RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Blood Analysis
2.3. Lipid Oxidation Measurement
2.4. RNA Isolation
2.5. Library Preparation, Sequencing, and RNA-Seq Analyses
2.6. IPA Analysis
2.7. Protein-Protein Interaction Analysis (STRING)
2.8. Statistics
3. Results
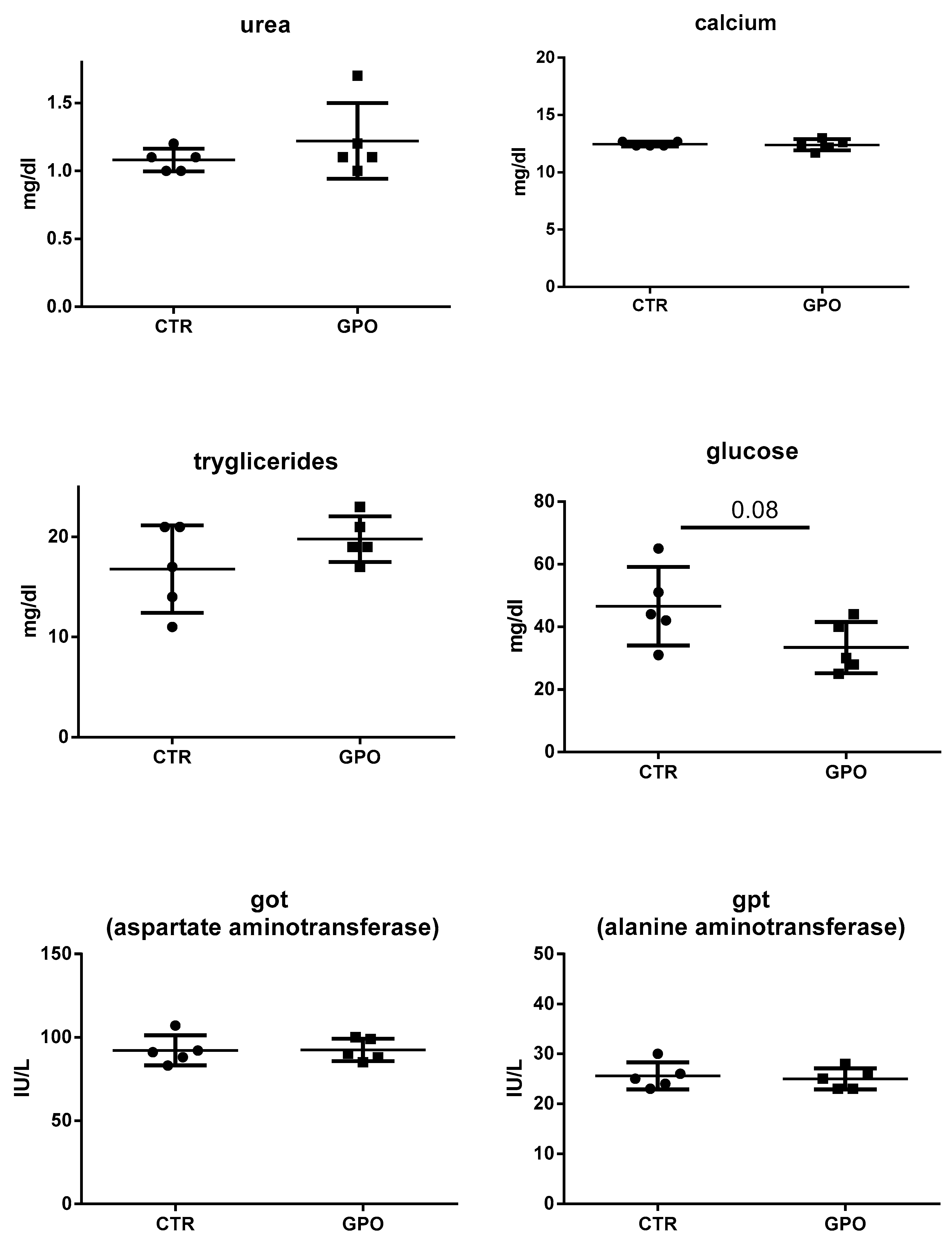
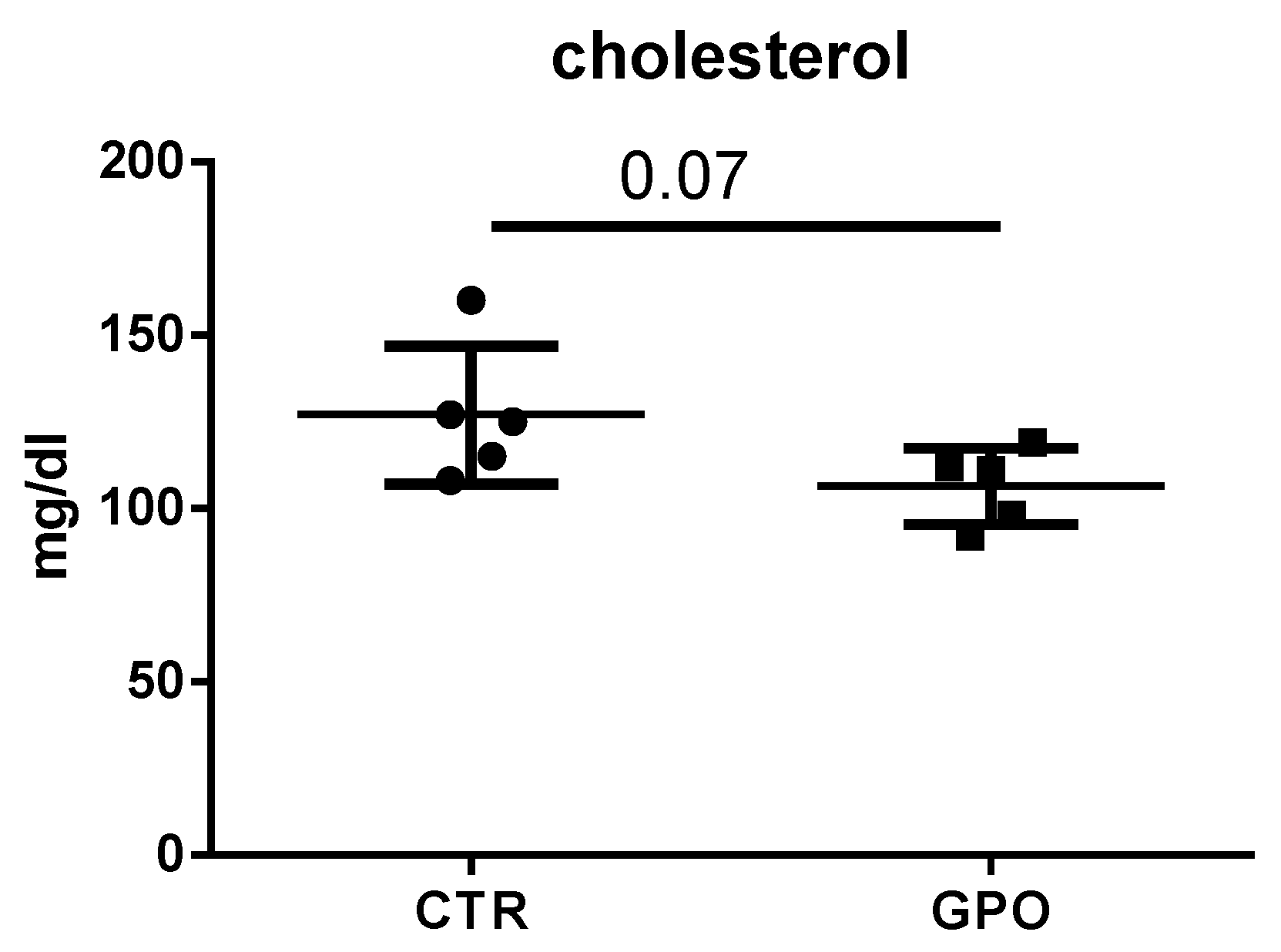
3.1. Effects of GPO-Supplemented Diet on Blood Biochemical Analysis
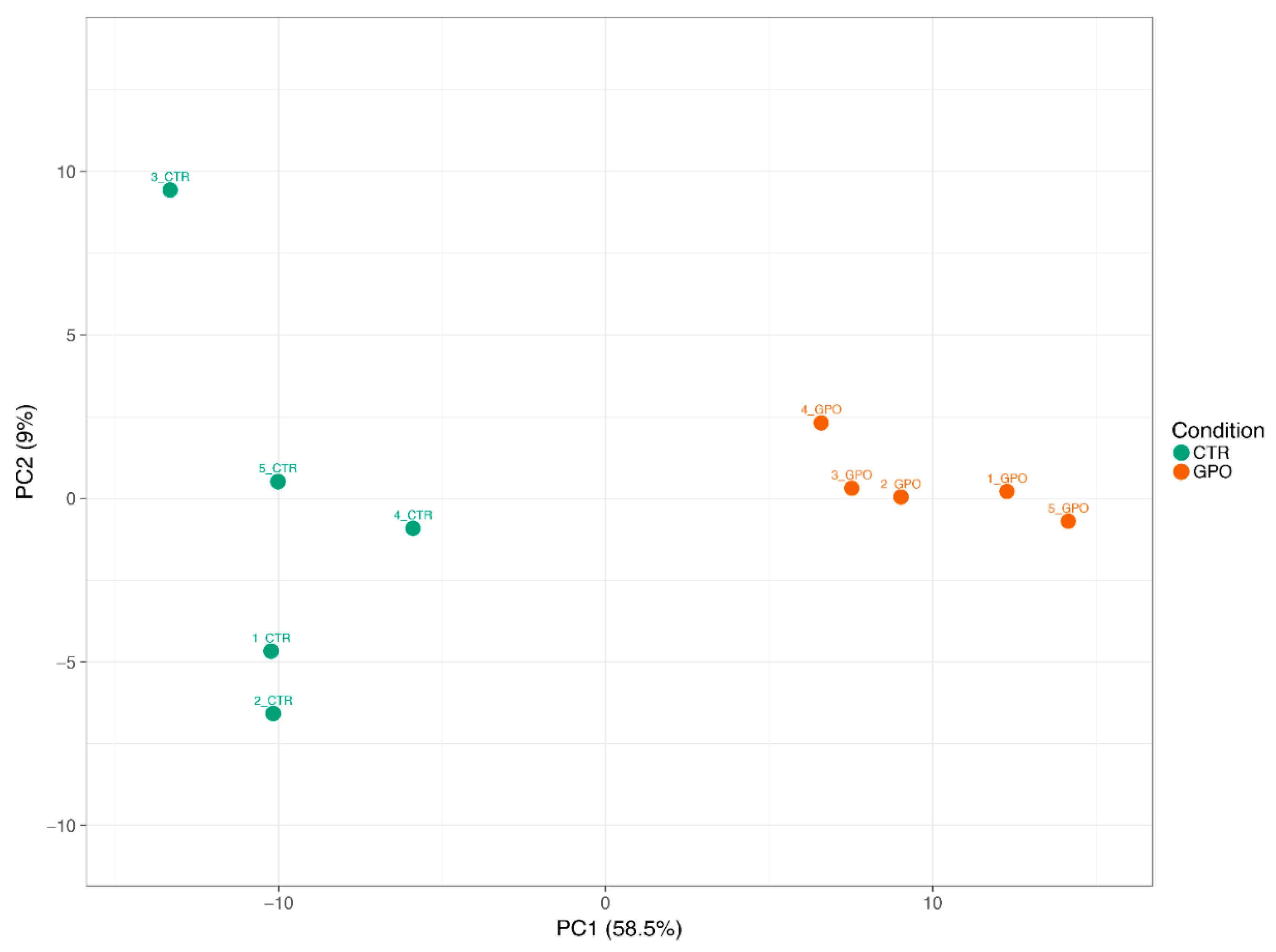
3.2. Influence of GPO-Supplemented Diet on Calves’ Blood Transcriptome
3.3. Effects of GPO-Supplemented Diet on Meat Cattle Lipid Oxidation
4. Discussion
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elgendy, R.; Giantin, M.; Castellani, F.; Grotta, L.; Palazzo, F.; Dacasto, M.; Martino, G. Transcriptomic signature of high dietary organic selenium supplementation in sheep: A nutrigenomic insight using a custom microarray platform and gene set enrichment analysis. J. Anim. Sci. 2016, 94, 3169–3184. [Google Scholar] [CrossRef] [PubMed]
- Elgendy, R.; Palazzo, F.; Castellani, F.; Giantin, M.; Grotta, L.; Cerretani, L.; Dacasto, M.; Martino, G. Transcriptome profiling and functional analysis of sheep fed with high zinc-supplemented diet: A nutrigenomic approach. Anim. Feed Sci. Technol. 2017, 234, 195–204. [Google Scholar] [CrossRef]
- Scalbert, A.; Manach, C.; Morand, C.; Rémésy, C.; Jiménez, L. Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 2005, 45, 287–306. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Ringseis, R.; Eder, K. Potential of plant polyphenols to combat oxidative stress and inflammatory processes in farm animals. J. Anim. Physiol. Anim. Nutr. 2017, 101, 605–628. [Google Scholar] [CrossRef] [PubMed]
