The First Identification and Antibiogram of Clostridium perfringens Type C Isolated from Soil and The Feces of Dead Foals in South Korea
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
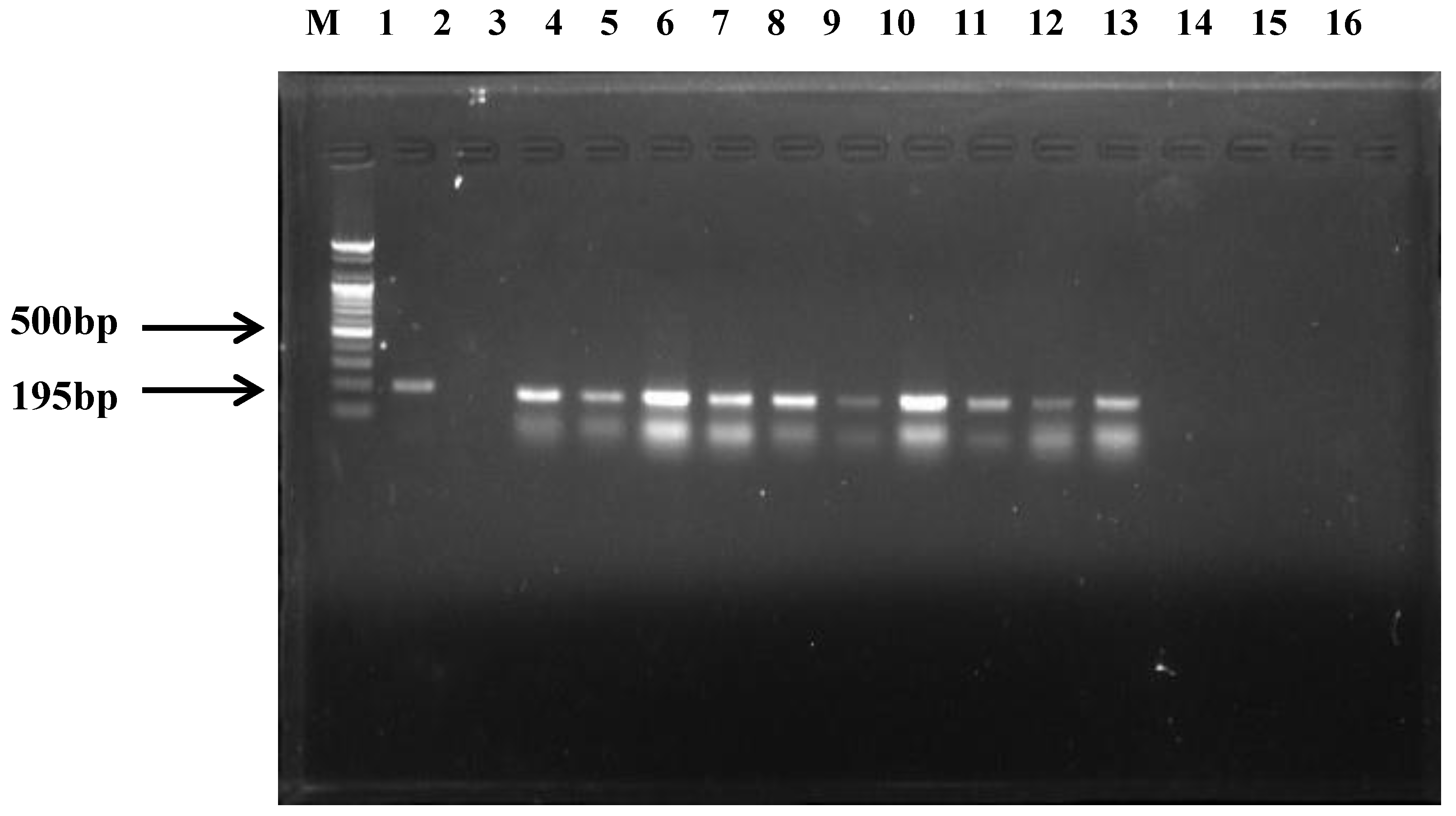
2.2. C. perfringens Detection
2.3. Preparation of Genomic DNA
2.4. 16S rRNA Gene Sequencing and Statistical Analysis
2.5. C. perfringens Toxin Detection
2.6. Antibiogram Test
3. Results
3.1. C. perfringens Detection
3.2. Toxins and Types of C. perfringens
3.3. Antibiogram Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Deguchi, A.; Miyamoto, K.; Kuwahara, T.; Miki, Y.; Kaneko, I.; Li, J.; McClane, B.A.; Akimoto, S. Genetic Characterization of Type A Enterotoxigenic Clostridium perfringens strains. PLoS ONE 2009, 4, e5598. [Google Scholar] [CrossRef] [PubMed]
- Diab, S.S.; Kind, H.; Moore, J.; Shahriar, M.F.; Odani, J.; Anthenill, L.; Songer, J.G.; Uzal, F.A. Pathology of Clostridium perfringens Type C Enterotoxemia in Horses. Vet. Pathol. 2012, 49, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Adams, V.; Bannam, T.L.; Miyamoto, K.; Garcia, J.P.; Uzal, F.A.; Rood, J.I. Toxin Plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 2013, 77, 208–233. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, M.; Ochi, S.; Oda, M.; Miyamoto, K.; Takehara, M.; Kobayashi, K. Recent Insights into Clostridium perfringens Beta-Toxin. Toxins 2015, 7, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Othani, K.; Shimizu, T. Regulation of Toxin Production in Clostridium perfringens. Toxins 2016, 8, 207. [Google Scholar]
- Uzal, F.A.; Vidal, J.E.; McClane, B.A.; Gurjar, A.A. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxinol. J. 2014, 2, 24. [Google Scholar] [CrossRef]
- Bokori-Brown, M.; Savva, C.G.; Fernandes da costa, S.P.; Naylor, C.E.; Basak, A.K.; Titball, R.W. Molecular basis of toxicity of Clostridium perfringens epsilon toxin. FEBS. J. 2011, 278, 4589–4601. [Google Scholar] [CrossRef]
- Nakano, V.; Ignacio, A.; Lianco, L.; Bueris, V.; Sircili, M.P.; Avila-Campos, M.J. Multiocus sequence typing analyses of Clostridium perfringens type A strains harboring tpeLand netB genes. Anaerobe 2017, 44, 99–105. [Google Scholar] [CrossRef]
- Coursodon, C.F.; Glock, R.D.; Moore, K.L.; Cooper, K.K.; Songer, J.G. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe 2012, 18, 117–121. [Google Scholar] [CrossRef]
