1. Introduction
It is well known that nutrition management strategies are closely related to growth performance and health, and the feeding frequency is one of the key factors affecting the growth of the body and feed efficiency [
1,
2]. Previous studies have reported that different feeding frequencies affect the feed intake, feed efficiency, body composition, and growth performance of pigs [
2,
3,
4,
5,
6]; however, due to the different trial conditions, conflicting results associated with changes in feed efficiency and body composition have been reported in previous studies [
2,
4]. Feeding once or twice per day are common feeding patterns used in the swine production of China, therefore, it is necessary to investigate and compare the responses on growth performance and the body metabolism of pigs under these two feeding patterns.
So far, the regulation mechanism of feeding frequency on the growth and body metabolism of pigs is not fully clear. Higher feeding frequency could improve the glucose clearance rate and prevent ruminants from accumulating fat in visceral adipose depots due to higher insulin sensitivity [
7]. Meal frequency could change the time-course profiles of plasma concentrations of glucose, insulin, and lactate in pig [
2]. A twelve meals regimen increased the liver concentration of total lipid, glycogen, and triglyceride than that in the two meals daily pigs [
8]. Compared with pigs fed ad libitum, the activities of citrate synthase, β-hydroxylacyl-CoA dehydrogenase, and lactate dehydrogenase were greater in the longissimus muscle of the two meals daily regimen [
9]. The intestine and liver are the major digestive, absorption, and metabolism organs of the body and play a key role in the metabolism of nutrients [
10,
11]. To date, information on the transcriptomic responses in the liver and jejunal mucosa of pigs under different feeding frequencies is not available.
The present study hypothesized that compared with feeding once daily, pigs feeding twice daily could regulate the gene expression of the intestine and liver, eventually affecting the body metabolism. Therefore, the objective of this study was to compare the growth performance, body metabolism, and transcriptional profiles in the livers and jejunal mucosa of pigs under different feeding frequencies.
2. Materials and Methods
2.1. Experimental Animals, Design, and Diet
This experiment was approved and conducted under the supervision of the Animal Care and Use Committee of Nanjing Agricultural University (Nanjing, Jiangsu Province, China). The ethic code is SYXK (SU) 2017–0007. All pigs were raised and maintained on a local commercial farm under the care of the Animal Care and Use Guidelines of Nanjing Agricultural University. In our study, all animals were individually housed in metal-floor cages (height, 0.85 m; length, 1.2 m; width, 0.70 m) with a smooth walled pan, a nipple drinker, and a feeder. The room temperature was maintained at 25 ± 2 °C during the experimental period. All pigs were kept at a 24 h light–dark cycle, with lights being turned on from 8:00 am to 20:00 pm. Twelve 42-d Duroc × Landrance × Yorkshire growing barrows (BW = 14.86 ± 0.20 kg) were randomly allocated to the one time feeding daily (M1) group and two times feeding daily (M2) group, and each group consisted of six replicates (pens) with one pig per pen. Pigs in the M1 group were fed the adequate diet on an ad libitum basis at 8:00 am on each experimental day. Pigs in the M2 group were fed half of the standard feeding requirement according to the National Research Council (NRC, 2012) at 8:00 am and adequate feed at 16:00 pm [
12], and the refusals were removed from the feeder and weighed at 20:00 pm. The composition and nutrient level of the diet in our study is shown in
Table 1, and all pigs had free access to water during the 30-day trial period. Individual initial and final body weight as well as feed intake were registered during the experiment to calculate the average daily gain (ADG) and average daily feed intake (ADFI). Feed efficiency (feed:gain) was expressed as F:G.
2.2. Sampling
After 12 h fasting, all pigs were euthanized at 8:00 am with a jugular vein injection of 4% sodium pentobarbital solution (40 mg/kg BW). Blood samples were collected and centrifuged at 2000× g for 10 min at 4 °C, and the supernatant stored at −70 °C until subsequent biochemical analysis. The animals were bled and opened immediately, and the liver, kidney, spleen, heart, and the entire gastrointestinal tract were removed and weighed. Tissues from the liver and jejunal mucosa were collected and kept in liquid nitrogen, then stored at −80 °C for further transcriptome analysis.
