Genetic Diversity Estimation and Genome-Wide Selective Sweep Analysis of the Bazhou Yak
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. SNP Calling and Annotation
2.3. Genetic Diversity Estimation and Phylogenetic Analysis
2.4. Selective Sweep Analysis
2.5. Gene Function Annotation
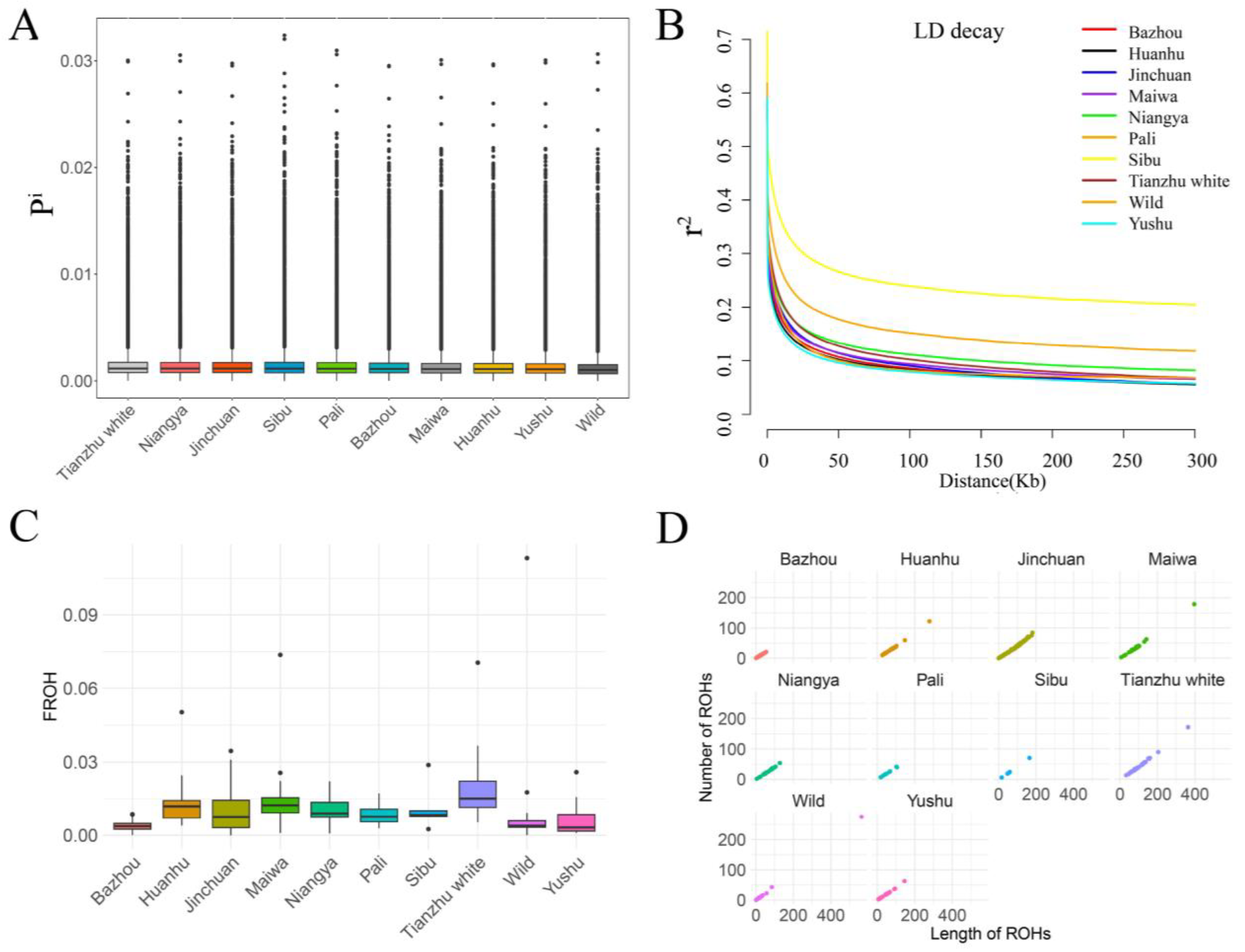
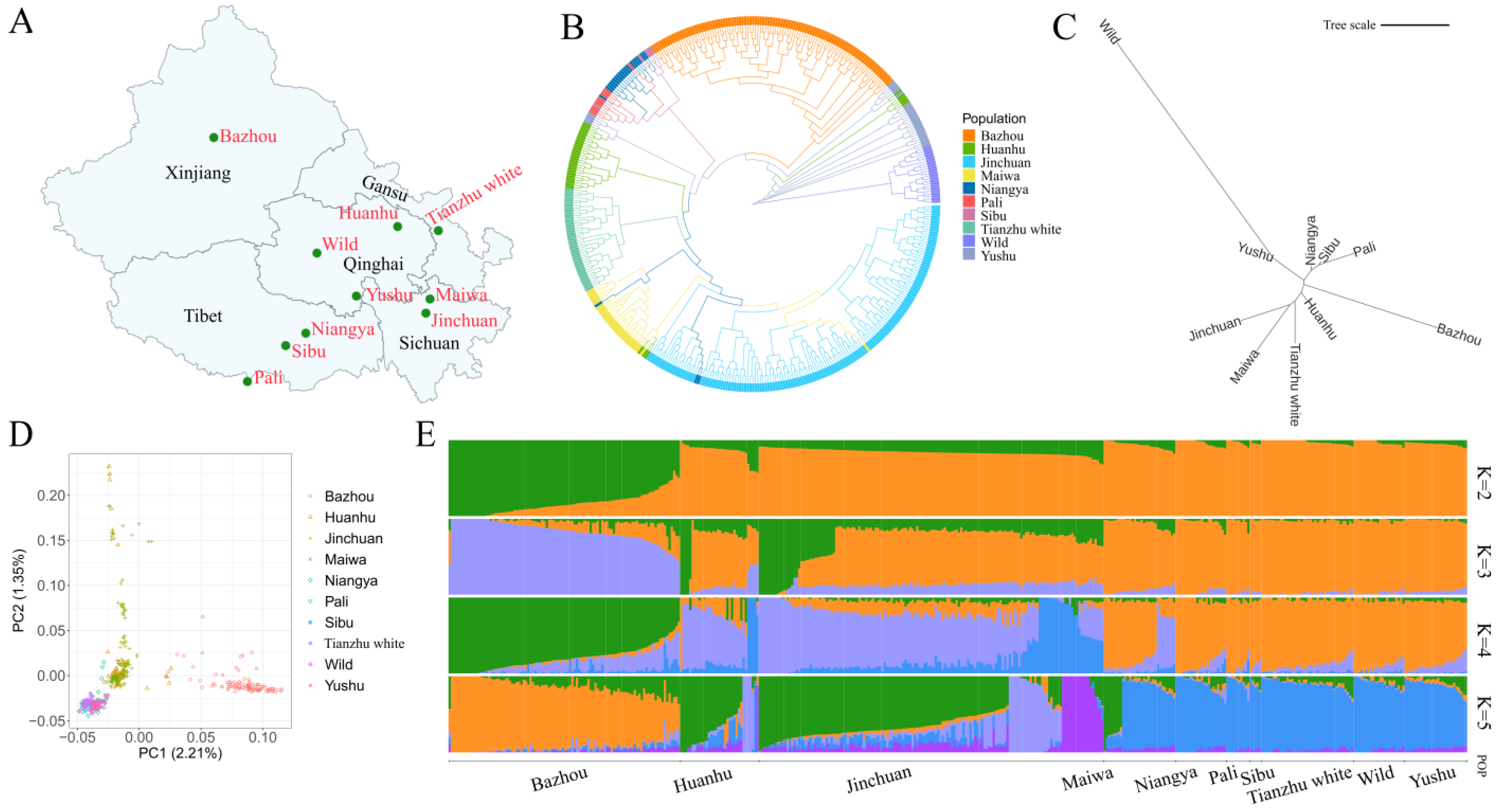
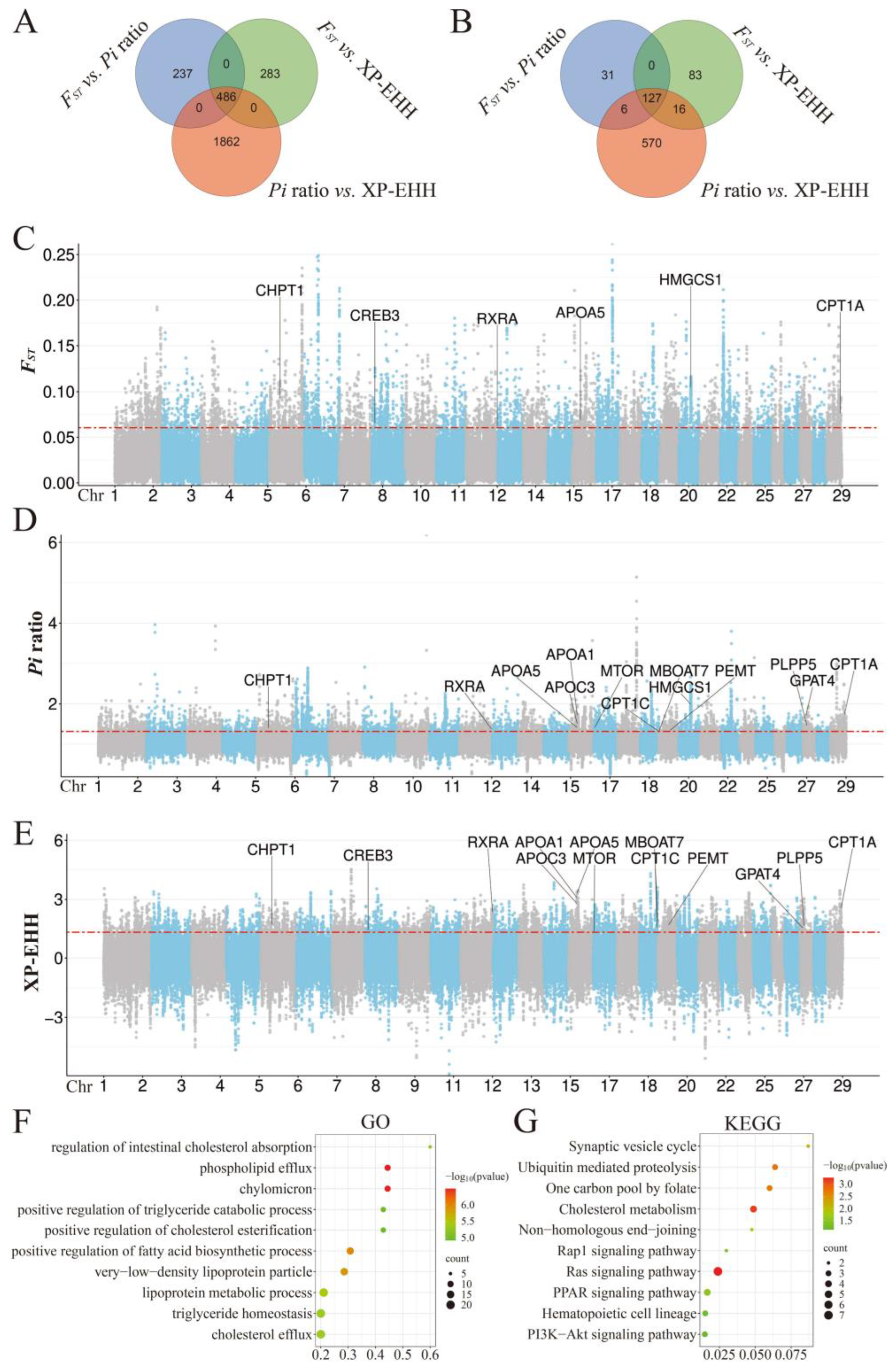
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDG | Candidate genes |
| CNV | Copy number variation |
| DAG | Diacylglycero |
| FAO | Fatty acid oxidation |
| FST | The fixation index |
| GO | Gene Ontology |
| GORs | Genomic overlapping regions |
| GWSA | Genome-wide selective sweep analysis |
| He | The expected heterozygosity |
| Ho | The observed heterozygosity |
| IMF | Intramuscular fat |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LD | Linkage disequilibrium |
| ML | Maximum likelihood |
| NJ | Neighbor-joining |
| PC | Phosphatidylcholine |
| PCA | Principal component analysis |
| Pi | The nucleotide diversity |
| PI3K | Phosphoinositide 3-kinase |
| ROH | The runs of homozygosity |
| XP-EHH | The cross-population extended haplotype homozygosity |
References
- Han, X.; Liang, D. Economic Analysis on Yak Industry in China. Food Nutr. China 2024, 30, 17–20. [Google Scholar] [CrossRef]
- Yan, X.; Ma, Z.; Geng, J.; Liu, J.; Yang, G.; Gao, L.; Haung, X. Current Situation, Existing Problems and Countermeasures of High-quality Development of injiang Beef Cattle Industry. Chin. Livest. Poult. Breed. 2024, 20, 142–148. (In Chinese) [Google Scholar]
- Li, H.; Zhang, J.; Yan, X.; Wang, Z.; Guan, Y.; Zhang, Y. Report on the Yak Industry in Bayingolin Mongol Autonomous Prefecture, Xinjiang. China Cattle Sci. 2017, 43, 65–68. (In Chinese) [Google Scholar]
- Gala; Guangtong, M.; Boyuan, Y.; Qinguo, X.; Yimin, L.; Xueguang, M. The Bazhou Yak in Xinjiang. Chin. Yak 1983, 02, 46–50. (In Chinese) [Google Scholar]
- Zhang, Q.; Hao, L.; Liu, S.; Chaishatuo; Niu, J.; Zhang, X.; Wang, X.; Sun, L.; Zhang, C.; Li, J. Comparison of nutritional components of adult yak meat from different regions. Sci. Technol. Food Ind. 2018, 39, 302–307+317. (In Chinese) [Google Scholar] [CrossRef]
- Hou, L.; Chai, S.; Liu, S.; Cui, Z.; Zhang, X.; Zhao, Y. Comparative Studies on Beef Amino Acid Composition and Fatty Acid Composition of Qinghai Yak and Qinchuan Cattle. Meat Res. 2013, 27, 30–36. (In Chinese) [Google Scholar]
