Genomic Insights into the Population Genetics and Adaptive Evolution of Yellow Seabream (Acanthopagrus latus) with Whole-Genome Resequencing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Genomic Library Construction and Sequencing
2.3. Sequencing Data Quality Control
2.4. Sequence Alignment
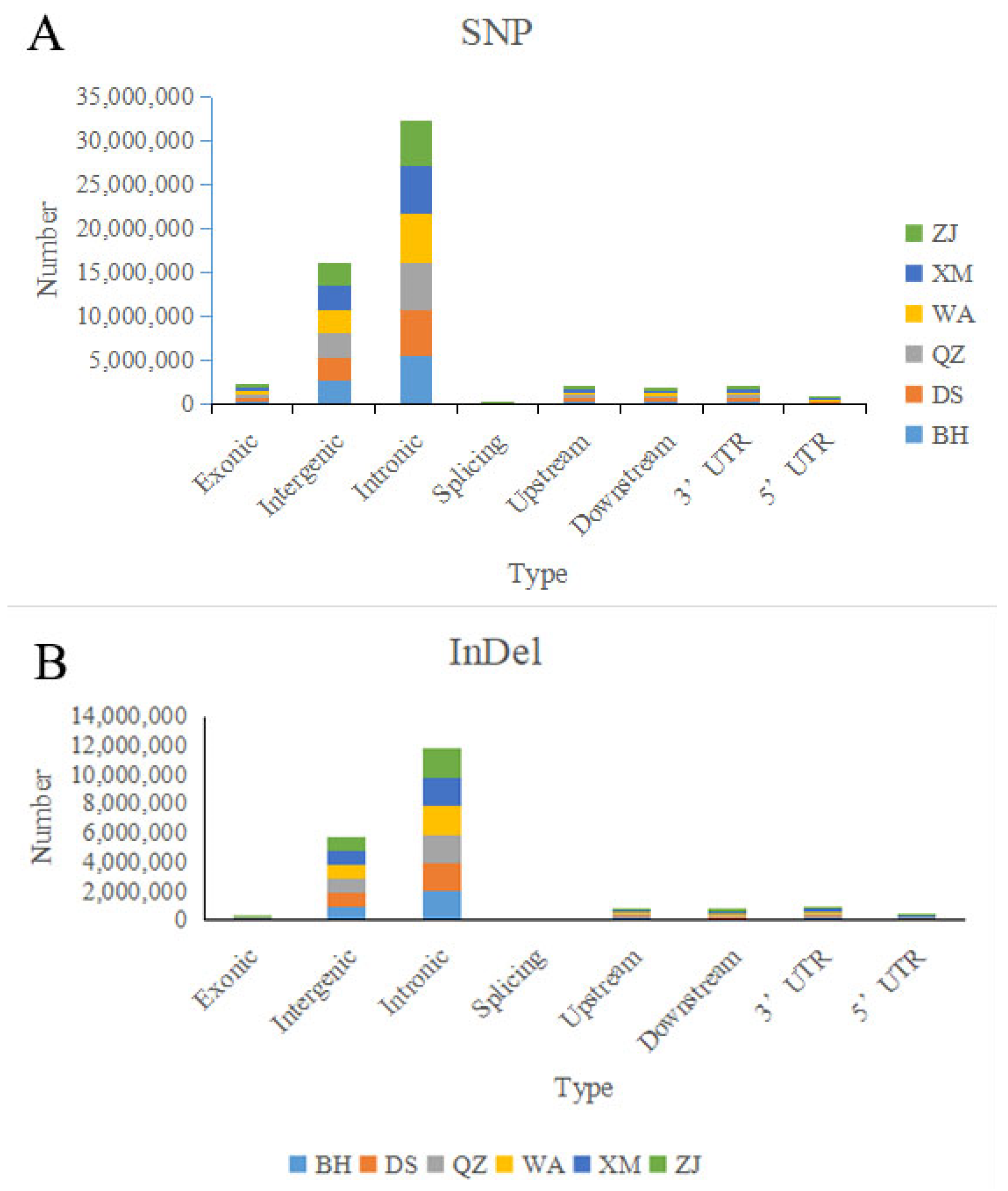
2.5. Variant Calling and Annotation
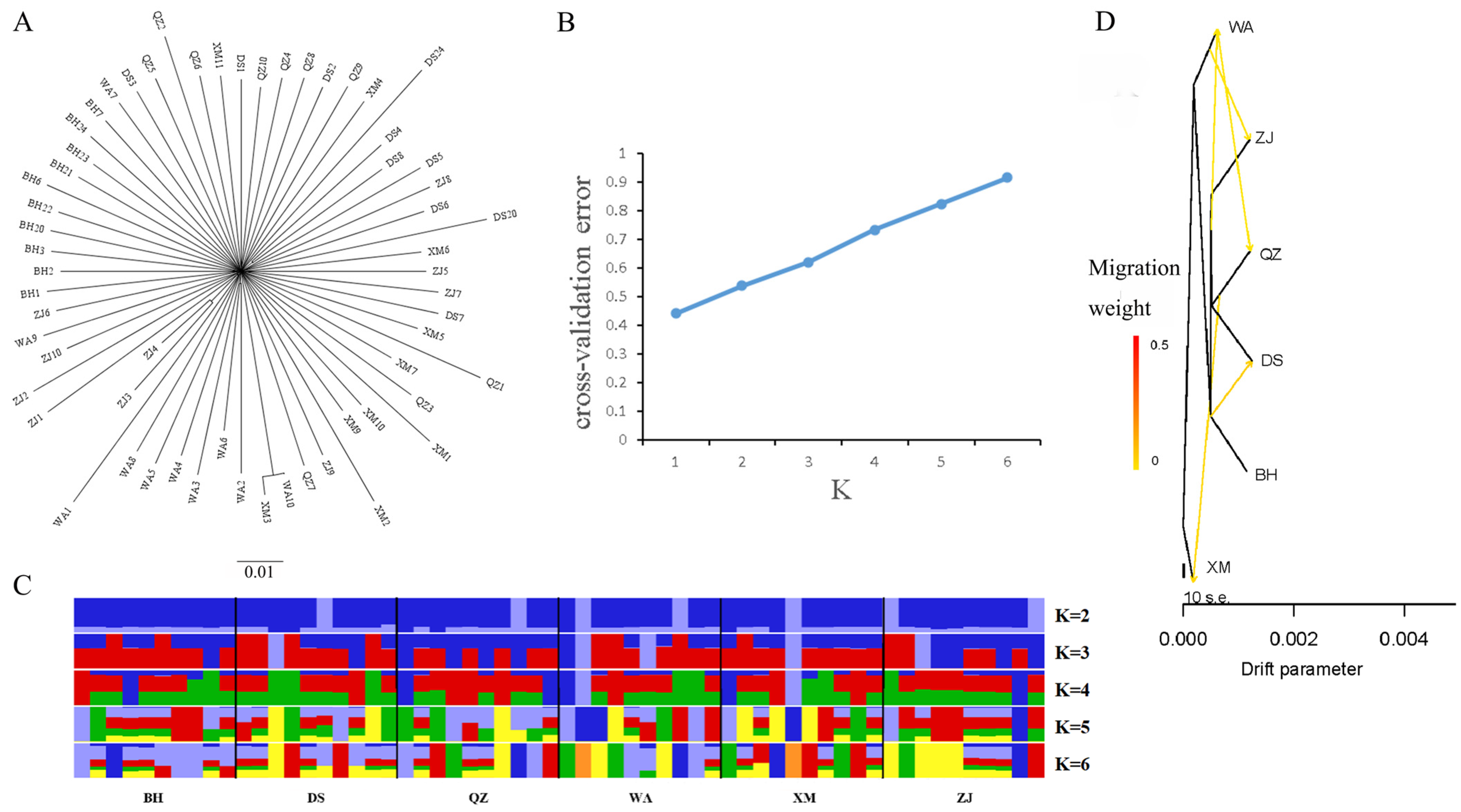
2.6. Population Genetic Structure Analysis
2.7. Population Genetic Diversity and Genetic Differentiation Analysis
2.8. Gene Flow Analysis
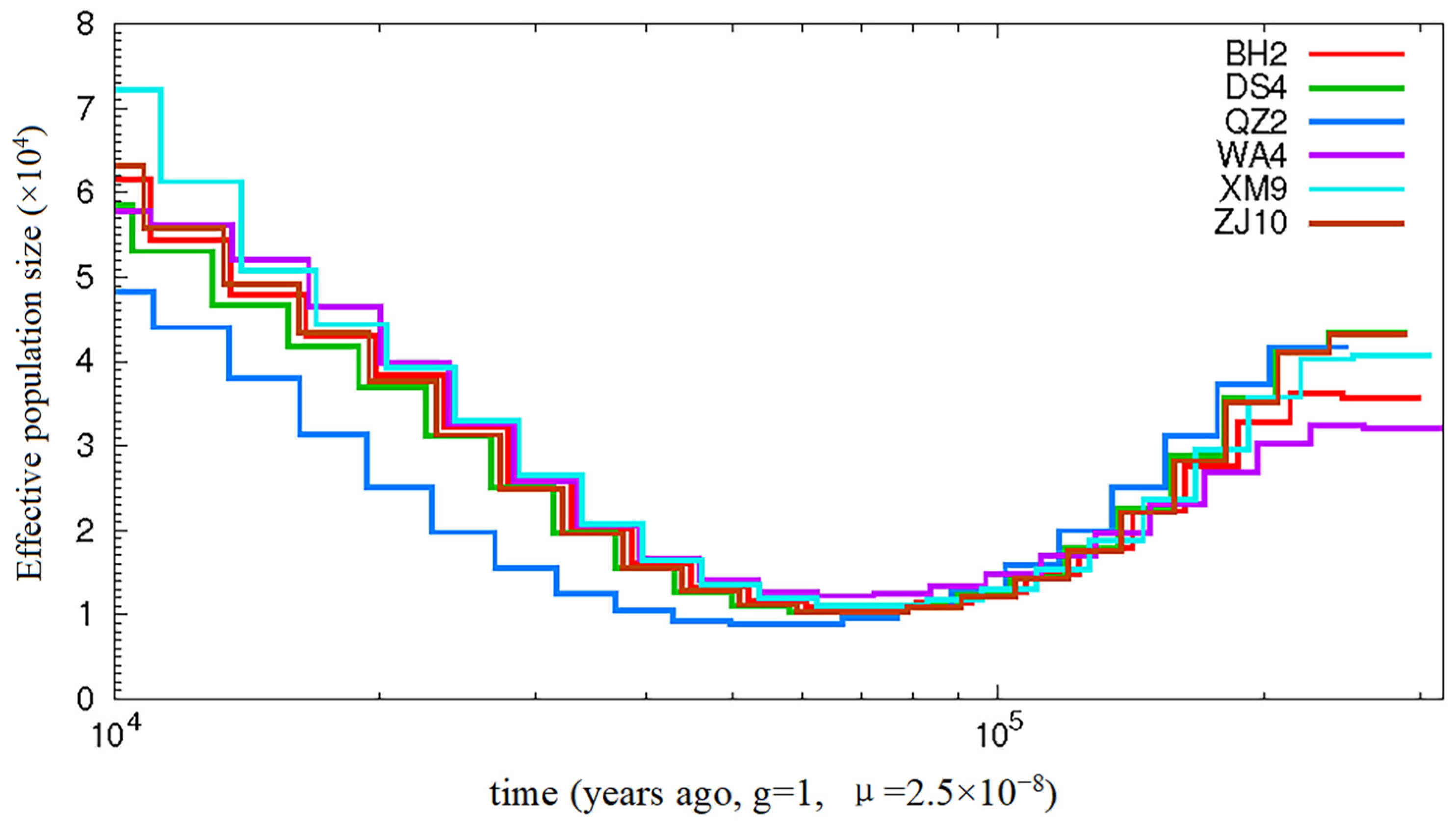
2.9. Effective Population Size Analysis of Evolutionary History
2.10. Detection of Selective Sweeps
3. Results
3.1. Data Quality Control
3.2. Variation Loci Distribution and Genetic Diversity
3.3. Population Genetic Structure and Gene Flow
3.4. Population Historical Dynamics
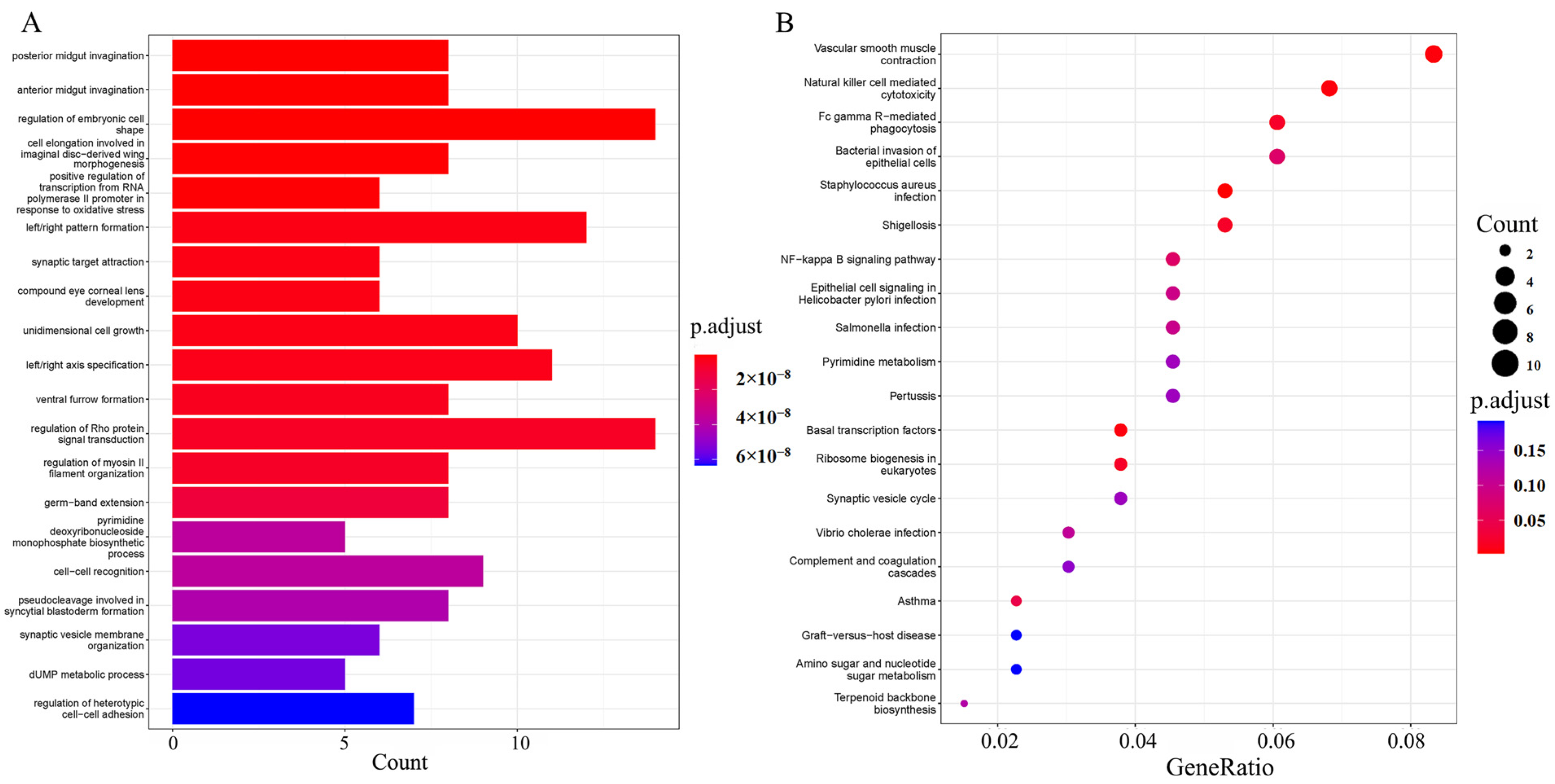
3.5. Screening and Enrichment Analysis of Selected Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iwatsuki, Y. Review of the Acanthopagrus latus complex (Perciformes: Sparidae) with descriptions of three new species from the Indo-West Pacific Ocean. J. Fish Biol. 2013, 83, 64–95. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Wang, X.; Li, C. Optimization of key procedures for fish tagging and releasing with its application to yellowfin seabream (Acanthopagrus latus). J. Fish. China 2019, 43, 584–592. [Google Scholar] [CrossRef]
