Single-Cell RNA Sequencing Reveals an Atlas of Meihua Pig Testis Cells
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Testicular Sample Collection
2.2. Histological Observation
2.3. Preparation of Single-Cell Suspension
2.4. Single-Cell Sequencing
2.5. Quality Control, Mapping and Clustering Analysis
2.6. Cell Trajectory Analysis
2.7. Differentially Expressed Gene (DEG) and Functional Enrichment Analysis
2.8. Immunohistochemistry and Immunofluorescence Analysis
3. Results
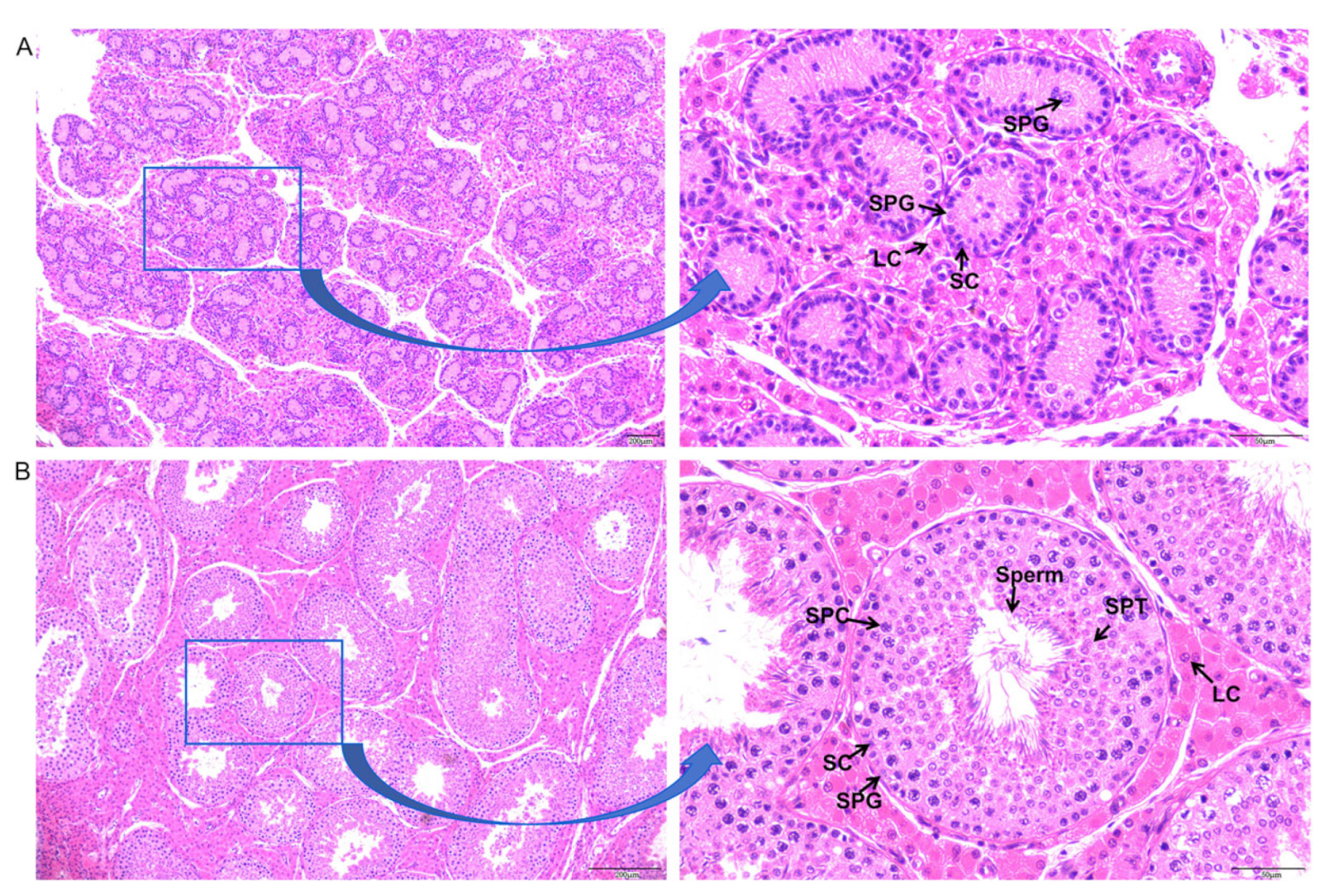
3.1. Histological Observation of Testicular Cell Types in Neonatal and Sexually Mature Meihua Pigs
3.2. Subset Analysis and Cell Type Identification of Testicular Cells in Meihua Pigs
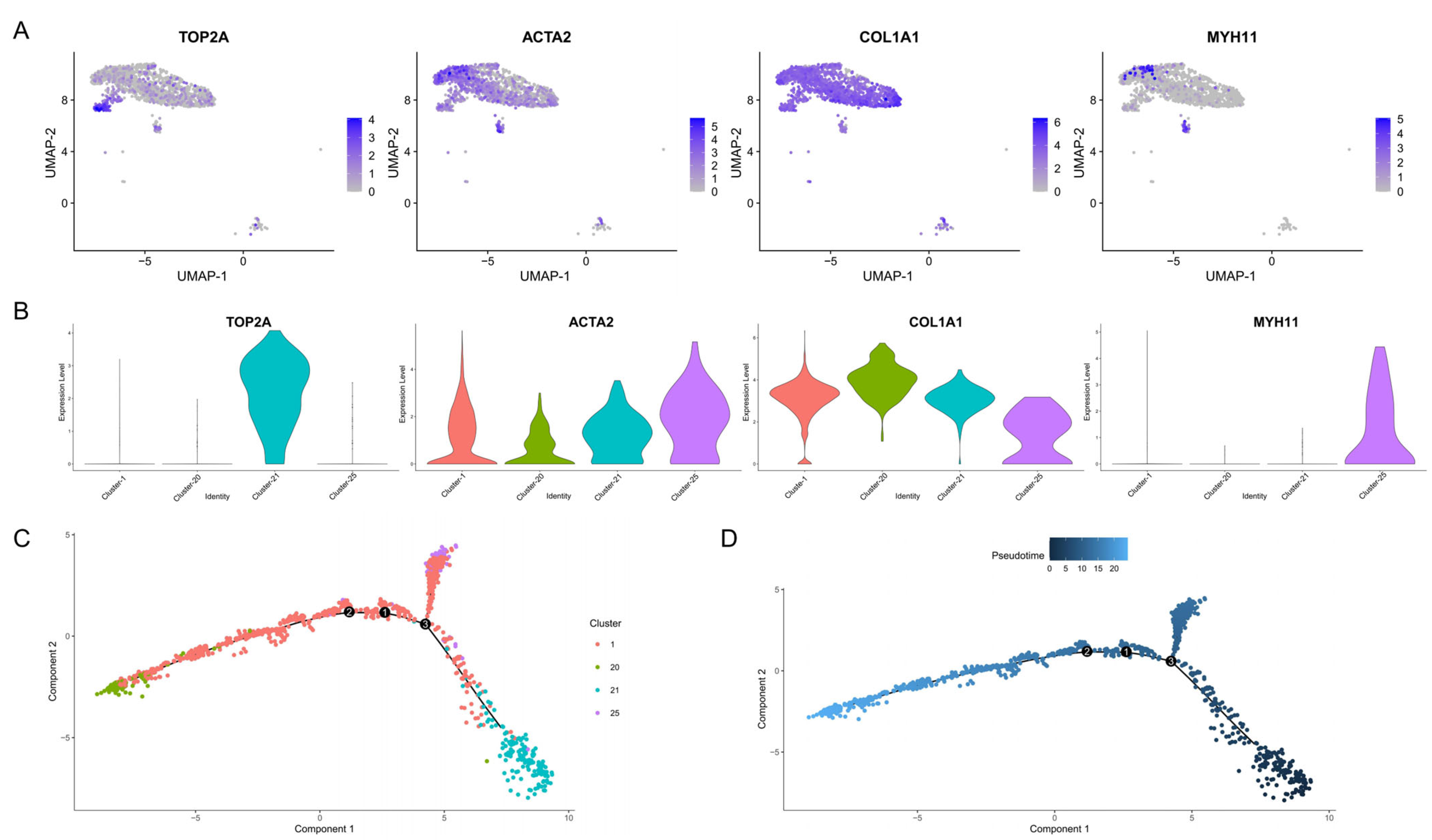
3.3. Cell Trajectory Analysis
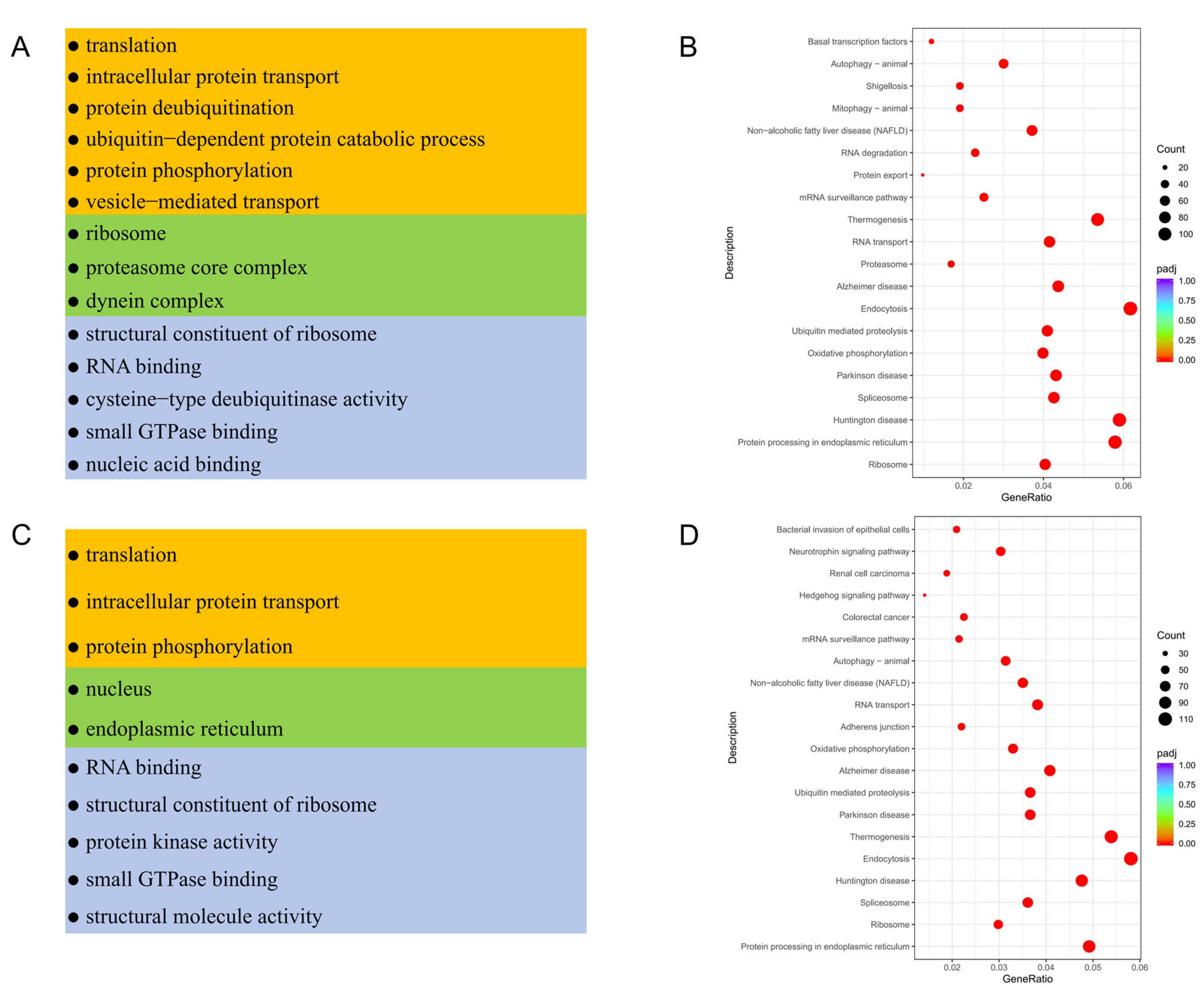
3.4. Functional Enrichment Analysis of Testicular Somatic Cells in Meihua Pigs
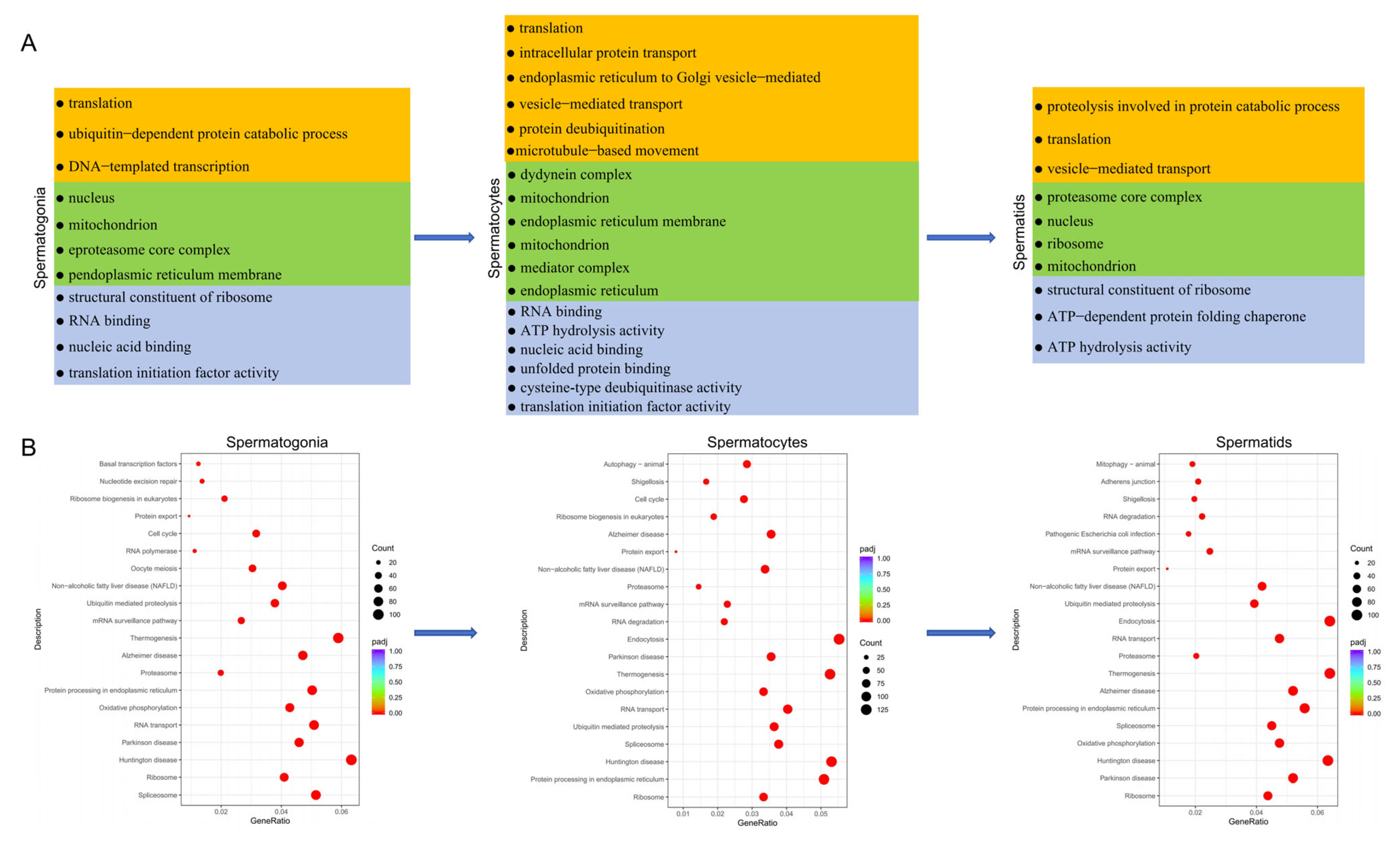
3.5. Functional Enrichment Analysis of Testicular Germ Cells
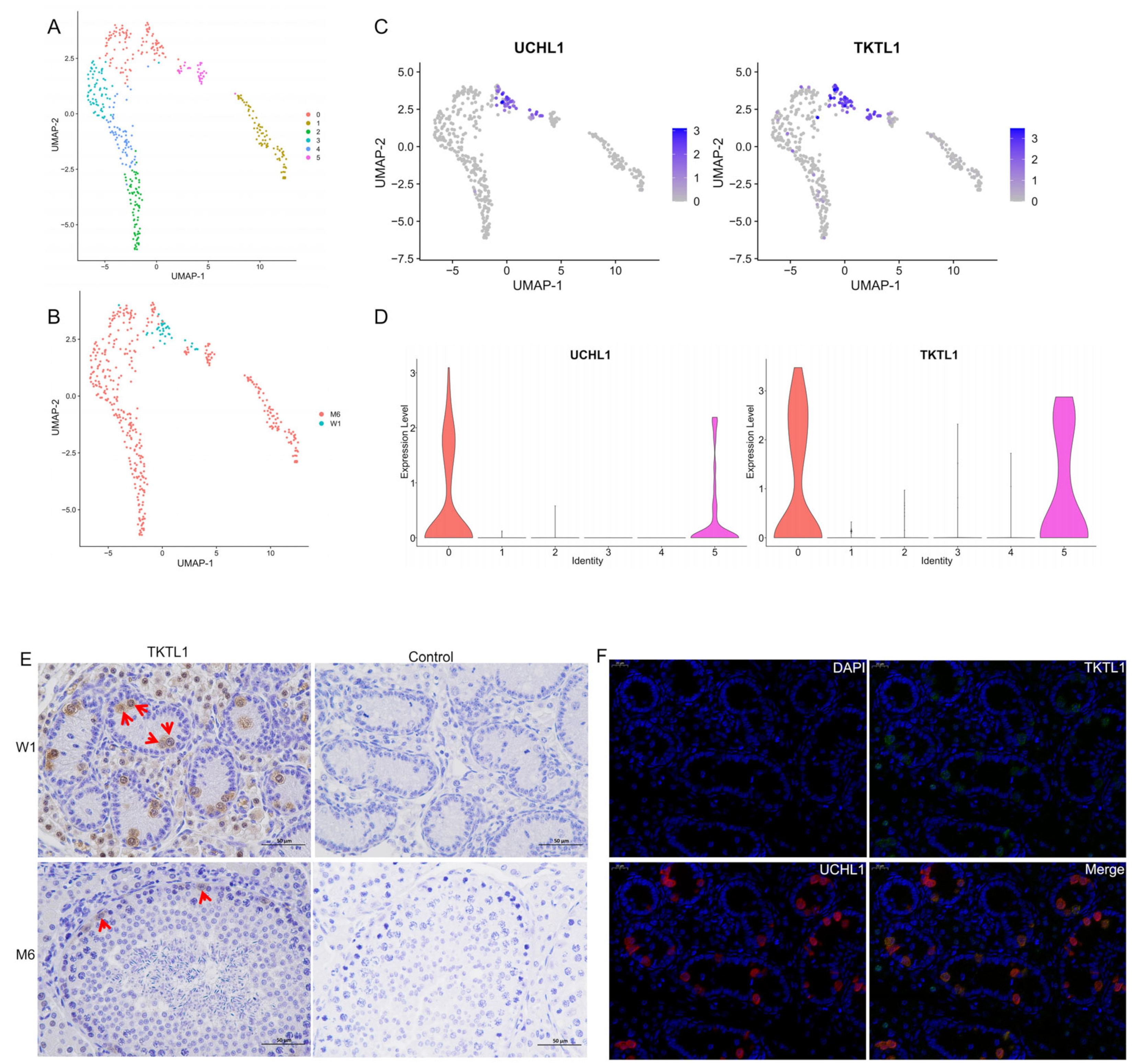
3.6. Re-Clustering of Spermatogonia and Screening of New Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, H.; Luo, Y.; Liu, M.; Huang, J.; Xu, D. Histological and transcriptome analyses of testes from duroc and meishan boars. Sci. Rep. 2016, 6, 20758. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lin, W.; Wang, Z.; Huang, R.; Xia, H.; Li, Z.; Deng, J.; Ye, T.; Huang, Y.; Yang, Y. Hormone regulation in testicular development and function. Int. J. Mol. Sci. 2024, 25, 5805. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, Y.; Qu, R.; He, Y.; Tian, X.; Zeng, W. Spermatogonial stem cells from domestic animals: Progress and prospects. Reproduction 2014, 147, R65–R74. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, D.G. The nature and dynamics of spermatogonial stem cells. Development 2017, 144, 3022–3030. [Google Scholar] [CrossRef]
- Kubota, H.; Brinster, R.L. Spermatogonial stem cells. Biol. Reprod. 2018, 99, 52–74. [Google Scholar] [CrossRef]
- Griswold, M.D. Spermatogenesis: The commitment to meiosis. Physiol. Rev. 2016, 96, 1–17. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Y. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by sertoli cell signaling. Reproduction 2015, 149, R159–R167. [Google Scholar] [CrossRef]
- Bhang, D.H.; Kim, B.; Kim, B.G.; Schadler, K.; Baek, K.; Kim, Y.H.; Hsiao, W.; Ding, B.; Rafii, S.; Weiss, M.J.; et al. Testicular endothelial cells are a critical population in the germline stem cell niche. Nat. Commun. 2018, 9, 4379. [Google Scholar] [CrossRef]
- Zheng, Y.; Gao, Q.; Li, T.; Liu, R.; Cheng, Z.; Guo, M.; Xiao, J.; Wu, D.; Zeng, W. Sertoli cell and spermatogonial development in pigs. J. Anim. Sci. Biotechnol. 2022, 13, 45. [Google Scholar] [CrossRef]
- Neto, F.T.L.; Flannigan, R.; Goldstein, M. Regulation of human spermatogenesis. Adv. Exp. Med. Biol. 2021, 1381, 255–286. [Google Scholar]
- Yang, H.; Ma, J.; Wan, Z.; Wang, Q.; Wang, Z.; Zhao, J.; Wang, F.; Zhang, Y. Characterization of sheep spermatogenesis through single-cell rna sequencing. FASEB J. 2021, 35, e21187. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Grow, E.J.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Nie, X.; Guo, Y.; Takei, Y.; Yun, J.; Cai, L.; et al. The adult human testis transcriptional cell atlas. Cell Res. 2018, 28, 1141–1157. [Google Scholar] [CrossRef]