- Zhao, H.; Xie, R.; An, T.; Li, H.; An, D.; Luo, X. Analysis of the meat quality of Jinchuan yak. Heilongjiang Anim. Sci. Vet. 2018, 19, 197–200. (In Chinese) [Google Scholar]
- Qiu, X.; Zhang, L.; Wen, Y.; Wang, J.; Liu, L.; Ma, L.; Wu, X.; Jin, J. Nutritional Composition Analysis of Meat from Yak and Yellow Cattle in Sichuan. Food Sci. 2010, 31, 112–116. [Google Scholar] [CrossRef]
- Ji, Q.; Pu, Q.; Dawa, Y.; Ciren, D.; Dawa, Q.; Zhang, Y.; Luo, S. Analysis on meat production performance and meat quality of three superior groups of yaks in Tibet. China Herbiv. Sci. 2000, 05, 3–6. (In Chinese) [Google Scholar]
- Wang, J.; Huang, H.; Tong, W. Characteristic analysis of physical and chemical indexes of white yak meat in Tianzhu County. Gansu Anim. Vet. Sci. 2019, 49, 62–64. (In Chinese) [Google Scholar] [CrossRef]
- Liu, H. Study on the meat quality characteristics of Qinghai yak and Tibetan sheep. Master’s Thesis, Gansu Agricultural University, Lanzhou, China, 2005. (In Chinese). [Google Scholar]
- Nguyen, D.V.; Nguyen, O.C.; Malau-Aduli, A.E. Main regulatory factors of marbling level in beef cattle. Vet. Anim. Sci. 2021, 14, 100219. [Google Scholar] [CrossRef] [PubMed]
- O’Quinn, T.G.; Brooks, J.C.; Polkinghorne, R.J.; Garmyn, A.J.; Johnson, B.J.; Starkey, J.D.; Rathmann, R.J.; Miller, M.F. Consumer assessment of beef strip loin steaks of varying fat levels. J. Anim. Sci. 2012, 90, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Qiu, Q.; Wang, L.; Wang, K.; Yang, Y.; Ma, T.; Wang, Z.; Zhang, X.; Ni, Z.; Hou, F.; Long, R. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 2015, 6, 10283. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, K.; Wang, L.; Yang, Y.; Ni, Z.; Xie, X.; Shao, X.; Han, J.; Wan, D.; Qiu, Q. Genome-wide patterns of copy number variation in the Chinese yak genome. BMC Genom. 2016, 17, 379. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chai, Z.; Hu, D.; Ji, Q.; Xin, J.; Zhang, C.; Zhong, J. A global analysis of CNVs in diverse yak populations using whole-genome resequencing. BMC Genom. 2019, 20, 61. [Google Scholar] [CrossRef]
- Lan, D.; Ji, W.; Xiong, X.; Liang, Q.; Yao, W.; Mipam, T.D.; Zhong, J.; Li, J. Population genome of the newly discovered Jinchuan yak to understand its adaptive evolution in extreme environments and generation mechanism of the multirib trait. Integr. Zool. 2021, 16, 685–695. [Google Scholar] [CrossRef]