- Ye, Y.; Huang, Z.; Xu, A.; Wang, J.; Li, Z. Genetic diversity and genetic structure of Acanthopagrus latus populations in the southeastern coast of China based on the mitochondrial DNA control region analysis. J. Appl. Oceanogr. 2022, 41, 583–590. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, J.; Liu, S.; Song, P.; Guan, Y.; Shan, B.; Li, Y.; Lin, L. Genetic diversity of the yellowfin seabream, Acanthopagrus latus (Actinopterygii: Perciformes: Sparidae)-an enhancement species in Dongshan Bay. Acta Ichthyol. Pisc. 2021, 51, 281–287. [Google Scholar] [CrossRef]
- Song, P.; Liu, C.; Cai, Z.; Liu, S.; Xiang, J.; Shan, B.; Ji, D.; Li, Y.; Lin, L. Genetic characteristics of yellow seabream Acanthopagrus latus (Houttuyn, 1782) (Teleostei: Sparidae) after stock enhancement in southeastern China coastal waters. Reg. Stud. Mar. Sci. 2021, 48, 102065. [Google Scholar] [CrossRef]
- He, Z.; Xiao, S.; Wang, S.S.; Chen, H.-F.; Su, S.; Zhao, Y.; Yang, C.; Zeng, D.; Zhu, W.; Chen, X.; et al. Genetic structure of D-loop sequence in Acanthopagrus latus. J. Fish. China 2021, 45, 345–356. [Google Scholar] [CrossRef]
- North, H.L.; McGaughran, A.; Jiggins, C.D. Insights into invasive species from whole-genome resequencing. Mol. Ecol. 2021, 30, 6289–6308. [Google Scholar] [CrossRef]
- Wang, J.; Hu, Z.; Liao, X.; Wang, Z.; Li, W.; Zhang, P.; Cheng, H.; Wang, Q.; Bhat, J.A.; Wang, H. Whole-genome resequencing reveals signature of local adaptation and divergence in wild soybean. Evol. Appl. 2022, 15, 1820–1833. [Google Scholar] [CrossRef]
- Zheng, J.; Zhao, L.; Zhao, X.; Gao, T.; Song, N. High genetic connectivity inferred from whole-genome resequencing provides insight into the phylogeographic pattern of Larimichthys polyactis. Mar. Biotechnol. 2022, 24, 671–680. [Google Scholar] [CrossRef]