- Goñi, I.; Arija, I.; Estevez, R. Effect of dietary grape pomace and vitamin E on growth performance, nutrient digestibility, and susceptibility to meat lipid oxidation in chickens effect of dietary grape pomace and vitamin E on growth performance, nutrient digestibility, and suscepti. Poult. Sci. 2007, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Kim, I.H. Effect of dietary grape pomace fermented by Saccharomyces boulardii on the growth performance, nutrient digestibility and meat quality in finishing pigs. Asian-Australas J. Anim. Sci. 2011, 24, 1763–1770. [Google Scholar] [CrossRef]
- Garrido, M.D.; Auqui, M.; Martí, N.; Linares, M.B. Effect of two different red grape pomace extracts obtained under different extraction systems on meat quality of pork burgers. LWT–Food Sci. Technol. 2011, 44, 2238–2243. [Google Scholar] [CrossRef]
- Baumgärtel, T.; Kluth, H.; Epperlein, K.; Rodehutscord, M. A note on digestibility and energy value for sheep of different grape pomace. Small Rumin. Res. 2007, 67, 302–306. [Google Scholar] [CrossRef]
- Santos, N.W.; Santos, G.T.D.; Silva-Kazama, D.C.; Grande, P.A.; Pintro, P.M.; de Marchi, F.E.; Jobim, C.C.; Petit, H.V. Production, composition and antioxidants in milk of dairy cows fed diets containing soybean oil and grape residue silage. Livest. Sci. 2014, 159, 37–45. [Google Scholar] [CrossRef]
- Chedea, V.S.; Pelmus, R.S.; Lazar, C.; Pistol, G.C.; Calin, L.G.; Toma, S.M.; Dragomir, C.; Taranu, I. Effects of a diet containing dried grape pomace on blood metabolites and milk composition of dairy cows. J. Sci. Food Agric. 2017, 97, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Chatterton, D.E.W.; Smithers, G.; Roupas, P.; Brodkorb, A. Bioactivity of β-lactoglobulin and α-lactalbumin-Technological implications for processing. Int. Dairy J. 2006, 16, 1229–1240. [Google Scholar] [CrossRef]
- Loor, J.J.; Mccann, J.C.; Zhou, Z.; Bionaz, M. Triennial Lactation Symposium: Nutrigenomics in livestock: Systems biology meets nutrition 1. J. Anim. Sci. 2015, 93, 5554–5574. [Google Scholar] [CrossRef] [PubMed]