- Tan, K.; Song, H.; Wilkinson, M.F. Single-cell rnaseq analysis of testicular germ and somatic cell development during the perinatal period. Development 2020, 147, dev183251. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pei, J.; Xiong, L.; Guo, S.; Cao, M.; Kang, Y.; Ding, Z.; La, Y.; Liang, C.; Yan, P.; et al. Single-cell rna sequencing reveals atlas of yak testis cells. Int. J. Mol. Sci. 2023, 24, 7982. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Diaz, V.D.; Hermann, B.P. What has single-cell rna-seq taught us about mammalian spermatogenesis? Biol. Reprod. 2019, 101, 617–634. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-cell rna-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef]
- Tan, H.; Wang, W.; Zhou, C.; Wang, Y.; Zhang, S.; Yang, P.; Guo, R.; Chen, W.; Zhang, J.; Ye, L.; et al. Single-cell rna-seq uncovers dynamic processes orchestrated by rna-binding protein ddx43 in chromatin remodeling during spermiogenesis. Nat. Commun. 2023, 14, 2499. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, M.; Liu, Z.; Liu, R.; Zheng, Y.; Yu, T.; Lv, Y.; Lu, H.; Zeng, W.; Zhang, T.; et al. Single-cell rna-seq analysis of testicular somatic cell development in pigs. J. Genet. Genomics 2022, 49, 1016–1028. [Google Scholar] [CrossRef]