- Saha, A.; Andersson, A.; Kurland, S.; Keehnen, N.L.; Kutschera, V.E.; Hössjer, O.; Ekman, D.; Karlsson, S.; Kardos, M.; Ståhl, G. Whole-genome resequencing confirms reproductive isolation between sympatric demes of brown trout (Salmo trutta) detected with allozymes. Mol. Ecol. 2022, 31, 498–511. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, T.; Gao, T.; Song, N. Whole-genome resequencing reveals genetic diversity and selection signals in warm temperate and subtropical Sillago sinica populations. BMC Genom. 2023, 24, 547. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Q.; Guo, X.Y.; Liu, Q.; Liu, S.S.; Zhang, Z.X.; Xiao, S.J.; Gao, T.X. Whole-genome resequencing of Japanese whiting (Sillago japonica) provide insights into local adaptations. Zool. Res. 2021, 42, 548–561. [Google Scholar] [CrossRef] [PubMed]
- She, Z.; Li, L.; Meng, J.; Jia, Z.; Que, H.; Zhang, G. Population resequencing reveals candidate genes associated with salinity adaptation of the Pacific oyster Crassostrea gigas. Sci. Rep. 2018, 8, 8683. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Yang, M.-j.; Hu, Z.; Shi, P.; Li, Y.-r.; Guo, Y.-j.; Zhang, T.; Song, H. Molecular evidence for the adaptive evolution in euryhaline bivalves. Mar. Environ. Res. 2023, 192, 106240. [Google Scholar] [CrossRef]
- Shan, X.; Hu, Z.; Shao, C. Progress in the study of fishing-induced evolution of fish biological characteristics. Prog. Fish. Sci. 2020, 41, 165–175. [Google Scholar] [CrossRef]