- Lan, D.; Xiong, X.; Mipam, T.D.; Fu, C.; Li, Q.; Ai, Y.; Hou, D.; Chai, Z.; Zhong, J.; Li, J. Genetic Diversity, Molecular Phylogeny, and Selection Evidence of Jinchuan Yak Revealed by Whole-Genome Resequencing. G3 Genes Genomes Genet. 2018, 8, 945–952. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Lenstra, J.A.; Zheng, Z.; Wu, X.; Yang, J.; Li, B.; Yang, Y.; Qiu, Q.; Liu, H.; et al. Evolutionary origin of genomic structural variations in domestic yaks. Nat. Commun. 2023, 14, 5617. [Google Scholar] [CrossRef]
- Wu, X.; Xu, L.; Zhang, H.; Zhu, Y.; Zhang, Q.; Zhang, C.E.G. Genome-Wide Selection Sweep Analysis to Identify Candidate Genes with Black and Brown Color in Tibetan Sibu Yaks. Animals 2024, 14, 2458. [Google Scholar] [CrossRef]
- Peng, W.; Fu, C.; Shu, S.; Wang, G.; Wang, H.; Yue, B.; Zhang, M.; Liu, X.; Liu, Y.; Zhang, J.; et al. Whole-genome resequencing of major populations revealed domestication-related genes in yaks. BMC Genom. 2024, 25, 69. [Google Scholar] [CrossRef] [PubMed]
- Gangwar, M.; Ahmad, S.F.; Ali, A.B.; Kumar, A.; Kumar, A.; Gaur, G.K.; Dutt, T. Identifying low-density, ancestry-informative SNP markers through whole genome resequencing in Indian, Chinese, and wild yak. BMC Genom. 2024, 25, 1043. [Google Scholar] [CrossRef]
- Xiong, L.; Pei, J.; Chu, M.; Wu, X.; Kalwar, Q.; Yan, P.; Guo, X. Fat deposition in the muscle of female and male yak and the correlation of yak meat quality with fat. Animals 2021, 11, 2142. [Google Scholar] [CrossRef]
- Xiong, L.; Pei, J.; Wang, X.; Guo, S.; Guo, X.; Yan, P. Lipidomics and Transcriptome Reveal the Effects of Feeding Systems on Fatty Acids in Yak’s Meat. Foods 2022, 11, 2582. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, H.; Zhang, J.; Yixi, K.; Shu, S.; Fu, C.; Zhong, J.; Peng, W. IMF deposition ceRNA network analysis and functional study of HIF1a in yak. Front. Vet. Sci. 2023, 10, 1272238. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Sun, Y.; Han, Y.; Liu, Y.; Jin, S. Transcriptome comparison revealed the difference in subcutaneous fat metabolism of Qinghai yak under different feeding conditions. PLoS ONE 2024, 19, e0311224. [Google Scholar] [CrossRef]
- Xu, F.; Wang, H.; Qin, C.; Yue, B.; Yang, Y.; Wang, J.; Zhong, J.; Wang, H. Combined Multi-Omics Analysis Reveals the Potential Role of ACADS in Yak Intramuscular Fat Deposition. Int. J. Mol. Sci. 2024, 25, 9131. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; pp. 314–324. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Meyermans, R.; Gorssen, W.; Buys, N.; Janssens, S. How to study runs of homozygosity using PLINK? A guide for analyzing medium density SNP data in livestock and pet species. BMC Genom. 2020, 21, 94. [Google Scholar] [CrossRef]
