolium dye as an indicator of viability in anaerobic bacteria. J. Microbiol. Methods 1999, 37, 231–243. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Lindstrom, M.E.; Chistoserdova, L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2008, 2, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Kotelnikova, S.; Pedersen, K. Evidence for methanogenic Archaea and homoacetogenic Bacteria in deep granitic rock aquifers. FEMS Microbiol. Rev. 1997, 20, 339–349. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y.; Imelfort, M.; Keller, J.; Hugenholtz, P.; Yuan, Z.; Tyson, G.W. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 500, 567–570. [Google Scholar] [CrossRef] [PubMed]
- McMahon, P.B.; Chapelle, F.H. Microbial production of organic acids in aquitard sediments and its role in aquifer geochemistry. Nature 1991, 349, 233–235. [Google Scholar] [CrossRef]
- Lin, L.H.; Hall, J.; Lippmann-Pipke, J.; Ward, J.A.; Lollar, B.S.; DeFlaun, M.; Rothmel, R.; Moser, D.; Gihring, T.M.; Mislowack, B.; et al. Radiolytic H2 in continental crust: Nuclear power for deep subsurface microbial communities. Geochem. Geophys. Geosyst. 2005, 6, 1–13. [Google Scholar] [CrossRef]
- Lin, L.H.; Slater, G.F.; Sherwood Lollar, B.; Lacrampe-Couloume, G.; Onstott, T.C. The yield and isotopic composition of radiolytic H2, a potential energy source for the deep subsurface biosphere. Geochim. Cosmochim. Acta 2005, 69, 893–903. [Google Scholar] [CrossRef]
- Blomberg, P.; Itävaara, M.; Marjamaa, K.; Salavirta, H.; Arvas, M.; Miettinen, H.; Vikman, M. Metabolic Pathways of Deep Groundwater Microbiomes and Sulphide Formation at Olkiluoto, 1st ed.; Posiva Ltd.: Eurajoki, Finland, 2017; p. 150. Available online: www.posiva.fi/files/4688/WR_2017-11_web.pdf (accessed on 30 October 2018).
- Miettinen, H.; Bomberg, M.; Nyyssönen, M.; Salavirta, H.; Sohlberg, E.; Vikman, M.; Itävaara, M. The Diversity of Microbial Communities in Olkiluoto Bedrock Groundwaters 2009–2013; Posiva Ltd.: Eurajoki, Finland, 2015; p. 160. Available online: http://www.posiva.fi/files/4125/WR_2015-12.pdf (accessed on 30 October 2018).
- Bomberg, M.; Nyyssönen, M.; Pitkänen, P.; Lehtinen, A.; Itävaara, M. Active microbial communities inhabit sulphate-methane interphase in deep bedrock fracture fluids in Olkiluoto, Finland. BioMed Res. Int. 2015, 2015, 979530. [Google Scholar] [CrossRef] [PubMed]
- Bomberg, M.; Lamminmäki, T.; Itävaara, M. Estimation of microbial metabolism and co-occurrence patterns in fracture groundwaters of deep crystalline bedrock at Olkiluoto, Finland. Biogeosci. Discuss. 2015, 12, 13819–13857. [Google Scholar] [CrossRef]
- Weber, H.S.; Habicht, K.; Thamdrup, B. Anaerobic methanotrophic archaea of the ANME-2d cluster are active in a low-sulfate, iron-rich freshwater sediment. Front. Microbiol. 2017, 8, 619. [Google Scholar] [CrossRef] [PubMed]
- Oni, O.; Miyatake, T.; Kasten, S.; Richter-Heitmann, T.; Fischer, D.; Wagenknecht, L.; Kulkarni, A.; Blumers, M.; Shylin, S.I.; Ksenofontov, V.; et al. Distinct microbial populations are tightly linked to the profile of dissolved iron in the methanic sediments of the Helgoland mud area, North Sea. Front. Microbiol. 2015, 6, 365. [Google Scholar] [CrossRef] [PubMed]
- Purkamo, L.; Bomberg, M.; Nyyssönen, M.; Kukkonen, I.; Ahonen, L.; Kietäväinen, R.; Itävaara, M. Dissecting the deep biosphere: Retrieving authentic microbial communities from packer-isolated deep crystalline bedrock fracture zones. FEMS Microbiol. Ecol. 2013, 85, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Tsitko, I.; Lusa, M.; Lehto, J.; Parviainen, L.; Ikonen, A.T.K.; Lahdenperä, A-M.; Bomberg, M. The variation of microbial communities in a depth profile of an acidic, nutrient-poor boreal bog in Southwestern Finland. Open J. Ecol. 2014, 4, 832–859. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16s rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [PubMed]
- Bano, N.; Ruffin, S.; Ransom, B.; Hollibaugh, J.T. Phylogenetic composition of Arctic ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 2004, 70, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Barns, S.M.; Fundyga, R.E.; Jeffries, M.W.; Pace, N.R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 1994, 91, 1609–1613. [Google Scholar] [CrossRef] [PubMed]
- Rajala, P.; Bomberg, M.; Kietäväinen, R.; Kukkonen, I.; Ahonen, L.; Nyyssö∂nen, M.; Itävaara, M. Rapid reactivation of deep subsurface microbes in the presence of C-1 compounds. Microorganisms 2015, 3, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Kuever, J. The Family Desulfobacteraceae. In The Prokaryotes, 4th ed.; Rodenberg, E., DeLong, E.F., Loy, S., Stackebrndt, E., Thompson, F., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2014; pp. 45–74. ISBN 978-3-642-39044-9. [Google Scholar]
- Sun, H.; Spring, S.; Lapidus, A.; Davenport, K.; Glavina Del Rio, T.; Tice, J.; Nolan, M.; Copeland, A.; Cheng, J-F.; Lucas, S.; et al. Complete genome sequence of Desulfarculus baarsii type strain (2st14T). Stand. Genomic Sci. 2010, 3, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Labrenz, M.; Grote, J.; Mammitzsch, K.; Boschker, H.T.S.; Laue, M.; Jost, G.; Glaubitz, S.; Jörgens, K. Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2013, 63, 4141–4148. [Google Scholar] [CrossRef] [PubMed]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Müller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Saw, J.H.; Lind., A.E.; Lazar, C.S.; Hinrichs, K.-U.; Teske, A.P.; Ettema, T.J.G. Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat. Microbiol. 2016, 1, 16002. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, V.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434. [Google Scholar] [CrossRef] [PubMed]
- Lamminmäki, T.; (Posiva Ltd., Eurajoki, Finland). Personal communication, 2018.