2.3. Biochemical Parameters of Serum and Liver
Glucose, total bile acid (TBA), triglyceride (TG), total cholesterol (T-CHO), high-density lipoprotein-cholesterol (HDL-C), and low-density lipoprotein-cholesterol (LDL-C) in the serum of pigs were measured with an Olympus AU400 Automatic Biochemical Analyzer (Olympus Optical Co., ltd., Tokyo, Japan). The concentrations of liver T-CHO, TG, HDL-C, LDL-C, and TBA were determined by using commercial biochemical assay kits (Nanjing Jiancheng Bioengineering Institution, Nanjing, China).
2.4. Library Construction for RNA Sequencing and Sequencing Procedures
Total RNA of the liver tissues and jejunal mucosa were isolated using an RNeasy mini kit (Qiagen, Hilden, North Rhine-Westphalia, Germany). As there were six replicates in each group, three biological replicates were randomly selected for the RNA-Seq to reduce the costs of the experiment. A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. In order to select cDNA fragments of preferentially 240 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, MA, USA). Then, 3 μL USER Enzyme (New England BioLabs, Ipswich, MA, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA, USA), universal PCR primers, and index (X) Primer (New England BioLabs, Ipswich, MA, USA). Finally, the PCR products were purified (AMPure XP system) and the library quality was assessed on an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v4-cBot-HS (Illumia (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated.
2.5. Quantitative Real-Time PCR
The first-strand cDNA synthesis was performed using 1 μg of total RNA with a reverse transcription kit (Takara Bio, Shiga, Japan). The expression of genes
Apolipoprotein A1 (APOA1), Apolipoprotein A4 (APOA4), Apolipoprotein C3 (APOC3), Niemann-Pick C1-Like 1 (NPC1L1), Acetyl-CoA acyltransferase 1 (ACAA1), Fatty acid binding protein 1 (FABP1), and
Phosphoenolpyruvate carboxykinase 1 (PCK1) were measured by quantitative real-time PCR using an ABI 7300 sequence detector (SDS, Foster City, CA, US). The PCR reactions were performed in a final volume of 20 μL with the Roche SYBR Green PCR Kit (Roche, Hercules, CA, USA), according to the manufacturer’s instructions. The sequences of the primers are listed in
Appendix A Table A1. The expression of the genes was calculated relative to the expression of
β-actin with the formula 2
−ΔΔCt [
13].
2.6. Statistics
Growth performance, organ weight, serum metabolites, and hepatic metabolite data were analyzed by SPSS version 22.0 software (SPSS Inc., Chicago, IL, USA) as a randomized complete block design. The pen was used as the experimental unit (n = 6), and the significance of differences between the M1 and M2 groups was evaluated by a Student’s t test. Significant differences were declared when p < 0.05.
Genes with altered expression (FC > 1.5 or < 0.67;
p < 0.05) were selected for further study. Additionally, the genes identified between the two groups were mapped to the Gene Ontology (GO,
http://www.geneontology.org/, accessed 16 July 2018) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG,
http://www.genome.jp/kegg/, accessed on 16 July 2018) pathways to identify potential pathways associated with dietary treatment. GO enrichment analysis of the differentially expressed genes (DEGs) was implemented based Wallenius non-central hyper-geometric distribution [
14]. KOBAS [
15] software (
http://kobas.cbi.pku.edu.cn/) was used to test the statistical enrichment of the DEGs in the KEGG pathways. ClueGO (version 2.5.4) and CluePedia (version 1.5.4), a plugin for Cytoscape version 3.7.1 (NIGMS, Bethesda, MD, USA), were used for functionally grouped network analysis [
16,
17].