- Wilkinson, L. ggplot2: Elegant graphics for data analysis by WICKHAM, H. Biometrics 2011, 67, 678–679. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Francis, R.M. Pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Xu, S.; Li, L.; Luo, X.; Chen, M.; Tang, W.; Zhan, L.; Dai, Z.; Lam, T.T.; Guan, Y.; Yu, G. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. Imeta 2022, 1, e56. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Nei, M.; Chesser, R.K. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 1983, 47, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zheng, Y.; Li, S.; Niu, X.; Huang, S.; Long, Q.; Ran, X.; Wang, J. Identification of genomic characteristics and selective signals in Guizhou black goat. BMC Genom. 2024, 25, 164. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- Li, G.; Luo, J.; Wang, F.; Xu, D.; Ahmed, Z.; Chen, S.; Li, R.; Ma, Z. Whole-genome resequencing reveals genetic diversity, differentiation, and selection signatures of yak breeds/populations in Qinghai, China. Front. Genet. 2022, 13, 1034094. [Google Scholar] [CrossRef]
- Browning, B.L.; Tian, X.; Zhou, Y.; Browning, S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef] [PubMed]
- Szpiech, Z.A. selscan 2.0: Scanning for sweeps in unphased data. Bioinformatics 2024, 40, btae006. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.m.; Xin, J.w.; Chai, Z.x.; Zhang, C.f.; Dawa, Y.; Luo, S.; Zhang, Q.; Pingcuo, Z.; Peng, M.S.; Zhu, Y. A chromosome-scale reference genome and genome-wide genetic variations elucidate adaptation in yak. Mol. Ecol. Resour. 2021, 21, 201–211. [Google Scholar] [CrossRef]
- Hausman, G.; Dodson, M.; Ajuwon, K.; Azain, M.; Barnes, K.; Guan, L.; Jiang, Z.; Poulos, S.; Sainz, R.; Smith, S. Board-invited review: The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef]
- Smith, U.; Kahn, B.B. Adipose tissue regulates insulin sensitivity: Role of adipogenesis, de novo lipogenesis and novel lipids. J. Intern. Med. 2016, 280, 465–475. [Google Scholar] [CrossRef] [PubMed]
- de Souza, D.K.; Salles, L.P.; Camargo, R.; Gulart, L.V.M.; Costa, E.S.S.; de Lima, B.D.; Torres, F.A.G.; Rosa, E.S.A.A.M. Effects of PI3K and FSH on steroidogenesis, viability and embryo development of the cumulus-oocyte complex after in vitro culture. Zygote 2018, 26, 50–61. [Google Scholar] [CrossRef]