- Songer, J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996, 9, 216. [Google Scholar] [CrossRef]
- Gkiourtzidis, K.; Frey, J.; Bourtzi-Hatzopoulou, E.; IIiadis, N.; Sarris, K. PCR detectionand prevalence of alpha-, beta-, beta2-, epsilon-, iota-, and enterotoxin genes in Clostridium perfringens isolated from lambs with Clostridial dysentery. Vet. Microbiol. 2001, 82, 39. [Google Scholar] [CrossRef]
- Klassen, H.L.; Molkenboer, M.J.; Bakker, J.; Miserez, R.; Hani, H.; Frey, J.; Popoff, M.R.; Van den bosch, J.F. Detection of the beta2 toxin gene of Clostridium perfringens indiarrhoeic piglets in The Netherlands and Switzerland. FEMS. Immunol. Med. Microbiol. 1999, 24, 325–332. [Google Scholar]
- Mahony, D.E.; Clark, G.A.; Stringer, M.F.; MacDonald, M.C.; Duchesne, D.R.; Mader, J.A. Rapid extraction of plasmids from Clostridium perfringens. Appl. Environ. Microbiol. 1986, 51, 521–523. [Google Scholar] [PubMed]
- Chalmers, G.; Bruce, H.L.; Hunter, D.B.; Parreira, V.R.; Kulkarni, R.R.; Jiang, Y.F.; Prescott, J.F.; Boerlin, P. Multlocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J. Clin. Microbiol. 2008, 46, 3957–3964. [Google Scholar] [CrossRef] [PubMed]
- Howard-Martin, M.; Morton, R.; Qualls, J.C.; MacAllister, C.G. Clostridium perfringens type C enterotoxemia in a newborn foal. J. Am. Vet. Med. Assoc. 1986, 189, 564–565. [Google Scholar]
- Bukar, A.; Mukhtar, M.D.; Adam, S.A. Current trend in antimicrobial susceptibility pattern of Clostridium tetani isolated soil samples Kano. Bayero J. Pure Appl. Sci. 2008, 1, 112–115. [Google Scholar] [CrossRef]
- Sanada, I.; Nishida, S. Isolation of Clostridium tetani from soil. J. Bacteriol. 1965, 89, 626–629. [Google Scholar]
- Smith, L. Inhibition of Clostridium botulinum by Strains of Clostridium perfringens Isolated from Soil. Appl. Microbiol. 1975, 30, 319–323. [Google Scholar] [PubMed]
- Gohari, I.M.; Arroyo, L.; MacInnes, J.I.; Timoney, J.F. Characterization of Clostridium perfringens in the feces of adult horses and foals with acute enterocolitis. Can. J. Vet. Res. 2014, 78, 1–7. [Google Scholar]
- Murdoch, D.A. Gram-Positive Anaerobic Cocci. Clin. Microbiol. Rev. 1998, 11, 81–120. [Google Scholar] [CrossRef]
- Yoo, S.Y.; Lee, S.U.; Park, K.Y.; Park, Y.H. Molecular Typing and Epidemiological Survey of Prevalence of Clostridium perfringens Types by Multiplex PCR. J. Clin. Microbiol. 1997, 35, 228–232. [Google Scholar]
