Genome-Wide Microarray Analysis Suggests Transcriptomic Response May Not Play a Major Role in High- to Low-Altitude Acclimation in Harvest Mouse (Micromys minutus)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
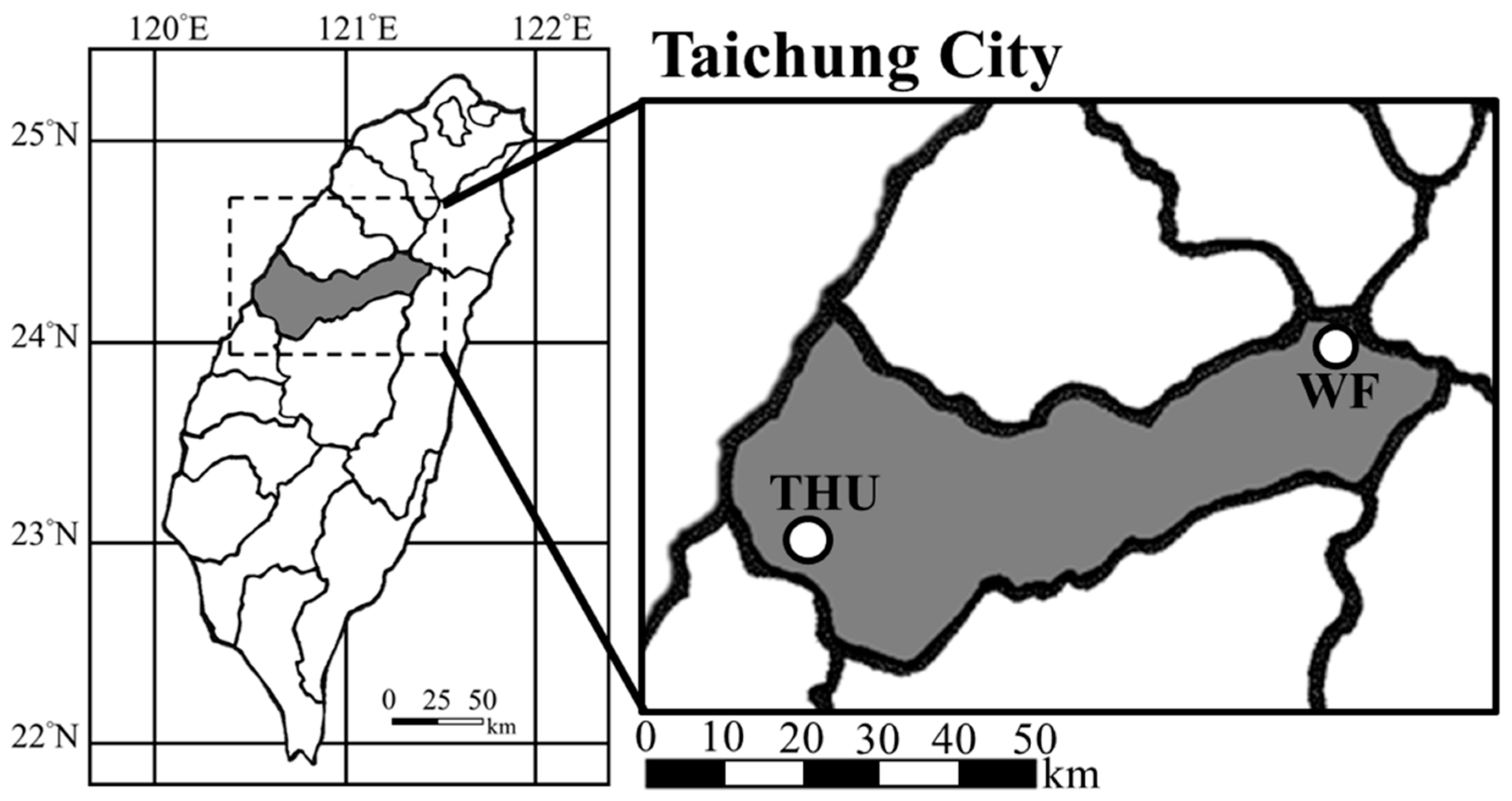
2.1. Sample Collection
2.2. Tissue Preparation and RNA Isolation
2.3. RNA Processing and Microarray Analysis
2.4. Statistical Analysis
2.5. Gene Ontology and Pathway Analyses
2.6. Quantitative Real-Time Polymerase Chain Reaction (qPCR) Confirmation
3. Results
3.1. High- to Low-Altitude Transfer
3.2. Microarray Analysis
3.3. qPCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Trout, R.C. A reviews of studies on populations of wild harvest mice (Mycromys minutes (Pallas)). Mammal Rev. 1978, 8, 143–158. [Google Scholar] [CrossRef]
- Lin, L.K.; Ma, G.C.; Chen, T.H.; Lin, W.H.; Lee, D.J.; Wen, P.Y.; Wu, S.H.; Chen, M. Genomic analyses of the Formosan harvest mouse (Micromys minutus) and comparisons to the brown Norway rat (Rattus norvegicus) and the house mouse (Mus musculus). Zoology 2013, 116, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Fabre, P.H.; Pagès, M.; Musser, G.G.; Fitriana, Y.S.; Fjeldså, J.; Jennings, A.; Jønsson, K.A.; Kennedy, J.; Michaux, J.; Semiadi, G.; et al. A new genus of rodent from Wallacea (Rodentia: Muridae: Murinae: Rattini), and its implication for biogeography and Indo-Pacific Rattini systematics. Zool. J. Linn. Soc. 2013, 169, 408–447. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Lin, Y.K. Polymorphic microsatellite markers for the harvest mouse (Micromys minutus) in Taiwan. Taiwania 2009, 54, 118–121. [Google Scholar] [CrossRef]
- Chen, Y.C. Comparative Thermoregulation of Field Rodents at Different Elevations in Central Taiwan. Master’s Thesis, Tunghai University, Taichung, Taiwan, 2004. [Google Scholar]
- Darinot, F.; Favier, C. Winter flooding and migrating moving in a harvest mouse population, Micromys minutus (Pallas, 1771). Bull. Mens. Soc. linn. Lyon. 2014, 83, 235–244, (In French with English abstract). [Google Scholar] [CrossRef]
- Sawabe, K.; Natuhara, Y. Effects of landscape structure on the distribution of harvest mice, Micromys minutus. Ecol. Civ. Eng. 2015, 18, 69–78, (In Japanese with English abstract). [Google Scholar] [CrossRef]
- Sutton, J.R.; Reeves, J.T.; Wagner, P.D.; Groves, B.M.; Cymerman, A.; Malconian, M.K.; Rock, P.B.; Young, P.M.; Walter, S.D.; Houston, C.S. Operation Everest II: Oxygen transport during exercise at extreme simulated altitude. J. Appl. Physiol. 1988, 64, 1309–1321. [Google Scholar] [CrossRef]
- Storz, J.F.; Runck, A.M.; Moriyama, H.; Weber, R.E.; Fago, A. Genetic differences in hemoglobin function between highland and lowland deer mice. J. Exp. Biol. 2010, 213, 2565–2574. [Google Scholar] [CrossRef]