- Savova, M.S.; Mihaylova, L.V.; Tews, D.; Wabitsch, M.; Georgiev, M.I. Targeting PI3K/AKT signaling pathway in obesity. Biomed. Pharmacother. 2023, 159, 114244. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, J.; Zhang, C.; Chai, Z.; Cao, H.; Wang, J.; Zhu, J.; Wang, J.; Ji, Q. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genom. 2020, 21, 347. [Google Scholar] [CrossRef]
- Qin, C.; Wang, H.; Zhong, J.; Ran, H.; Peng, W. miR-129 Regulates Yak Intramuscular Preadipocyte Proliferation and Differentiation through the PI3K/AKT Pathway. Int. J. Mol. Sci. 2024, 25, 632. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Richard, D.; Laplante, M. The roles of mTOR complexes in lipid metabolism. Annu. Rev. Nutr. 2015, 35, 321–348. [Google Scholar] [CrossRef]
- Soliman, G.A. The integral role of mTOR in lipid metabolism. Cell Cycle 2011, 10, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Hall, M.N. Regulation of mTORC2 signaling. Genes 2020, 11, 1045. [Google Scholar] [CrossRef]
- Jones, K.T.; Greer, E.R.; Pearce, D.; Ashrafi, K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009, 7, e1000060. [Google Scholar] [CrossRef]
- Guri, Y.; Colombi, M.; Dazert, E.; Hindupur, S.K.; Roszik, J.; Moes, S.; Jenoe, P.; Heim, M.H.; Riezman, I.; Riezman, H. mTORC2 promotes tumorigenesis via lipid synthesis. Cancer Cell 2017, 32, 807–823.e812. [Google Scholar] [CrossRef]
- Janani, C.; Kumari, B.R. PPAR gamma gene–a review. Diabetes Metab. Syndr. Clin. Res. Rev. 2015, 9, 46–50. [Google Scholar] [CrossRef]
- Hamza, M.S.; Pott, S.; Vega, V.B.; Thomsen, J.S.; Kandhadayar, G.S.; Ng, P.W.P.; Chiu, K.P.; Pettersson, S.; Wei, C.L.; Ruan, Y. De-novo identification of PPARγ/RXR binding sites and direct targets during adipogenesis. PLoS ONE 2009, 4, e4907. [Google Scholar] [CrossRef] [PubMed]
- Resnyk, C.W.; Carré, W.; Wang, X.; Porter, T.E.; Simon, J.; Le Bihan-Duval, E.; Duclos, M.J.; Aggrey, S.E.; Cogburn, L.A. Transcriptional analysis of abdominal fat in genetically fat and lean chickens reveals adipokines, lipogenic genes and a link between hemostasis and leanness. BMC Genom. 2013, 14, 557. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Li, X.; Wu, D.; Chen, X.; Zhang, C.; Jin, S.; Geng, Z. The Duck RXRA Gene Promotes Adipogenesis and Correlates with Feed Efficiency. Animals 2023, 13, 680. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zhao, J.; Yang, X.; Wu, S.; An, Y.; Qu, Y.; Li, Z.; Ge, H.; Li, E.; Qi, W. TET1 promotes RXRα expression and adipogenesis through DNA demethylation. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158919. [Google Scholar] [CrossRef]