- Kalia, V.C.; Mukherjee, T.; Bhushan, A.; Joshi, J.; Shankar, P.; Huma, N. Analysis of the unexplored features of rrs(16S rDNA) of the Genus Clostridium. BMC Genom. 2011, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Chan, K.M.; Fung, A.M.; Tang, B.S.; Yuen, K.Y. Clostridium bacteraemia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Pathol. 2005, 58, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Fallani, M.; Rigottier-Gois, L.; Aguliera, M.; Bridonneau, C.; Collignon, A.; Edwards, C.A.; Corthier, G.; Dore, J. Clostridium difficle and Clostridium perfringens species detected in infant faecal microbiota using 16S rRNA targeted probes. J. Microbiol. Methods. 2006, 67, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Cho, G.J. First Identification of Taylorella equgenitalis From Genital Tracts of Thoroughbred Horses from the Inland Area of South Korea by Multilocus Sequence Typing. J. Equine Vet. Sci. 2018, 60, 16–22. [Google Scholar] [CrossRef]
- Farzan, A.; Kircanski, J.; Delay, J. An investigation into the association between cpb2-encoding Clostridum perfringens type A and diarrhea in neonatal piglets. Can. J. Vet. Res. 2013, 77, 45–53. [Google Scholar] [PubMed]
- Andrews, J.M.; Howe, R.A. BSAC standardized disk susceptibility testing method (version 10). J. Antimicrob. Chemother. 2011, 66, 2726–2757. [Google Scholar] [CrossRef] [PubMed]
- Nachnani, S.; Scuteri, A.; Newman, M.G.; Avanessian, A.B.; Lomeli, S.L. E-test: A new technique for antimicrobial susceptibility testing for periodontal microorganisms. J. Periodontol. 1992, 63, 576–583. [Google Scholar] [CrossRef]
- Thornsberry, C. NCCLS Standards for Antimicrobial Susceptibility Tests. Lab. Med. 2016, 14, 549–553. [Google Scholar] [CrossRef]
- Feary, D.J.; Hassel, D.M. Enteritis and colitis in horses. Vet. Clin. North. Am. Equine Pract. 2006, 22, 437–479. [Google Scholar] [CrossRef]
- Lee, K.E.; Lim, S.I.; Shin, S.H.; Kwon, Y.K.; Kim, H.Y.; Song, J.Y.; An, D.J. Distribution of Clostridium perfringens isolates from piglets in South Korea. J. Vet. Med. Sci. 2014, 76, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J. Antimicrobial resistance and molecular characterization of Clostridium perfringens isolated from chicken. J. Prev. Vet. Med. 2016, 40, 71–79. [Google Scholar] [CrossRef]
- Slavic, D.; Boerlin, P.; Fabric, M.; Klotins, K.; Zothout, J.K.; Weir, P.E.; Bateman, D. Antimicrobial susceptibility of Clostridium perfringens isolates of bovine, chicken, porcine and turkey origin from Ontario. Can. J. Vet. Res. 2011, 75, 89. [Google Scholar] [PubMed]