- Kafantaris, I.; Kotsampasi, B.; Christodoulou, V.; Kokka, E.; Kouka, P.; Terzopoulou, Z.; Gerasopoulos, K. Grape pomace improves antioxidant capacity and faecal microflora of lambs. J. Anim. Physiol. Anim. Nutr. 2017, 101, e108–e121. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Kishi, Y.; Oishi, K.; Hirooka, H.; Kumagai, H. Effects of feeding polyphenol-rich winery wastes on digestibility, nitrogen utilization, ruminal fermentation, antioxidant status and oxidative stress in wethers. Anim. Sci. J. 2015, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Sapanidou, V.G.; Margaritis, I.; Siahos, N.; Arsenopoulos, K.; Dragatidou, E.; Taitzoglou, I.A.; Zervos, I.A.; Theodoridis, A.; Tsantarliotou, M.P. Antioxidant effect of a polyphenol-rich grape pomace extract on motility, viability and lipid peroxidation of thawed bovine spermatozoa. J. Biol. Res.-Thessalon. 2014, 21, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Grotta, L.; Castellani, F.; Palazzo, F.; Naceur Haouet, M.; Martino, G. Treatment optimisation and sample preparation for the evaluation of lipid oxidation in various meats through TBARs assays before analysis. Food Anal. Methods 2017, 10, 1870–1880. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2016, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucl. Acids Res. 2017, 45, D362–368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tsao, R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr. Opin. Food Sci. 2016, 8, 33–42. [Google Scholar] [CrossRef]
- Liew, C.C.; Ma, J.; Tang, H.C.; Zheng, R.; Dempsey, A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.; Liew, C. The peripheral-blood transcriptome: New insights into disease and risk assessment. Trends Mol. Med. 2007, 13, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.R.; Krych, L.; Ahmad, H.F.; Nejsum, P.; Skovgaard, K.; Nielsen, D.S.; Thamsborg, S.M. A polyphenol-enriched diet and Ascaris suum infection modulate mucosal immune responses and gut microbiota composition in