- Ma, X.; Bai, Y.; Liu, K.; Han, Y.; Zhang, J.; Liu, Y.; Hou, X.; Hao, E.; Hou, Y.; Bai, G. Ursolic acid inhibits the cholesterol biosynthesis and alleviates high fat diet-induced hypercholesterolemia via irreversible inhibition of HMGCS1 in vivo. Phytomedicine 2022, 103, 154233. [Google Scholar] [CrossRef]
- Dorado, M.; Gómez, E.M.n.; Jiménez-Colmenero, F.; Masoud, T. Cholesterol and fat contents of Spanish commercial pork cuts. Meat Sci. 1999, 51, 321–323. [Google Scholar] [CrossRef]
- Gu, D.; Ye, M.; Zhu, G.; Bai, J.; Chen, J.; Yan, L.; Yu, P.; Lu, F.; Hu, C.; Zhong, Y. Hypoxia upregulating ACSS2 enhances lipid metabolism reprogramming through HMGCS1 mediated PI3K/AKT/mTOR pathway to promote the progression of pancreatic neuroendocrine neoplasms. J. Transl. Med. 2024, 22, 93. [Google Scholar] [CrossRef]
- Cochran, B.J.; Ong, K.-L.; Manandhar, B.; Rye, K.-A. APOA1: A protein with multiple therapeutic functions. Curr. Atheroscler. Rep. 2021, 23, 11. [Google Scholar] [CrossRef]
- Guardiola, M.; Ribalta, J. Update on APOA5 genetics: Toward a better understanding of its physiological impact. Curr. Atheroscler. Rep. 2017, 19, 30. [Google Scholar] [CrossRef]
- Giammanco, A.; Spina, R.; Cefalù, A.B.; Averna, M. APOC-III: A gatekeeper in controlling triglyceride metabolism. Curr. Atheroscler. Rep. 2023, 25, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Duivenvoorden, I.; Teusink, B.; Rensen, P.C.; Romijn, J.A.; Havekes, L.M.; Voshol, P.J. Apolipoprotein C3 deficiency results in diet-induced obesity and aggravated insulin resistance in mice. Diabetes 2005, 54, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Nye, C.; Kim, J.; Kalhan, S.C.; Hanson, R.W. Reassessing triglyceride synthesis in adipose tissue. Trends Endocrinol. Metab. 2008, 19, 356–361. [Google Scholar] [CrossRef]
- Cooper, D.E.; Grevengoed, T.J.; Klett, E.L.; Coleman, R.A. Glycerol-3-phosphate acyltransferase isoform-4 (GPAT4) limits oxidation of exogenous fatty acids in brown adipocytes. J. Biol. Chem. 2015, 290, 15112–15120. [Google Scholar] [CrossRef]
- Yamashita, A.; Kawagishi, N.; Miyashita, T.; Nagatsuka, T.; Sugiura, T.; Kume, K.; Shimizu, T.; Waku, K. ATP-independent fatty acyl-coenzyme A synthesis from phospholipid: Coenzyme A-dependent transacylation activity toward lysophosphatidic acid catalyzed by acyl-coenzyme A: Lysophosphatidic acid acyltransferase. J. Biol. Chem. 2001, 276, 26745–26752. [Google Scholar] [CrossRef]