- Wang, Z.; Jin, C.; Li, P.; Li, Y.; Tang, J.; Yu, Z.; Jiao, T.; Ou, J.; Wang, H.; Zou, D.; et al. Identification of quiescent foxc2(+) spermatogonial stem cells in adult mammals. Elife 2023, 12, RP85380. [Google Scholar] [CrossRef]
- Li, S.; Yan, R.; Gao, X.; He, Z.; Wu, S.; Wang, Y.; Zhang, Y.; Tao, H.; Zhang, X.; Jia, G.; et al. Single-cell transcriptome analyses reveal critical regulators of spermatogonial stem cell fate transitions. BMC Genomics 2024, 25, 138. [Google Scholar] [CrossRef]
- Guo, J.; Nie, X.; Giebler, M.; Mlcochova, H.; Wang, Y.; Grow, E.J.; Kim, R.; Tharmalingam, M.; Matilionyte, G.; Lindskog, C.; et al. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell 2020, 26, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell rna-seq. Dev. Cell 2018, 46, 651–667. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Shen, X.; Luo, C.; Sun, F. Comparative analysis of the testes from wild-type andalkbh5 -knockout mice using single-cell rna sequencing. G3 Genes 2022, 12, jkac130. [Google Scholar]
- Grive, K.J.; Hu, Y.; Shu, E.; Grimson, A.; Elemento, O.; Grenier, J.K.; Cohen, P.E. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet. 2019, 15, e1007810. [Google Scholar] [CrossRef]
- Shami, A.N.; Zheng, X.; Munyoki, S.K.; Ma, Q.; Manske, G.L.; Green, C.D.; Sukhwani, M.; Orwig, K.E.; Li, J.Z.; Hammoud, S.S. Single-cell rna sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis. Dev. Cell 2020, 54, 529–547. [Google Scholar] [CrossRef]