- Wilson, A.D.; Binder, T.R.; McGrath, K.P.; Cooke, S.J.; Godin, J.-G.J. Capture technique and fish personality: Angling targets timid bluegill sunfish, Lepomis macrochirus. J. Fish. Res. Board Can. 2011, 68, 749–757. [Google Scholar] [CrossRef]
- Peng, C.; Luo, C.; Xiang, G.; Huang, J.; Shao, L.; Huang, H.; Fan, S. Genome-Wide Microsatellites in Acanthopagrus latus: Development, Distribution, Characterization, and Polymorphism. Animals 2024, 14, 3709. [Google Scholar] [CrossRef]
- Lu, J.; Gao, D.; Sims, Y.; Fang, W.; Collins, J.; Torrance, J.; Lin, G.; Xie, J.; Liu, J.; Howe, K. Chromosome-level genome assembly of Acanthopagrus latus provides insights into salinity stress adaptation of Sparidae. Mar. Biotechnol. 2022, 24, 655–660. [Google Scholar] [CrossRef]
- Wang, W.; Huang, J.; Hu, Y.; Feng, J.; Gao, D.; Fang, W.; Xu, M.; Ma, C.; Fu, Z.; Chen, Q. Seascapes Shaped the Local Adaptation and Population Structure of South China Coast Yellowfin Seabream (Acanthopagrus latus). Mar. Biotechnol. 2024, 26, 60–73. [Google Scholar] [CrossRef]
- Yan, C. Analysis and Evaluation of Protection and Management of the Chinese White Dolphins (Sousa chinensis) in Xiamen Rare Marine Species National Nature Reserve. Master’s Thesis, Shandong University, Jinan, China, 2012. [Google Scholar]
- Yu, Z.; Yu, J. Preliminary Study on Ecological Compensation Standard of Marine Reserve: A case of Xiamen Rare Marine Species National Nature Reserve. Mar. Environ. Sci. 2017, 36, 202–208. [Google Scholar] [CrossRef]