- Bell, E.; Lamminmäki, T.; Alneberg, J.; Andersson, A.F.; Qian, C.; Xiong, W.; Hettich, R.L.; Balmer, L.; Frutschi, M.; Sommer, G.; et al. Biogehochemmical cycling by a low-diversity microbial community in deep groundwater. Front. Microbiol. 2018, 9, 2129. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Arlinger, J.; Eriksson, S.; Hallbeck, A.; Hallbeck, L.; Johansson, J. Numbers, biomass and cultivable diversity of microbial populations relate to depth and borehole-specific conditions in groundwater from depths of 4–450 m in Olkiluoto, Finland. ISME J. 2008, 2, 760–775. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Purkamo, L.; Bomberg, M.; Nyyssönen, M.; Ahonen, L.; Kukkonen, I.; Itävaara, M. Response of deep subsurface microbial community to different carbon sources and electron acceptors during ∼2 months incubation in microcosms. Front. Microbiol. 2017, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.H.; Park, M.; Lee, J.R.; Yun, P.-Y.; Jeon, C.O. Brevundimonas aveniformis sp. nov., a stalked species isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2007, 57, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Ino, K.; Hernsdorf, AW.; Konno, U.; Kouduka, M.; Yanagawa, K.; Kato, S.; Sunamura, M.; Hirota, A.; Togo, Y.; Ito, K.; et al. Ecological and genomic profiling of anaerobic methane-oxidizing archaea in a deep granitic environment. ISME J. 2018, 12, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Scheller, S.; Yu, H.; Chadwick, G.L.; McGlynn, S.E.; Orphan, V.J. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 2016, 351, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Rasigraf, O.; Sapart, C.J.; Jilbert, T.; Jetten, M.S.; Röckmann, T.; van der Veen, C.; Bândă, N.; Kartal, B.; Ettwig, K.F.; et al. Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ. Sci. Technol. 2015, 49, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Beal, E.J.; House, C.H.; Orphan, V.J. Manganese- and iron-dependent marine methane oxidation. Science 2009, 325, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Hatzinger, P.B.; Palmer, P.; Smith, R.L.; Peñrrieta, C.T.; Yoshinari, T. Applicability of tetrazolium salts for the measurement of respiratory activity and viability of groundwater bacteria. J. Microbiol. Methods 2003, 52, 47–58. [Google Scholar] [CrossRef]
- Bomberg, M.; Raulio, M.; Jylhä, S.; Mueller, C.W.; Höschen, C.; Rajala, P.; Purkamo, L.; Kietäväinen, R.; Ahonen, L.; Itävaara, M. CO2 and carbonate as substrate for the activation of the microbial community in 180 m deep bedrock fracture fluid of Outokumpu Deep Drill Hole, Finland. AIMS Microbiol. 2017, 3, 846–871. [Google Scholar] [CrossRef]
- Rajala, P.; Bomberg, M. Reactivation of deep subsurface microbial community in response to methane or methanol amendment. Front. Microbiol. 2017, 8, 431. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Perner, M. The globally widespread genus Sulfurimonas: Versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 2015, 6, 989. [Google Scholar] [CrossRef] [PubMed]
- Sias, S.R.; Ingraham, J.L. Isolation and analysis of mutants of Pseudomonas aeruginosa unable to assimilate nitrate. Arch. Microbiol. 1979, 122, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Bedzyk, L.; Wang, T.; Ye, R.W. The periplasmic nitrate reductase in Pseudomonas sp. G-179 catalyzes the first step of denitrification. J. Bacteriol. 1999, 181, 2801–2806. [Google Scholar]
- Silver, B.J.; Raymond, R.; Sigman, D.M.; Prokopeko, M.; Sherwood Lollar, B.; Lacrampe-Couloume, G.; Fogel, M.L.; Pratt, L.M.; Lefticariu, L.; Onstott, T.C. The origin of NO3− and N2 in deep subsurface fracture water of South Africa. Chem. Geol. 2012, 294, 51–62. [Google Scholar] [CrossRef]
- Kutvonen, H.; Rajala, P.; Carpén, L.; Bomberg, M. Nitrate and ammonia as nitrogen sources for deep subsurface microorganisms. Front. Microbiol. 2015, 6, 1079. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Hug, L.A.; Brown, C.T.; Wilkins, M.J. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 2015, 25, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Miller, C.S.; Castelle, C.J.; VerBerkmoes, N.C.; Wilkins, M.J.; Hettich, R.L.; Lipton, M.S.; Williams, K.H.; et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 2012, 337, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, T.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.C.; Stegen, J.C. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 2015, 6, 713. [Google Scholar] [CrossRef] [PubMed]
- Kizina, J. Insights into the Biology of Candidate Division OP3 LiM Populations. Ph.D. Thesis, University of Bremen, Bremen, Germany, 18 September 2017. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).