4. Discussion
Restricted meal frequency is considered a potentially effective treatment for metabolic disease besides limited caloric intake [
18,
19,
20]. A previous study showed that the liver fat content of pigs fed two meals daily was lower than that in pigs with twelve meals [
8]. In the present study, compared with the M1 group, the M2 regimen significantly decreased the concentrations of serum T-CHO, TG, HDL-C, LDL-C, and liver TG, although the growth performance of pigs was not affected. In order to explore the underlying mechanisms, transcriptomic responses in the livers and jejunal mucosa of growing pigs were investigated following two different feeding frequency regimens.
By recording the feeding intake at different time points in one day, we found that pigs in the M1 group consumed 75–85% of the total diet before 16:00 pm every day, while the M2 group only consumed half of the total diet at 16:00 pm every day, which indicates that the feeding patterns impacted the meal regimen. However, across the one-month trial, the two different feeding regimens had no significant effect on the growth performance of the growing pig. The average daily feed intake (ADFI) was correlated with the time of eating, therefore, no difference in ADFI in our study was likely due to the same duration of eating time in the M2 group when compared to free access. Colpoys et al. observed a decrease in ADFI in gilts fed two meals daily when compared to free access [
5]. Newman et al. observed a tendency for boars fed two 1-h meals to eat less than ad libitum [
4]; however, the ADFI was not significantly changed when boars were fed two 90-min meals. The inconsistent results may partly be due to the differences in the sex of the pigs used in different studies. In addition, the different growth period of pigs used in this study may partly explain the difference between earlier studies [
2,
3,
4]. Schneider et al. pointed out that feeding two or six meals daily had the same effect on the growth performance of gestating gilts [
21], but the average daily gain (ADG) of gilts was increased from day 0 to 42. In pigs maintained on a high fat dietary regimen, two meals per day decreased the fat deposition content when compared to twelve meals per day [
22]. However, Hatori et al. showed that an increase in the feeding frequency significantly increased the ADG of rats fed a normal diet or high-fat diet [