- Zhuang, Q. Study on the Gap Analysis and Establishment Management of Xiamen Marine Protected Area. Master’s Thesis, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, China, 2020. [Google Scholar]
- Hesp, S.A. Biology of Two Species of Sparid on the West Coast of Australia. Ph.D. Thesis, Murdoch University, Perth, Australia, 2003. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Durvasula, A.; Fulgione, A.; Gutaker, R.M.; Alacakaptan, S.I.; Flood, P.J.; Neto, C.; Tsuchimatsu, T.; Burbano, H.A.; Pico, F.X.; Alonso-Blanco, C. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 5213–5218. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Cornelis, A.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.; Pritchard, J. Inference of population splits and mixtures from genome-wide allele frequency data. Nat. Preced. 2012, 1-1. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 2011, 475, 493–496. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhu, W.; Xiao, S.; Yang, C.; Chen, H.; He, T.; Zeng, D.; Chen, X.; Peng, M. Genetic structure analysis of Acanthopagrus latus populations along South China coast based on mitochondrial Cytb gene sequences. CABI Databases 2021, 34, 446–454. [Google Scholar] [CrossRef]
- Zhao, H.; Huang, L.; Zhang, J.; You, S.; Zeng, Q.; Liu, X. Development of SNP parentage assignment techniques in the yellowfin seabream Acanthopagrus latus. Acta Oceanolog. Sin. 2024, 43, 151–155. [Google Scholar] [CrossRef]
- Xia, J.H.; Huang, J.H.; Gong, J.B.; Jiang, S.G. Significant population genetic structure of yellowfin seabream Acanthopagrus latus in China. J. Fish Biol. 2008, 73, 1979–1992. [Google Scholar] [CrossRef]