pigs. PLoS ONE 2017, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Chedea, V.S.; Palade, L.M.; Marin, D.E.; Pelmus, R.S.; Habeanu, M.; Rotar, M.C.; Gras, M.A.; Pistol, G.C.; Taranu, I. Intestinal absorption and antioxidant activity of grape pomace polyphenols. Nutrients 2018, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Rivas, C.; Gallardo, B.; Mantecón, Á.R.; del Álamo-Sanza, M.; Manso, T. Evaluation of grape pomace from red wine by-product as feed for sheep. J. Sci. Food Agric. 2017, 97, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Roland, L.; Drillich, M.; Iwersen, M. Hematology as a diagnostic tool in bovine medicine. J. Vet. Diagn. Investig. 2014, 26, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Roopchand, D.E.; Kuhn, P.; Krueger, C.G.; Moskal, K.; Lila, M.A.; Raskin, I. Concord grape pomace polyphenols complexed to soy protein isolate are stable and hypoglycemic in diabetic mice. J. Agric. Food Chem. 2013, 61, 11428–11433. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Kwon, S.; Hwang, J.H.; Park, D.H.; Kim, T.W.; Kang, D.G.; Yu, G.E.; Park, H.C.; An, S.M.; Kim, C.W. Squalene epoxidase plays a critical role in determining pig meat quality by regulating adipogenesis, myogenesis, and ROS scavengers. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bartley, G.E.; Arvik, T.; Lipson, R.; Nah, S.Y.; Seo, K.; Yokoyama, W. Dietary supplementation of chardonnay grape seed flour reduces plasma cholesterol concentration, hepatic steatosis, and abdominal fat content in high-fat diet-induced obese hamsters. J. Agric. Food Chem. 2014, 62, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Abarghuei, M.J.; Rouzbehan, Y.; Salem, A.Z.M.; Zamiri, M.J. Nitrogen balance, blood metabolites and milk fatty acid composition of dairy cows fed pomegranate-peel extract. Livest. Sci. 2014, 164, 72–80. [Google Scholar] [CrossRef]