- Cheviron, Z.A.; Whitehead, A.; Brumfield, R.T. Transcriptomic variation and plasticity rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Mol. Ecol. 2008, 17, 4556–4569. [Google Scholar] [CrossRef]
- Storz, J.F.; Scott, G.R.; Cheviron, Z.A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 2010, 213, 4125–4136. [Google Scholar] [CrossRef]
- Cheviron, Z.A.; Bachman, G.C.; Storz, J.F. Contributions of phenotypic plasticity to differences in thermogenic performance between highland and lowland deer mice. J. Exp. Biol. 2013, 216, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Reeves, J.T.; Leon-Velarde, F. Chronic mountain sickness: Recent studies of the relationship between hemoglobin concentration and oxygen transport. High Alt. Med. Biol. 2004, 5, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Windsor, J.S.; Rodway, G.W. Caudwell Xtreme Everest Research Group. Supplemental oxygen effects on ventilation in acclimatized subjects exercising at 5700 m altitude. Aviat. Space Environ. Med. 2007, 78, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Storz, J.F.; Hoffmann, F.G.; Opazo, J.C.; Moriyama, H. Adaptive functional divergence among triplicated alpha-globin genes in rodents. Genetics 2008, 178, 1623–1638. [Google Scholar] [CrossRef] [PubMed]
- Stobdan, T.; Karar, J.; Pasha, M.A. High altitude adaptation: Genetic perspectives. High Alt. Med. Biol. 2008, 9, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Gavin, T.P.; Drew, J.L.; Kubik, C.J.; Pofahl, W.E.; Hickner, R.C. Acute resistance exercise increases skeletal muscle angiogenic growth factor expression. Acta Physiol. 2007, 191, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Viganò, A.; Ripamonti, M.; De Palma, S.; Capitanio, D.; Vasso, M.; Wait, R.; Lundby, C.; Cerretelli, P.; Gelfi, C. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics 2008, 8, 4668–4679. [Google Scholar] [CrossRef]
- Murray, A.J. Metabolic adaptation of skeletal muscle to high altitude hypoxia: How new technologies could resolve the controversies. Genome Med. 2009, 1, 117. [Google Scholar] [CrossRef]
- Cheviron, Z.A.; Connaty, A.D.; McClelland, G.B.; Storz, J.F. Functional genomics of adaptation to hypoxic cold-stress in high-altitude deer mice: Transcriptomic plasticity and thermogenic performance. Evolution 2014, 68, 48–62. [Google Scholar] [CrossRef]
- Oufara, S.; Barré, H.; Rouanet, J.L.; Chatonnet, J. Adaptation to extreme ambient temperatures in cold-acclimated gerbils and mice. Am. J. Physiol. 1987, 253, R39–R45. [Google Scholar] [CrossRef]
- Van Sant, M.J.; Hammond, K.A. Contribution of shivering and nonshivering thermogenesis to thermogenic capacity for the deer mouse (Peromyscus maniculatus). Physiol. Biochem. Zool. 2008, 81, 605–611. [Google Scholar] [CrossRef] [PubMed]