- Lau, X.; Munusamy, P.; Ng, M.J.; Sangrithi, M. Single-cell rna sequencing of the cynomolgus macaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis. Dev. Cell 2020, 54, 548–566. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, P.; Liu, W.X.; Shan, L.Y.; Hu, Y.T.; Fan, H.T.; Shen, W.; Liu, Y.B.; Zhou, Y.; Zhang, T. Single-cell rna sequencing of the mongolia sheep testis reveals a conserved and divergent transcriptome landscape of mammalian spermatogenesis. FASEB J. 2022, 36, e22348. [Google Scholar] [CrossRef]
- Ren, F.; Xi, H.; Qiao, P.; Li, Y.; Xian, M.; Zhu, D.; Hu, J. Single-cell transcriptomics reveals male germ cells and sertoli cells developmental patterns in dairy goats. Front. Cell Dev. Biol. 2022, 10, 944325. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, J.; Zhang, P.; Huang, X.; Yang, W.; Liu, R.; Sun, Q.; Lu, Y.; Zhang, M.; Fu, Q. Single-cell rna sequencing uncovers dynamic roadmap and cell-cell communication during buffalo spermatogenesis. iScience 2023, 26, 105733. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, S.; Wang, G.; Wan, R.; Yang, Q. Single-cell analysis identifies critical regulators of spermatogonial development and differentiation in cattle-yak bulls. J. Dairy. Sci. 2024, 107, 7317–7336. [Google Scholar] [CrossRef]
- Zhang, L.; Li, F.; Lei, P.; Guo, M.; Liu, R.; Wang, L.; Yu, T.; Lv, Y.; Zhang, T.; Zeng, W.; et al. Single-cell rna-sequencing reveals the dynamic process and novel markers in porcine spermatogenesis. J. Anim. Sci. Biotechnol. 2021, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, C.; Yan, S.; Yang, A.; Deng, Y.; Chen, B.; Gu, J. Single-nucleus rna-seq reveals spermatogonial stem cell developmental pattern in shaziling pigs. Biomolecules 2024, 14, 607. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, P.; Yang, Q.; Gun, S. Single-cell rna sequencing reveals an atlas of hezuo pig testis cells. Int. J. Mol. Sci. 2024, 25, 9786. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, M.; Li, X.; Zhang, L.; Zhao, B.; Wang, N.; Dugarjaviin, M. Single-cell transcriptome sequencing reveals molecular expression differences and marker genes in testes during the sexual maturation of mongolian horses. Animals 2024, 14, 1258. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Feng, T.; Zhang, P.; Lei, P.; Li, F.; Zeng, W. Establishment of cell lines with porcine spermatogonial stem cell properties. J. Anim. Sci. Biotechnol. 2020, 11, 33. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Wang, Y.; Guo, Y.; Zong, R.; Hu, S.; Yue, J.; Yao, J.; Han, C.; Guo, J.; et al. Single-cell transcriptomic and cross-species comparison analyses reveal distinct molecular changes of porcine testes during puberty. Commun. Biol. 2024, 7, 1478. [Google Scholar] [CrossRef]
- Parrish, J.J.; Willenburg, K.L.; Gibbs, K.M.; Yagoda, K.B.; Krautkramer, M.M.; Loether, T.M.; Melo, F.C.S.A. Scrotal insulation and sperm production in the boar. Mol. Reprod. Dev. 2017, 84, 969–978. [Google Scholar] [CrossRef]
- Gao, Y.; Bai, F.; Zhang, Q.; An, X.; Wang, Z.; Lei, C.; Dang, R. Dynamic transcriptome profiles and novel markers in bovine spermatogenesis revealed by single-cell sequencing. J. Integr. Agric. 2024, 23, 2362–2378. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, J.K. Dissecting cellular heterogeneity using single-cell rna sequencing. Mol. Cells 2019, 42, 189–199. [Google Scholar]