- Dávalos, A.; Fernández-Hernando, C.; Cerrato, F.; Martínez-Botas, J.; Gómez-Coronado, D.; Gómez-Cordovés, C.; Lasunción, M.A. Red grape juice polyphenols alter cholesterol homeostasis and increase LDL-receptor activity in human cells in vitro. J. Nutr. 2006, 136, 1766–1773. [Google Scholar] [CrossRef] [PubMed]
- Dohadwala, M.M.; Vita, J. A Grapes and Cardiovascular Disease 1–3. Clin. Res. 2009, 139, 1788S–1793S. [Google Scholar] [CrossRef] [PubMed]
- Bloch, K. The biological synthesis of cholesterol. Science 1965, 150, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schulte, P.; Godin, D.V.; Cheng, K.M. Differential mRNA expression of seven genes involved in cholesterol metabolism and transport in the liver of atherosclerosis-susceptible and -resistant Japanese quail strains. Genet. Sel. Evol. 2012, 44, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Do, R.; Kiss, R.S.; Gaudet, D.; Engert, J.C. Squalene synthase: A critical enzyme in the cholesterol biosynthesis pathway. Clin. Genet. 2009, 75, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-Y.; Liu, Y.; Yan, Y.-M.; Lu, X.-F.; Cheng, Y.-X. Phenolic compounds from Belamcanda chinensis seeds. Molecules 2018, 23, 580. [Google Scholar] [CrossRef] [PubMed]
- Trapani, L.; Segatto, M.; Ascenzi, P.; Pallottini, V. Potential role of nonstatin cholesterol lowering agents. IUBMB Life 2011, 63, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, J.; Gu, Y.; Deng, D.; Wu, Z.; Bao, L.; Li, M.; Yao, Z. Child syndrome mimicking verrucous nevus in a Chinese patient responded well to the topical therapy of compound of simvastatin and cholesterol. J. Eur. Acad. Dermatol. Venereol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Frisso, G.; Gelzo, M.; Procopio, E.; Sica, C.; Lenza, M.P.; Dello Russo, A.; Donati, M.A.; Salvatore, F.; Corso, G. A rare case of sterol-C4-methyl oxidase deficiency in a young Italian male: Biochemical and molecular characterization. Mol. Genet. Metab. 