18]. This suggests that different feeding regimens may have different effects on the growth performance of animals from different species, different genders, and different physiological stages. Both the diet composition and frequency of daily feeding could affect growth performance.
The accumulation of triglyceride-rich lipoproteins (TRLs) in blood is believed to be related to the occurrence of atherosclerotic dyslipidemia. Blood TG level and/or the remnant-cholesterol (remnant-cholesterol = T-CHO – LDL-C – HDL-C) reflect the level of TRL [
23]. In our study, the two times feeding regimen significantly decreased the concentrations of serum T-CHO, HDL-C, and LDL-C. In addition, pigs on the M2 regimen showed a decreased TG concentration in the serum and liver, which is consistent with the results of previous studies on pigs or rodents [
2,
18,
19]. Our study supports the idea that different feeding regimens can affect lipid metabolism.
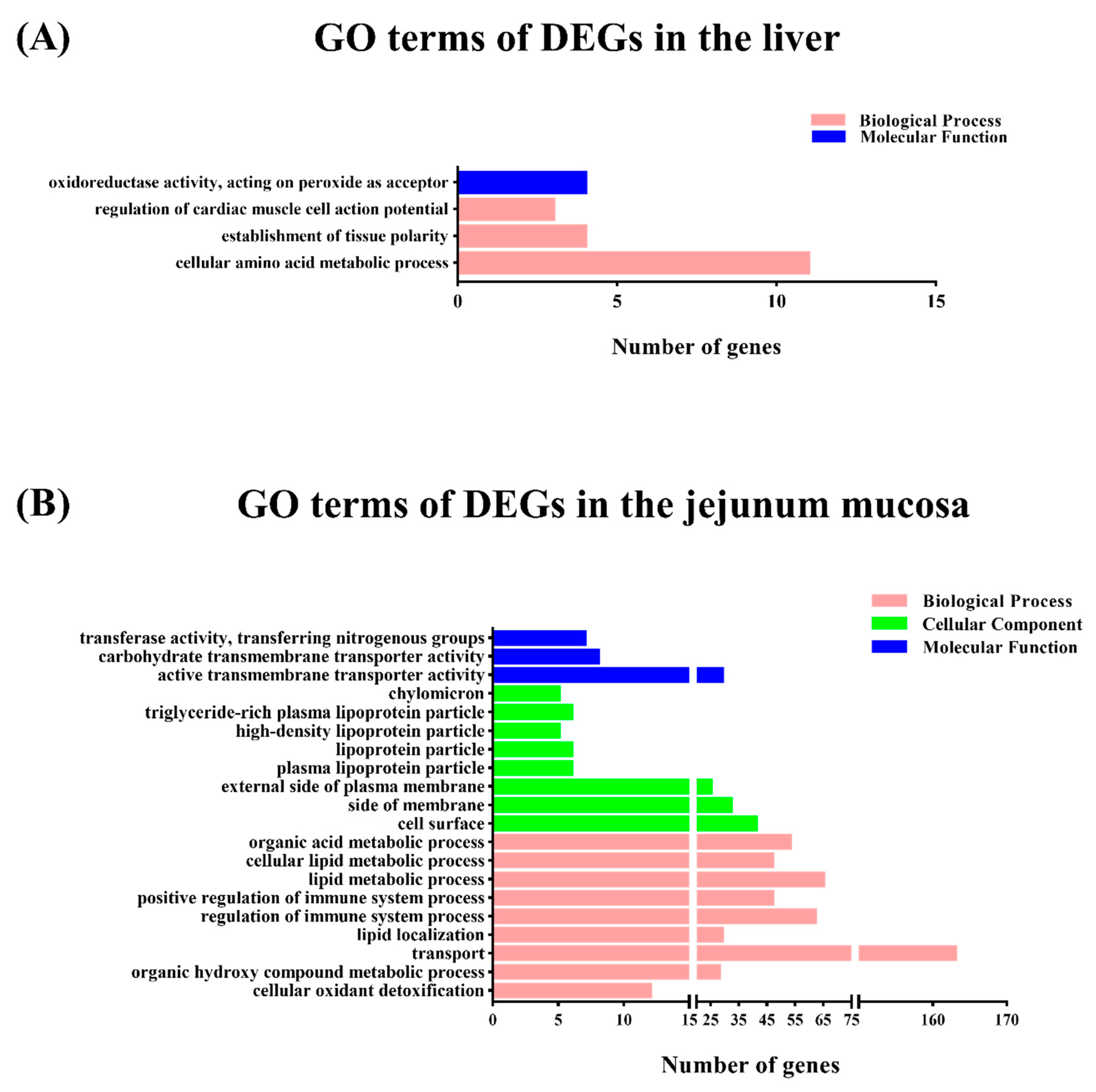
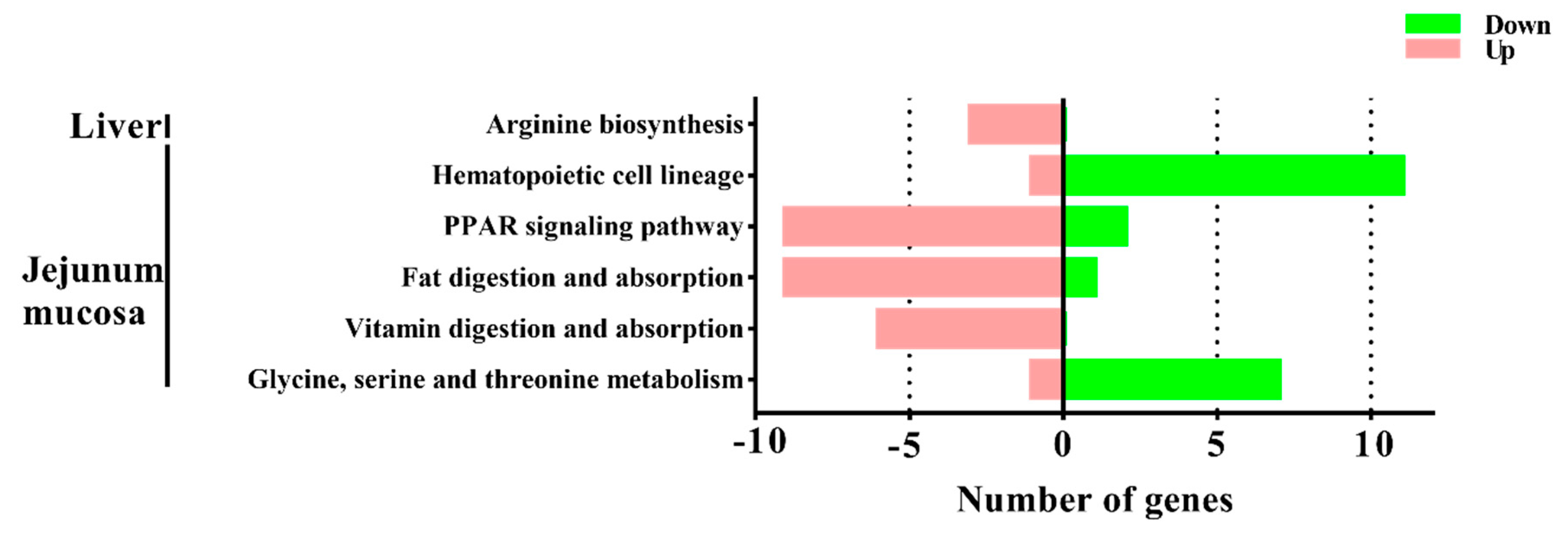
To gain insights into the mechanisms underlying this process, transcriptomic analysis was conducted to identify the DEGs between the M1 and M2 regimens. In the liver, only the arginine biosynthesis pathway was