- Koskenniemi, J.J.; Virtanen, H.E.; Toppari, J. Testicular growth and development in puberty. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 215–224. [Google Scholar] [CrossRef]
- Mäkelä, J.; Koskenniemi, J.J.; Virtanen, H.E.; Toppari, J. Testis development. Endocr. Rev. 2019, 40, 857–905. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Hou, J.; Liu, Y.; Liu, Y.; Yang, H.; Li, Z.; He, Z. The roles and regulation of sertoli cells in fate determinations of spermatogonial stem cells and spermatogenesis. Semin. Cell Dev. Biol. 2014, 29, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Lindahl, M.; Hyvönen, M.E.; Parvinen, M.; de Rooij, D.G.; Hess, M.W.; Raatikainen-Ahokas, A.; Sainio, K.; Rauvala, H.; Lakso, M.; et al. Regulation of cell fate decision of undifferentiated spermatogonia by gdnf. Sci. (Am. Assoc. Adv. Sci.) 2000, 287, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, A.; DeFalco, T. Essential roles of interstitial cells in testicular development and function. Andrology 2020, 8, 903–914. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, P.; Dong, W.; Zeng, W.; Pan, C. Identification of stem leydig cells derived from pig testicular interstitium. Stem Cells Int. 2017, 2017, 2740272. [Google Scholar] [CrossRef]
- Potter, S.J.; DeFalco, T. Role of the testis interstitial compartment in spermatogonial stem cell function. Reproduction 2017, 153, R151–R162. [Google Scholar] [CrossRef]
- Yu, X.; Li, T.; Du, X.; Shen, Q.; Zhang, M.; Wei, Y.; Yang, D.; Xu, W.; Chen, W.; Bai, C.; et al. Single-cell rna sequencing reveals atlas of dairy goat testis cells. Zool. Res. 2021, 42, 401–405. [Google Scholar] [CrossRef]