2017, 121, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Bunel, A.; Nivet, A.L.; Blondin, P.; Vigneault, C.; Richard, F.J.; Sirard, M.A. Cumulus cell gene expression associated with pre-ovulatory acquisition of developmental competence in bovine oocytes. Reprod. Fertil. Dev. 2014, 26, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhang, J.; Li, S.; Ji, S.; Cao, Z.; Zhang, H.; Wang, Y. Effects of a wide range of dietary forage-to-concentrate ratios on nutrient utilization and hepatic transcriptional profiles in limit-fed Holstein heifers. BMC Genom. 2018, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.; He, X.; Li, H.; Wu, S.; Li, S.; Deng, G. Biological activities of polyphenols from grapes. Polyphenols Hum. Heal. Dis. 2013, 1, 47–58. [Google Scholar] [CrossRef]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Kafantaris, I.; Stagos, D.; Kotsampasi, B.; Hatzis, A.; Kypriotakis, A.; Gerasopoulos, K.; Makri, S.; Goutzourelas, N.; Mitsagga, C.; Giavasis, I.; et al. Grape pomace improves performance, antioxidant status, fecal microbiota and meat quality of piglets. Animal 2018, 12, 246–255. [Google Scholar] [CrossRef] [PubMed]

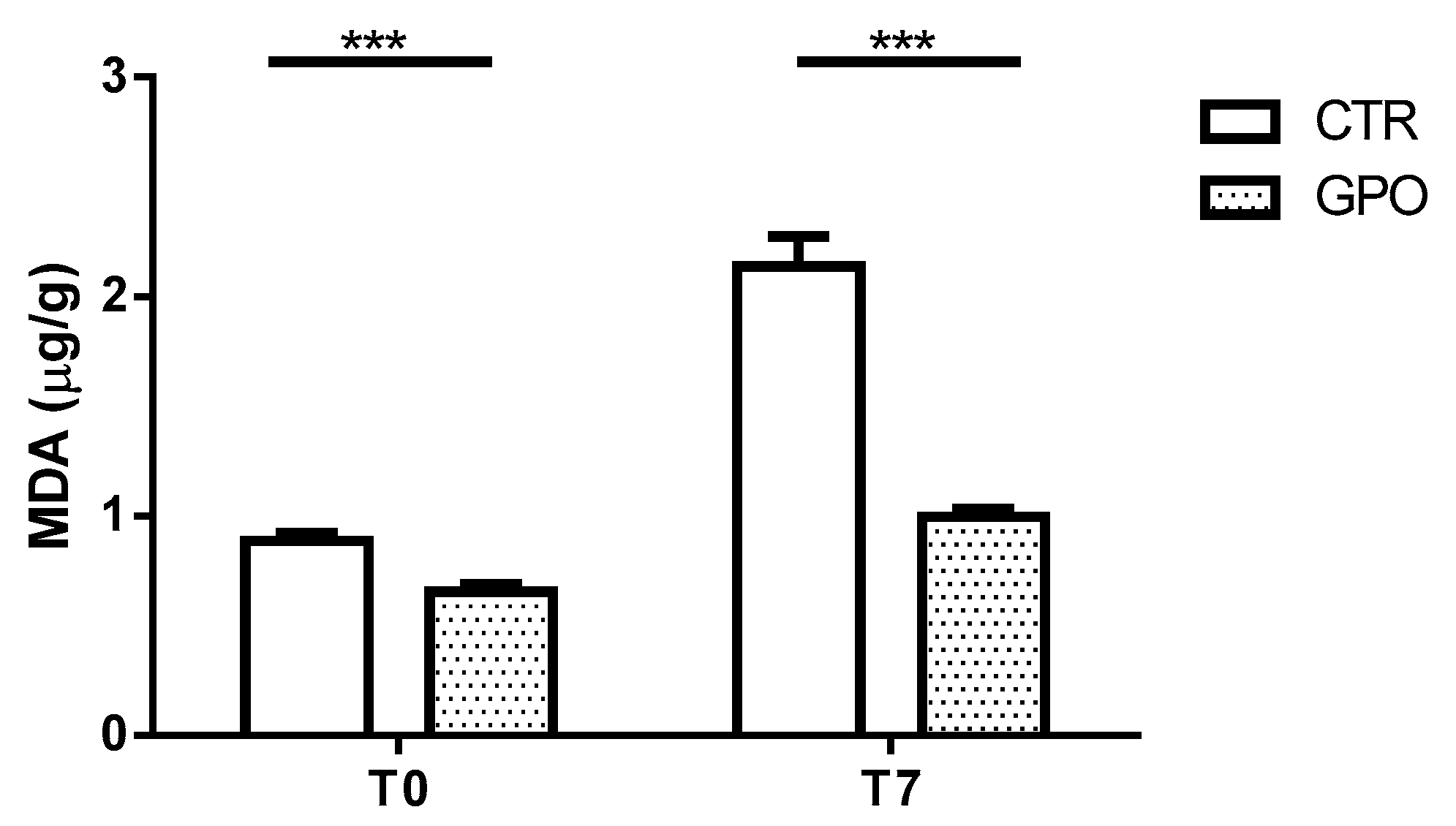
| Parameters | % |
|---|---|
| Dry matter | 87.13 |
| Crude protein | 16.35 |
| Ash | 9.47 |
| Ether extract | 1.21 |
| Neutral detergent fiber | 30.75 |
| Acid detergent fiber | 21.43 |
| Acid detergent lignin | 6.78 |
| Ingredient | Composition % | |
|---|---|---|
| Control Group | Grape Pomace Group | |
| Corn | 22 | 34 |
| Grain dust | 14 | 22 |
| Grape pomace flour | 0 | 10 |
| Fine bran | 9.6 | 3.4 |
| Sunflower seed | 10 | 4.4 |
| Faba bean | 7.5 | 0 |
| Biscuits waste | 5.3 | 6.4 |
| Distillers mais | 5 | 1 |
| Corn gluten feed | - | 9 |
| Barley | 8.2 | 4 |
| Bran wheat | 5 | 0 |
| Orange pulp dried | 3.7 | 0 |
| Soybean meal | 2.9 | 0 |
| Calcium carbonate | 25 | 1.6 |
| Molasses | 1 | 1 |
| Soybean hull | 0.6 | 0 |
| Common salt | 0.6 | 0.6 |