enriched by the M2 regimen, however, in the jejunal mucosa, the significant enriched pathways included glycine, serine, and threonine metabolism, the PPAR signaling pathway, fat digestion and absorption, vitamin digestion and absorption, and hematopoietic cell lineage, which suggests that the different feeding frequencies in our experiment appeared to have a more significant effect on bay metabolism at the intestinal level.
The NPC1L1 transmembrane protein in the intestinal villi is responsible for the efficient and specific transport of cholesterol into the absorbing cells. Previous studies showed that
NPC1L1 gene ablation reduced the absorption of cholesterol by the small intestine, and increased cholesterol synthesis in the liver [
24,
25,
26]. A previous study found that the concentration of cholesterol was significantly increased in transgenic mice for a human
NPC1L1 gene [
27], while
NPC1L1-deficient mice exhibited a drastic reduction of dietary cholesterol absorption [
28,
29]. The
GOT2 gene, known as plasma membrane-associated fatty acid-binding protein (FABPpm), can promote the transport and metabolism of fatty acids. It was reported that FABPpm participated in fatty acid metabolism by transporting long-chain fatty acids into the cell [
30]. Acetyl-CoA acetyltransferase 2 (ACAT2) can catalyze the absorption of cholesterol absorbed in cells to form cholesteryl esters [
31]. ACAT2-deficient in the intestine reduced the efficiency of the absorption and transportation of cholesterol by chyle particles and the absorption of cholesterol in the diet [
32]. Under the action of apolipoprotein APOB48 and MTTP, cholesteryl esters were assembled together with triglycerides, phospholipids, and a small portion of free cholesterol to form chylomicrons, which could be secreted into lymphatic circulation through basement membrane [
33,
34,
35]. In the present study, genes
NPC1L1,
FABP1,
MOGAT2, and apolipoprotein (
APOA1, APOA4, and
APOB) were upregulated by the M2 regimen in fat digestion and absorption pathway, combined with the upregulation of genes
PLB1,
RBP2, and apolipoprotein (
APOA1, APOA4, and
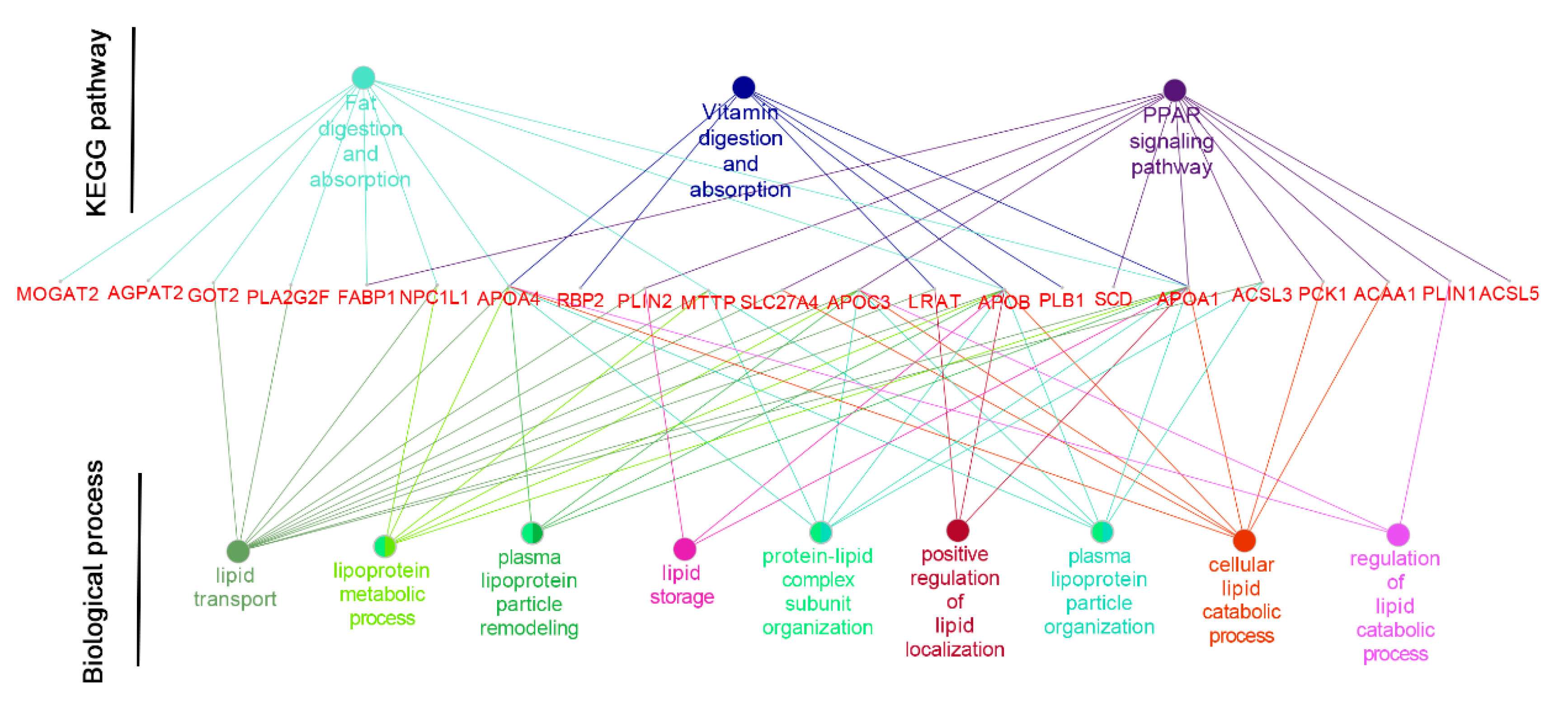
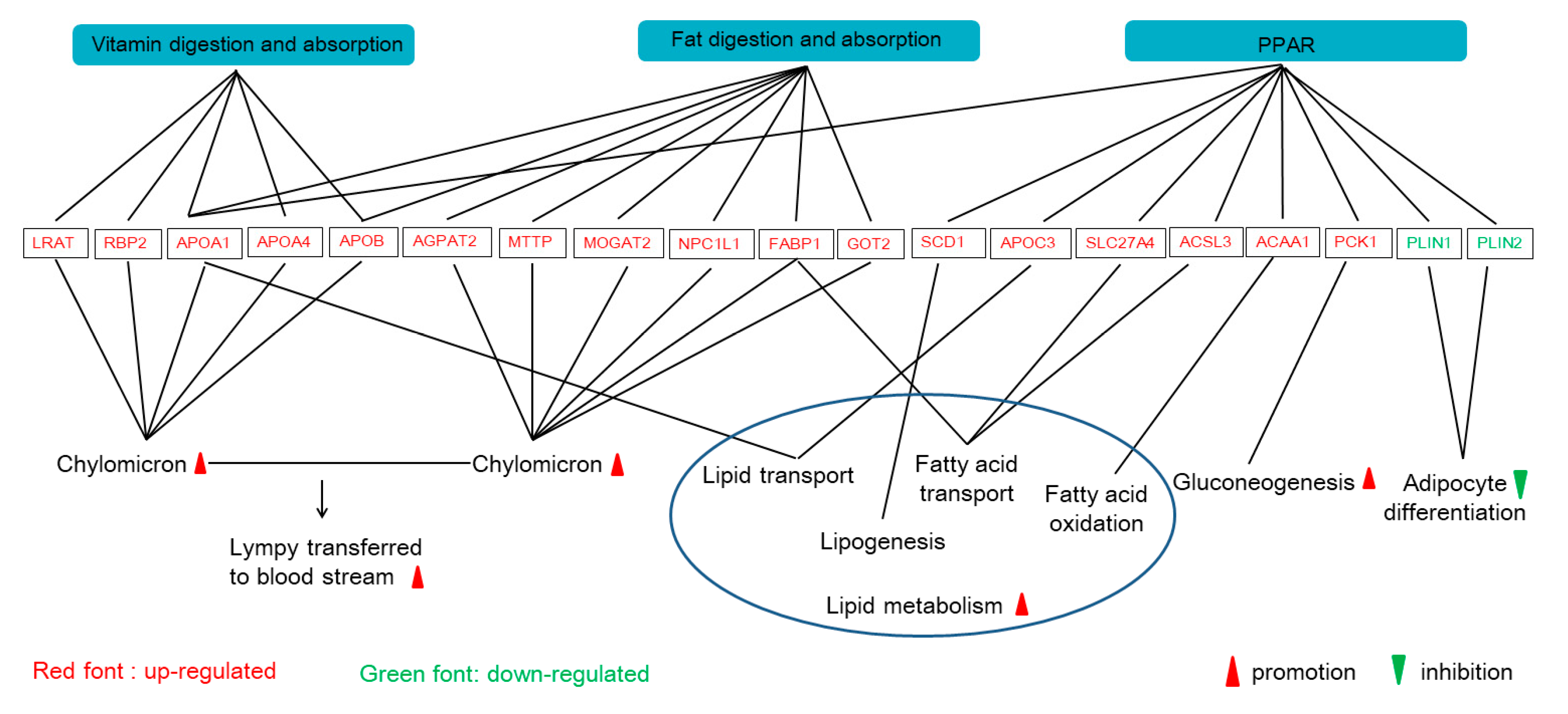
APOB) in the vitamin digestion and absorption pathway, which indicates that the M2 regimen promoted the formation and transport of chylomicrons into the bloodstream, and the digestion of fat in the small intestine (
Figure 4).