| Tract | No. of Farms | No. (%) of C. perfringens Identified Farms | No. of C. perfringens Isolates | No. of C. perfringens Type (A/C) |
|---|---|---|---|---|
| Jeju-Island * | 17 | 4 (23.53) | 9 | 4/5 |
| Gyeongsang-Province * | 5 | 4 (80.00) | 6 | 5/1 |
| Chungcheong-Province | 2 | 1 (50.00) | 1 | 0/1 |
| Gyunggi-Province | 10 | 4 (40.00) | 3 | 2/1 |
| Jeolla-province | 10 | 2 (20.00) | 3 | 2/1 |
| Gangwon-Province * | 1 | 1 (100.0) | 3 | 2/1 |
| Total | 45 | 16 (35.56) | 25 | 15/10 |
| Types | No. (%) of C. perfringens Isolates | No. of Isolates from Dead Foals and Tracks | cpa | cpb | cpe/etx/itx/NetF | cpb2 |
|---|---|---|---|---|---|---|
| A | 15 (60.0) | 0/0 | 15/15 | 0/15 | 0/15 | 10/15 |
| C | 10 (40.0) | 7/3 | 10/10 | 10/10 | 0/10 | 5/10 |
| Total | 25 (100) | 7/3 | 25/25 | 10/25 | 0/25 | 15/25 |
| Antibiotics | No. (%) of Isolates | ||
|---|---|---|---|
| Resistance | Intermediate | Susceptible | |
| Penicillin | 0(0) | 25(100) | 0(0) |
| Amoxicillin/clavulanic acid | 3(12) | 1(4) | 21(84) |
| Ampicillin | 0(0) | 2(8) | 23(92) |
| Vancomycin | 1(4) | 24(96) | 0(0) |
| Tetracycline | 0(0) | 23(92) | 2(8) |
| Clindamycin | 0(0) | 25(100) | 0(0) |
| Metronidazole | 0(0) | 25(100) | 0(0) |
| Meropenem | 1(4) | 0(0) | 24(96) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, C.S.; Hwang, J.Y.; Cho, G.J. The First Identification and Antibiogram of Clostridium perfringens Type C Isolated from Soil and The Feces of Dead Foals in South Korea. Animals 2019, 9, 579. https://doi.org/10.3390/ani9080579
Park CS, Hwang JY, Cho GJ. The First Identification and Antibiogram of Clostridium perfringens Type C Isolated from Soil and The Feces of Dead Foals in South Korea. Animals. 2019; 9(8):579. https://doi.org/10.3390/ani9080579
Chicago/Turabian StylePark, Chul Song, Ji Yong Hwang, and Gil Jae Cho. 2019. "The First Identification and Antibiogram of Clostridium perfringens Type C Isolated from Soil and The Feces of Dead Foals in South Korea" Animals 9, no. 8: 579. https://doi.org/10.3390/ani9080579
APA StylePark, C. S., Hwang, J. Y., & Cho, G. J. (2019). The First Identification and Antibiogram of Clostridium perfringens Type C Isolated from Soil and The Feces of Dead Foals in South Korea. Animals, 9(8), 579. https://doi.org/10.3390/ani9080579