- Tang, X.; Brindley, D.N. Lipid phosphate phosphatases and cancer. Biomolecules 2020, 10, 1263. [Google Scholar] [CrossRef]
- Nagle, C.A.; Vergnes, L.; DeJong, H.; Wang, S.; Lewin, T.M.; Reue, K.; Coleman, R.A. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 2008, 49, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Dorighello, G.; McPhee, M.; Halliday, K.; Dellaire, G.; Ridgway, N.D. Differential contributions of phosphotransferases CEPT1 and CHPT1 to phosphatidylcholine homeostasis and lipid droplet biogenesis. J. Biol. Chem. 2023, 299, 104578. [Google Scholar] [CrossRef]
- Tso, P.; Scobey, M. The role of phosphatidylcholine in the absorption and transport of dietary fat. Fat Absorpt. 2018, 1, 177–196. [Google Scholar] [CrossRef]
- Gong, X.; Zheng, M.; Zhang, J.; Ye, Y.; Duan, M.; Chamba, Y.; Wang, Z.; Shang, P. Transcriptomics-based study of differentially expressed genes related to fat deposition in Tibetan and Yorkshire pigs. Front. Vet. Sci. 2022, 9, 919904. [Google Scholar] [CrossRef]
- Vance, D.E. Physiological roles of phosphatidylethanolamine N-methyltransferase. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2013, 1831, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Presa, N.; Dominguez-Herrera, A.; van der Veen, J.N.; Vance, D.E.; Gómez-Muñoz, A. Implication of phosphatidylethanolamine N-methyltransferase in adipocyte differentiation. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165853. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Chandrasekaran, P.; Rong, S.; Fu, X.; Mitsche, M.A. Hepatic deletion of Mboat7 (LPIAT1) causes activation of SREBP-1c and fatty liver. J. Lipid Res. 2021, 62, 100031. [Google Scholar] [CrossRef] [PubMed]
- Caddeo, A.; Hedfalk, K.; Romeo, S.; Pingitore, P. LPIAT1/MBOAT7 contains a catalytic dyad transferring polyunsaturated fatty acids to lysophosphatidylinositol. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158891. [Google Scholar] [CrossRef]
- Massey, W.J.; Varadharajan, V.; Banerjee, R.; Brown, A.L.; Horak, A.J.; Hohe, R.C.; Jung, B.M.; Qiu, Y.; Chan, E.R.; Pan, C. MBOAT7-driven lysophosphatidylinositol acylation in adipocytes contributes to systemic glucose homeostasis. J. Lipid Res. 2023, 64, 100349. [Google Scholar] [CrossRef]
- Abu-Elheiga, L.; Matzuk, M.M.; Abo-Hashema, K.A.; Wakil, S.J. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 2001, 291, 2613–2616. [Google Scholar] [CrossRef]