- Zhang, P.; Qin, Y.; Zheng, Y.; Zeng, W. Phospholipase d family member 6 is a surface marker for enrichment of undifferentiated spermatogonia in prepubertal boars. Stem Cells Dev. 2018, 27, 55–64. [Google Scholar] [CrossRef]
- Yuan, X.; Kang, H.; Liu, B.; Yu, X.; Zhu, H.; Chen, S.; Du, X. Molecular characteristics and expression oftktl1in germ cells: Implications for nontumour cell research. Reprod. Domest. Anim. 2024, 59, e14723. [Google Scholar] [CrossRef]
- Zheng, X.; Li, H. Tktl1 modulates the response of paclitaxel-resistant human ovarian cancer cells to paclitaxel. Biochem. Biophys. Res. Commun. 2018, 503, 572–579. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Yan, Y.; Peng, G.; Gao, S.; Li, H.; Li, Y. Single-Cell RNA Sequencing Reveals an Atlas of Meihua Pig Testis Cells. Animals 2025, 15, 752. https://doi.org/10.3390/ani15050752
Zhang M, Yan Y, Peng G, Gao S, Li H, Li Y. Single-Cell RNA Sequencing Reveals an Atlas of Meihua Pig Testis Cells. Animals. 2025; 15(5):752. https://doi.org/10.3390/ani15050752
Chicago/Turabian StyleZhang, Mao, Yiming Yan, Guoliang Peng, Shuang Gao, Hongyi Li, and Yuan Li. 2025. "Single-Cell RNA Sequencing Reveals an Atlas of Meihua Pig Testis Cells" Animals 15, no. 5: 752. https://doi.org/10.3390/ani15050752
APA StyleZhang, M., Yan, Y., Peng, G., Gao, S., Li, H., & Li, Y. (2025). Single-Cell RNA Sequencing Reveals an Atlas of Meihua Pig Testis Cells. Animals, 15(5), 752. https://doi.org/10.3390/ani15050752