- León-Velarde, F.; Sanchez, J.; Bigard, A.X.; Brunet, A.; Lesty, C.; Monge, C. High altitude tissue adaptation in Andean coots: Capillarity, fiber area, fiber type and enzymatic activities of skeletal muscle. J. Comp. Physiol. B 1993, 163, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Hepple, R.T.; Agey, P.J.; Hazelwood, L.; Szewczak, J.M.; MacMillen, R.E.; Mathieu-Costello, O. Increased capillarity in leg muscle of finches living at altitude. J. Appl. Physiol. 1998, 85, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
- Cheviron, Z.A.; Brumfield, R.T. Genomic insights into adaptation to high-altitude environments. Heredity 2012, 108, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 25, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Bar-Or, C.; Czosnek, H.; Koltai, H. Cross-species microarray hybridizations: A developing tool for studying species diversity. Trends Genet. 2007, 23, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.R.; Elogio, T.S.; Lui, M.A.; Storz, J.F.; Cheviron, Z.A. Adaptive modifications of muscle phenotype in high-altitude deer mice are associated with evolved changes in gene regulation. Mol. Biol. Evol. 2015, 32, 1962–1976. [Google Scholar] [CrossRef]
- Kerr, M.K.; Martin, M.; Churchill, G.A. Analysis of variance for gene expression microarray data. J. Comput. Biol. 2000, 7, 819–837. [Google Scholar] [CrossRef]
- Gracey, A.Y.; Cossins, A.R. Application of microarray technology in environmental and comparative physiology. Annu. Rev. Physiol. 2003, 65, 231–259. [Google Scholar] [CrossRef]
- Whitehead, A.; Crawford, D.L. Variation within and among species in gene expression: Raw material for evolution. Mol. Ecol. 2006, 15, 1197–1211. [Google Scholar] [CrossRef]
- Brodsky, L.I.; Jacob-Hirsch, J.; Avivi, A.; Trahtenbrot, L.; Zelingson, S.; Amariglio, N.; Paz, A.; Korol, A.B.; Band, M.; Rechavi, G.; et al. Evolutionary regulation of the blind subterranean mole rat, Spalax, revealed by genome-wide gene expression. Proc. Natl. Acad. Sci. USA 2005, 47, 17047–17052. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A. Comparative genomics in ecological physiology: Toward a more nuanced understanding of acclimation and adaptation. J. Exp. Biol. 2012, 215, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller, O.; Minko, T.; Qualls, C.; Pozharov, V.; Gamboa, J.; Gamboa, A.; Wang, Y. Gene expression, autonomic function and chronic hypoxia: Lessons from the Andes. Clin. Auton. Res. 2006, 16, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Everett, M.V.; Antal, C.E.; Crawford, D.L. The effect of short-term hypoxic exposure on metabolic gene expression. J. Exp. Zool. A Ecol. Genet. Physiol. 2012, 317, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Melius, J.; Cirilo, E.; Coort, S.L.; Digles, D.; et al. WikiPathway: A multifaceted pathway database bridging metabolomics to others omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef] [PubMed]