| Sodium bicarbonate 27% | 0.5 | 0.8 |
| Soybean oil | 0.4 | |
| Dicalcium phosphate | 0 | 0.6 |
| Magnesium oxide | 0.4 | 0.4 |
| Vitamin Premix | 0.8 | 0.8 |
| Chemical composition of the concentrate | ||
| Dry matter | 88.5 | 88.6 |
| Crude protein | 16.0 | 15.91 |
| Ash | 7.5 | 7.53 |
| Ether extract | 3.75 | 4.0 |
| Crude Fiber | 7.65 | 7.24 |
| Starch | 31.0 | 33.5 |
| Neutral detergent fiber | 20.37 | 18.90 |
| Acid detergent fiber | 7.95 | 8.32 |
| Acid detergent lignin | 3.50 | 3.86 |
| Canonical Pathways | -Log (p-Value) | Ratio | Molecules |
|---|---|---|---|
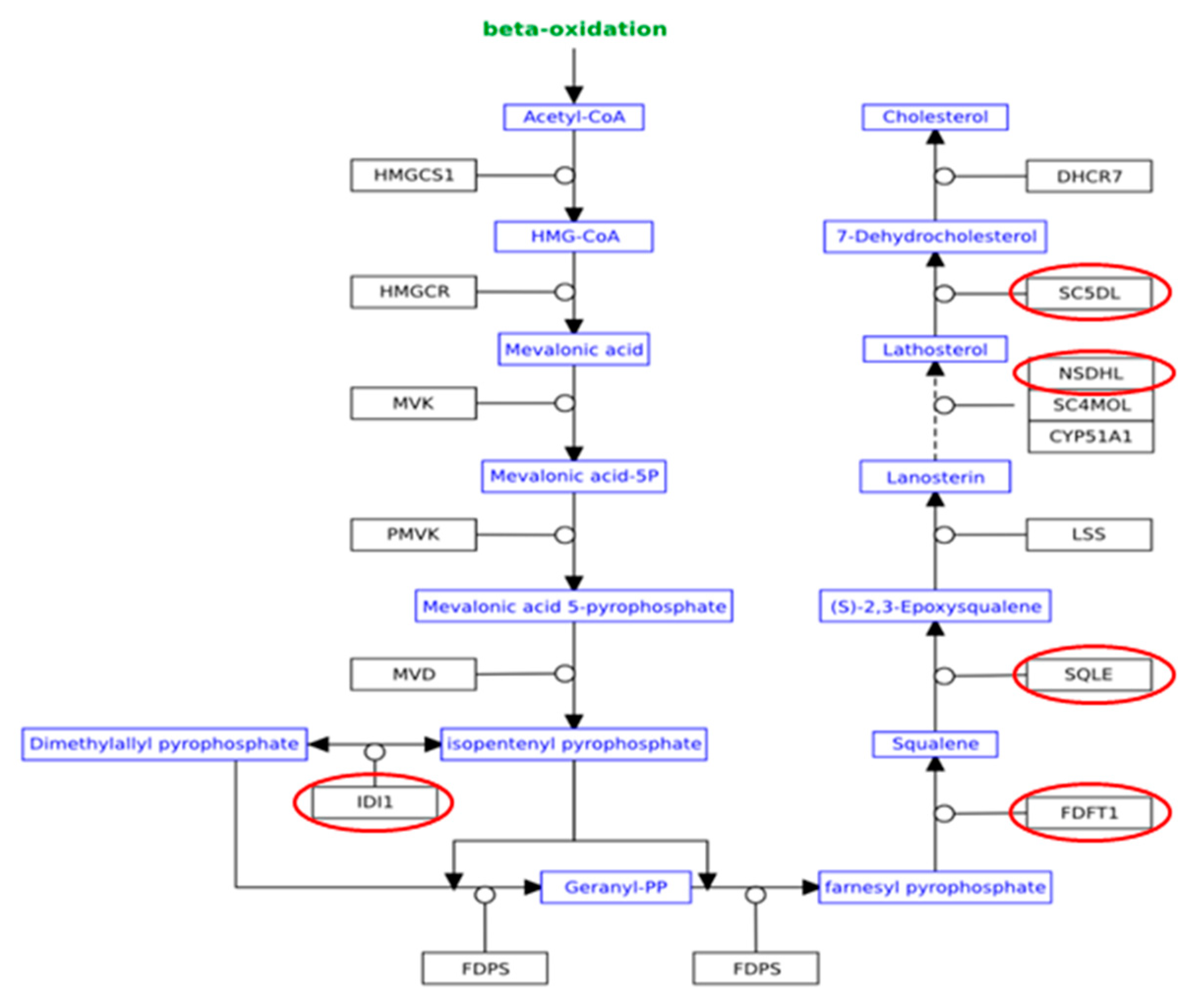
| Cholesterol Biosynthesis I | 5.76 | 0.385 | FDFT1,SQLE,NSDHL,MSMO1,SC5D |
| Cholesterol Biosynthesis II (via 24,25-dihydrolanosterol) | 5.76 | 0.385 | FDFT1,SQLE,NSDHL,MSMO1,SC5D |
| Cholesterol Biosynthesis III (via Desmosterol) | 5.76 | 0.385 | FDFT1,SQLE,NSDHL,MSMO1,SC5D |
| Superpathway of Cholesterol Biosynthesis | 5.25 | 0.222 | FDFT1,SQLE,NSDHL,IDI1,MSMO1,SC5D |
| Epoxysqualene Biosynthesis | 3.53 | 1 | FDFT1,SQLE |
| Zymosterol Biosynthesis | 2.37 | 0.333 | NSDHL,MSMO1 |
| IL-1 Signaling | 2.33 | 0.0667 | GNAI3,IL1A,PRKAR2B,MAPK8,GNA13,IRAK4 |
| PPAR Signaling | 2.26 | 0.0645 | SRA1,IL1A,PDGFA,IL1RL1,PDGFRA,PTGS2 |
| Toll-like Receptor Signaling | 2.09 | 0.0694 | UBD,IL1A,IL1RL1,MAPK8,IRAK4 |
| Role of JAK family kinases in IL-6-type Cytokine Signaling | 2.05 | 0.12 | SOCS1,MAPK8,OSM |
| NF-κB Signaling | 1.97 | 0.0462 | IL1A,BCL10,MAPK8,FCER1G,PDGFRA,TBK1,MAP3K8,IRAK4 |
| VDR/RXR Activation | 1.95 | 0.0641 | PDGFA,IL1RL1,HR,THBD,KLF4 |
| Semaphorin Signaling in Neurons | 1.94 | 0.0784 | MET,RHOB,DPYSL3,PLXNB1 |
| LXR/RXR Activation | 1.91 | 0.0545 | FDFT1,IL1A,LDLR,SREBF1,IL1RL1,PTGS2 |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 1.88 | 0.0444 | MET,IL1A,MYH9,PDGFA,IL1RL1,PDGFRA,SMAD7,COL18A1 |
| Hepatic Cholestasis | 1.78 | 0.0461 | IL1A,PRKAR2B,SREBF1,IL1RL1,MAPK8,OSM,IRAK4 |