Long-chain acyl-coenzyme a synthase (ACSL) catalyzes the first step in the activation of intracellular fatty acid metabolism by converting long-chain fatty acids into long-chain acyl-CoAs [
36]. The
ACSL3 gene can promote the synthesis of lecithin and the formation of intracellular lipid droplets, participate in fatty acid oxidation, and maintain lipid droplet formation in two metabolic pathways [
37]. ACSL5 is a key enzyme in the process of fatty acid beta oxidation and triglyceride synthesis and metabolism in the animal body. Cao et al. upregulated the transcription of
ACSL3 gene by using cytokine tumor suppressor in HepG2 cells, enhanced β-oxidation, and reduced the TG content of cells [
38]. Acetyl-CoA acyltransferase (ACAA), also known as 3-Ketoacyl-CoA thiolase, includes two subtypes, ACAA1 and ACAA2, which catalyze the last step of fatty acid beta oxidation. Previous studies have shown that the increase of ACAA activity promotes the oxidation of fatty acids [
39,
40]. The
ACAA1 gene is mainly involved in physiological and biochemical processes such as the oxidation of very long-chain fatty acids, bile acid metabolism, and regulation of peroxisome proliferation [
41]. In the present study, we found that the M2 regimen significantly upregulated the expression of genes
ACSL3 and
ACSL5 in pig jejunum, which suggests that two times feeding daily could increase fatty acid β-oxidation (
Figure 4).
The
FABP1 (L-FABP) could be affected by PPAR and in turn affect PPARα [
42].
FABP can assist in the transport of fatty acids to the mitochondria into the β-oxidation pathway and transport to the endoplasmic reticulum to enter the triglyceride esterification pathway. A previous study on mice fed a high-fat diet indicated that the
L-FABP gene promoted long-chain fatty acid oxidation and inhibited weight gain and obesity [
43]. The over-expression of
L-FABP could significantly increase the intake of long-chain fatty acid oxygen and medium-chain fatty acid in the nucleus [
44]. When L-FABP binds to PPARα, it can enhance the transcriptional activity of PPAR to long-chain fatty acid oxidase, regulate the utilization and metabolism of intracellular fatty acid, and maintain the relative balance of fatty acid metabolism in vivo [
44]. In adipocytes, Perilipin (PLIN) anchors on the surface of the lipid droplets to produce a barrier effect, which blocks TG from lipohydrolase [
45,
46]. In the present study, the
PLIN gene was downregulated, and its barrier effect was weakened with the increase of lipohydrolase activity, then lipid droplets became significantly smaller, which accelerated fat decomposition and reduced paper deposition in cells. In addition, phosphoenolpyruvate carboxykinase (PCK), a rate-limiting enzyme of hepatic glycogenesis, was also upregulated in the M2 group. Therefore, the results suggest that the two times feeding regimen can increase fatty acid β-oxidation and reduce fatty acid accumulation (
Figure 4). However, Liu et al. found that the protein abundance of PCK2 in glucose and energy metabolism, the protein abundance of APOA1, APOB, FABP1, MTTP, and GOT2 in the lipid metabolism of pigs with two meals a day were significantly higher than that of pigs with twelve meals a day [
8]. The inconsistent results between the two studies may be partly due to the different feeding regimens and physiological stages of the pigs.
Interestingly, the transcriptome date of this study revealed significant changes in the arginine biosynthesis pathway in the livers of pigs. In the enriched arginine biosynthesis pathway,
ARG2 and
NAGS genes in the M2 group were upregulated when compared with the M1 group. These genes contribute to the synthesis of glutamic acid, glutamine (glutamate), and proline in the small intestine of pigs, which are precursors for the synthesis of citrulline and arginine [
47]. Arginine can detoxify through the urea cycle, promote insulin and growth hormone secretion, and regulate nutrient metabolism, and has been widely used as an immune nutrient in clinical practice [
48]. However, there was no difference in the growth performance between the two groups, and the influence of feeding frequency on the immune function needs further study. A study of proteomics in the liver revealed 35 differentially expressed proteins in the liver between pigs fed two and twelve meals per day, which were involved in the regulation of glucose and energy metabolism, lipid metabolism, protein and amino acid metabolism, and other general biological process [
8]. The inconsistent results between the two studies may be partly due to the different feeding frequency and diet composition used in the pig trials. In addition, the present study mainly compared the body response to different feeding frequencies at the transcriptomic level, therefore, the enzyme activity or protein expression are needed to be determined in further study.