- Valadan, R.; Amjadi, O.; Tehrani, M.; Rafiei, A.; Hedayatizadeh-Omran, A.; Alizadeh-Navaei, R. Pseudogene-free amplification of HPRT1 in quantitative reverse transcriptase polymerase chain reaction. Anal. Biochem. 2015, 485, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, K.; Natuhara, Y. Extensive distribution models of the harvest mouse (Micromys minutus) in different landscapes. Glob. Ecol. Conserv. 2016, 8, 108–115. [Google Scholar] [CrossRef]
- Sawabe, K.; Hata, S.; Natuhara, Y. Influence of bank-grass mowing on the nest building of harvest mouse. J. Jpn. Inst. Landsc. Arch. 2005, 68, 571–574, (In Japanese with English abstract). [Google Scholar] [CrossRef]
- Hata, S.; Sawabe, K.; Natuhara, Y. A suitable embankment mowing strategy for habitat conservation of the harvest mouse. Landsc. Ecol. Eng. 2010, 6, 133–142. [Google Scholar] [CrossRef]
- Wiley, S.R.; Cassiano, L.; Lofton, T.; Davis-Smith, T.; Winkles, J.A.; Lindner, V.; Liu, H.; Daniel, T.O.; Smith, C.A.; Fanslow, W.C. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity 2001, 15, 837–846. [Google Scholar] [CrossRef]
- Wajant, H. The TWEAK-Fn14 as a potential drug target. Br. J. Pharmacol. 2013, 170, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Girgenrath, M.; Weng, S.; Kostek, C.A.; Browning, B.; Wang, M.; Brown, S.A.; Winkles, J.A.; Michaelson, J.S.; Allaire, N.; Schneider, P.; et al. TWEAK, via its receptor Fn14, is a novel regulator of mesenchymal progenitor cells and skeletal muscle regeneration. EMBO J. 2006, 13, 5826–5839. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [PubMed]
- Tajrishi, M.M.; Zheng, T.S.; Burkly, L.C.; Kumar, A. The TWEAK-Fn14 pathway: A potent regulator of skeletal muscle biology in health and disease. Cytokine Growth Factor Rev. 2014, 25, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Raue, U.; Trappe, T.A.; Estrem, S.T.; Qian, H.R.; Helvering, L.M.; Smith, R.C.; Trappe, S. Transcriptome signature of resistance exercise adaptations: Mixed muscle and fiber type specific profiles in young and old adults. J. Appl. Physiol. 2012, 112, 1625–1636. [Google Scholar] [CrossRef]
- Raue, U.; Jemiolo, B.; Yang, Y.; Trappe, S. TWEAK-Fn14 pathway activation after exercise in human skeletal muscle: Insights from two exercise modes and a time course investigation. J. Appl. Physiol. 2015, 118, 569–578. [Google Scholar] [CrossRef]