| IL-8 Signaling | 1.75 | 0.0421 | GNAI3,MTOR,RHOB,MAPK8,PTGS2,GNA13,IRAK4,MYL12B |
| Nucleotide Excision Repair Pathway | 1.72 | 0.0909 | ERCC4,XPC,POLR2J |
| Gene Symbol | Log2FC | p-Value |
|---|---|---|
| FDFT1 | −0.31606 | 0.00267 |
| SQLE | −0.2705 | 0.001057 |
| NSDHL | −0.2293 | 0.016672 |
| MSMO1 | −0.26965 | 0.012593 |
| SC5D | −0.27862 | 0.002411 |
| SREBF-1 | −0.16536 | 0.02396 |
| LDLR | −0.37172 | 0.001701 |
| Upstream Regulator | Molecule Type | Predicted State | p-Value |
|---|---|---|---|
| SREBF2 | Transcription regulator | Inhibited | 0.000339911 |
| SIRT2 | Transcription regulator | Inhibited | 0.027754644 |
| SREBF1 | Transcription regulator | Inhibited | 0.0015 |
| CYP51A1 | Enzyme | Activated | 6.15 × 10−5 |
| IL1B | Cytokine | Inhibited | 1.15 × 10−5 |
| POR | Enzyme | Activated | 9.32 × 10−5 |
| TLR9 | Transmembrane receptor | Inhibited | 0.00105 |
| IL5 | Cytokine | Inhibited | 0.00315 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iannaccone, M.; Elgendy, R.; Giantin, M.; Martino, C.; Giansante, D.; Ianni, A.; Dacasto, M.; Martino, G. RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet. Animals 2018, 8, 188. https://doi.org/10.3390/ani8110188
Iannaccone M, Elgendy R, Giantin M, Martino C, Giansante D, Ianni A, Dacasto M, Martino G. RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet. Animals. 2018; 8(11):188. https://doi.org/10.3390/ani8110188
Chicago/Turabian StyleIannaccone, Marco, Ramy Elgendy, Mery Giantin, Camillo Martino, Daniele Giansante, Andrea Ianni, Mauro Dacasto, and Giuseppe Martino. 2018. "RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet" Animals 8, no. 11: 188. https://doi.org/10.3390/ani8110188
APA StyleIannaccone, M., Elgendy, R., Giantin, M., Martino, C., Giansante, D., Ianni, A., Dacasto, M., & Martino, G. (2018). RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet. Animals, 8(11), 188. https://doi.org/10.3390/ani8110188