- Locke, B.; Campbell, E.; Lu, R. CREB3 mediates the transcriptional regulation of PGC-1α, a master regulator of energy homeostasis and mitochondrial biogenesis. FEBS Lett. 2024, 598, 1730–1739. [Google Scholar] [CrossRef]
- Cheng, C.-F.; Ku, H.-C.; Lin, H. PGC-1α as a Pivotal Factor in Lipid and Metabolic Regulation. Int. J. Mol. Sci. 2018, 19, 3447. [Google Scholar] [CrossRef]
- Smith, B.S.; Diaguarachchige De Silva, K.H.; Hashemi, A.; Duncan, R.E.; Grapentine, S.; Bakovic, M.; Lu, R. Transcription factor CREB3 is a potent regulator of high-fat diet-induced obesity and energy metabolism. Int. J. Obes. 2022, 46, 1446–1455. [Google Scholar] [CrossRef]
- Schlaepfer, I.R.; Joshi, M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology 2020, 161, bqz046. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, W.; Wang, Y.; Li, H.; Zhang, C.; Wang, Y.; Lin, Y.; Shi, H.; Xiang, H.; Huang, L. Expression variation of CPT1A induces lipid reconstruction in goat intramuscular precursor adipocytes. Int. J. Mol. Sci. 2023, 24, 13415. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, W.; Kong, X.; Xu, A.; Wang, Z.; Sweeney, G.; Wu, D. Enhanced susceptibility of Cpt1c knockout mice to glucose intolerance induced by a high-fat diet involves elevated hepatic gluconeogenesis and decreased skeletal muscle glucose uptake. Diabetologia 2009, 52, 912–920. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Zhang, H.; Feng, X.; Yu, Z.; Cao, J.; Niu, Y.; Wan, P.; Liu, G.; Zhao, X. Genetic Diversity Estimation and Genome-Wide Selective Sweep Analysis of the Bazhou Yak. Animals 2025, 15, 849. https://doi.org/10.3390/ani15060849
Yang B, Zhang H, Feng X, Yu Z, Cao J, Niu Y, Wan P, Liu G, Zhao X. Genetic Diversity Estimation and Genome-Wide Selective Sweep Analysis of the Bazhou Yak. Animals. 2025; 15(6):849. https://doi.org/10.3390/ani15060849
Chicago/Turabian StyleYang, Baigao, Hang Zhang, Xiaoyi Feng, Zhou Yu, Jianhua Cao, Yifan Niu, Pengcheng Wan, Gang Liu, and Xueming Zhao. 2025. "Genetic Diversity Estimation and Genome-Wide Selective Sweep Analysis of the Bazhou Yak" Animals 15, no. 6: 849. https://doi.org/10.3390/ani15060849
APA StyleYang, B., Zhang, H., Feng, X., Yu, Z., Cao, J., Niu, Y., Wan, P., Liu, G., & Zhao, X. (2025). Genetic Diversity Estimation and Genome-Wide Selective Sweep Analysis of the Bazhou Yak. Animals, 15(6), 849. https://doi.org/10.3390/ani15060849