- Lundby, C.; Sander, M.; van Hall, G.; Saltin, B.; Calbet, J.A.L. Maximal exercise and muscle oxygen extraction in acclimatizing lowlanders and high altitude natives. J. Physiol. 2006, 573, 535–547. [Google Scholar] [CrossRef]
- Bärtsch, P.; Saltin, B. General introduction to altitude adaptation and mountain sickness. Scand. J. Med. Sci. Sports 2008, 18 (Suppl. 1), 1–10. [Google Scholar] [CrossRef]
- Saltin, B.; Gollnick, P.D. Skeletal muscle adaptability: Significance for metabolism and performance. In Handbook of Physiology; Section 10: Skeletal Muscle; Peachey, L.D., Adrian, R.H., Geiger, S.R., Eds.; American Physiological Society: Bethesda, MD, USA, 1983; pp. 555–631. [Google Scholar]
- Lundby, C.; Pilegaard, H.; Andersen, J.L.; van Hall, G.; Sander, M.; Calbet, J.A.L. Acclimation to 4100 m does not change capillary density or mRNA expression of potential angiogenesis regulatory factors in human skeletal muscle. J. Exp. Biol. 2004, 207, 3865–3871. [Google Scholar] [CrossRef]
- Lundby, C.; Haslund, M.L.; Robach, P.; Calbet, J.A.L. Whole body protein turnover at altitude. FASEB J. 2006, 20, A1257. [Google Scholar]
- Ishiwaka, R.; Kinoshita, Y.; Satou, H.; Kakihara, H.; Masuda, Y. Overwintering in nests on the ground in the harvest mouse. Landsc. Ecol. Eng. 2010, 6, 335–342. [Google Scholar] [CrossRef]
- Górecki, A. Metabolism and energy budget in the harvest mouse. Acta Theriol. 1971, 15, 213–220. [Google Scholar] [CrossRef]
- Grodziński, W.H.; Böckler, H.; Heldmaier, G. Basal and cold-induced metabolic rates in the harvest mouse (Micromys minutus). Acta Theriol. 1988, 20, 283–291. [Google Scholar] [CrossRef]
| No. of Total Genes Examined | 23,188 |
|---|---|
| No. of filtered genes (% of examined genes) | 47 (0.20%) |
| No. of up-regulated genes (% of filtered genes) | 33 (70.21%) |
| No. of down-regulated genes (% of filtered genes) | 14 (29.79%) |
| No. of filtered genes with FDR p < 0.05 (% of filtered genes) | 1 (2.13%) |
| Pathway | No. of Differentially Expressed Genes | Up-Regulated Genes | Down-Regulated Genes | ||
|---|---|---|---|---|---|
| Number | List | Number | List | ||
| CDKN1A-EGF-CREB | 3 | 3 | Hbb, Cdkn1a, Hbb-b1 | 0 | |
| IL-2 Signaling Pathway | 2 | 2 | Il2rg, Cd53 | 0 | |
| MAPK Signaling Pathway | 2 | 0 | 2 | Nr4a1, Cacng6 | |
| B Cell Receptor Signaling Pathway | 2 | 2 | Blnk, Cd79a | 0 | |
| Tryptophan metabolism | 2 | 1 | Cyp2f4 | 1 | Cyp2e1 |
| Nuclear factor, erythroid-derived 2, like 2 signaling pathway | 2 | 1 | Cdkn1a | 1 | Cacng6 |
| Heme Biosynthesis | 1 | 1 | Alas2 | 0 | |
| IL-9 Signaling Pathway | 1 | 1 | Il2rg | 0 | |
| ATM Signaling Pathway | 1 | 1 | Cdkn1a | 0 | |
| Electron Transport Chain | 1 | 1 | Ucp2 | 0 | |
| Alpha6-Beta4 Integrin Signaling Pathway | 1 | 1 | Cdkn1a | 0 | |
| Hypertrophy Model | 1 | 0 | 1 | Dusp14l1 | |
| Cell cycle | 1 | 1 | Cdkn1a | 0 | |
| Ethanol metabolism resulting in the production of ROS by CYP2E1 | 1 | 0 | 1 | Cyp2e1 | |
| Inflammatory Response Pathway | 1 | 1 | Il2rg | 0 | |
| TGF-beta Receptor Signaling Pathway | 1 | 1 | Cdkn1a | 0 | |
| G1 to S cell cycle control | 1 | 1 | Cdkn1a | 0 | |
| Cytoplasmic Ribosomal Proteins | 1 | 1 | Rpl35 | 0 | |
| Relationship between glutathione and NADPH | 1 | 1 | Cdkn1a | 0 | |
| Spinal Cord Injury | 1 | 0 | 1 | Nr4a1 | |
| Nuclear Receptors | 1 | 0 | 1 | Nr4a1 | |
| Serotonin and anxiety | 1 | 1 | Plek | 0 | |
| IL-4 Signaling Pathway | 1 | 1 | Il2rg | 0 | |
| Adipogenesis | 1 | 1 | Cdkn1a | 0 | |
| Nuclear receptors in lipid metabolism and toxicity | 1 | 0 | 1 | Cyp2e1 | |
| Fatty Acid Omega Oxidation | 1 | 0 | 1 | Cyp2e1 | |
| Senescence and Autophagy | 1 | 1 | Cdkn1a | 0 | |
| ErbB signaling pathway | 1 | 1 | Cdkn1a | 0 | |
| Retinol metabolism | 1 | 0 | 1 | Cyp2e1 | |
| Metapathway biotransformation | 1 | 0 | 1 | Cyp2e1 | |
| IL-7 Signaling Pathway | 1 | 1 | Il2rg | 0 | |
| Sample | Tnfrsf12a (Average Cq) | Hprt1 (Average Cq) | ΔCq (Tnfrsf12a − Hprt1) | ΔΔCq (ΔCq Low altitude − ΔCq High altitude) | Fold Change (2−(ΔΔCq)) |
|---|---|---|---|---|---|
| High altitude (n = 4) | 28.46 ± 1.36 | 22.40 ± 1.24 | 6.05 ± 0.25 | - | - |
| Low altitude (n = 4) | 26.73 ± 2.71 | 23.63 ± 2.68 | 3.10 ± 0.11 | −2.95 ± 0.25 | 7.85 ± 1.32 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-H.; Ma, G.-C.; Lin, W.-H.; Lee, D.-J.; Wu, S.-H.; Liao, B.-Y.; Chen, M.; Lin, L.-K. Genome-Wide Microarray Analysis Suggests Transcriptomic Response May Not Play a Major Role in High- to Low-Altitude Acclimation in Harvest Mouse (Micromys minutus). Animals 2019, 9, 92. https://doi.org/10.3390/ani9030092
Chen T-H, Ma G-C, Lin W-H, Lee D-J, Wu S-H, Liao B-Y, Chen M, Lin L-K. Genome-Wide Microarray Analysis Suggests Transcriptomic Response May Not Play a Major Role in High- to Low-Altitude Acclimation in Harvest Mouse (Micromys minutus). Animals. 2019; 9(3):92. https://doi.org/10.3390/ani9030092
Chicago/Turabian StyleChen, Tze-Ho, Gwo-Chin Ma, Wen-Hsiang Lin, Dong-Jay Lee, Sheng-Hai Wu, Ben-Yang Liao, Ming Chen, and Liang-Kong Lin. 2019. "Genome-Wide Microarray Analysis Suggests Transcriptomic Response May Not Play a Major Role in High- to Low-Altitude Acclimation in Harvest Mouse (Micromys minutus)" Animals 9, no. 3: 92. https://doi.org/10.3390/ani9030092
APA StyleChen, T.-H., Ma, G.-C., Lin, W.-H., Lee, D.-J., Wu, S.-H., Liao, B.-Y., Chen, M., & Lin, L.-K. (2019). Genome-Wide Microarray Analysis Suggests Transcriptomic Response May Not Play a Major Role in High- to Low-Altitude Acclimation in Harvest Mouse (Micromys minutus). Animals, 9(3), 92. https://doi.org/10.3390/ani9030092