- Sun, C.-H.; Gozlan, R.E.; Wu, T.; Xue, D.; Lao, Y.-L.; Yu, J.-F.; Zeng, X.-S.; Li, S.; Hardouin, E.A.; Andreou, D. The role of ancestral seascape discontinuity and geographical distance in structuring rockfish populations in the Pacific Northwest. Front. Mar. Sci. 2022, 9, 1018864. [Google Scholar] [CrossRef]
- Akazaki, M.; Tokitô, A. Studies on the seedling production of yellowfin porgy (Kichinu), Acanthopagrus latus Houttuyn—II Egg development and metamorphoses of larvae. Aquac. Sci. 1982, 29, 218–228. [Google Scholar] [CrossRef]
- Blampied, S.R.; Sheehan, E.V.; Binney, F.C.T.; Attrill, M.J.; Rees, S.E. Value of coastal habitats to commercial fisheries in Jersey, English Channel, and the role of marine protected areas. Fish. Manag. Ecol. 2022, 29, 734–744. [Google Scholar] [CrossRef]
- Uusi-Heikkilä, S.; Whiteley, A.R.; Kuparinen, A.; Matsumura, S.; Venturelli, P.A.; Wolter, C.; Slate, J.; Primmer, C.R.; Meinelt, T.; Killen, S.S. The evolutionary legacy of size-selective harvesting extends from genes to populations. Evol. Appl. 2015, 8, 597–620. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, C.; Tan, Y.; Li, L.; Zhang, H.; Liang, Y.; Zeng, J.; Zou, H. Transcription levels and prognostic significance of the NFI family members in human cancers. PeerJ 2020, 8, e8816. [Google Scholar] [CrossRef]
- Steele-Perkins, G.; Butz, K.G.; Lyons, G.E.; Zeichner-David, M.; Kim, H.-J.; Cho, M.-I.; Gronostajski, R.M. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol. Cell. Biol. 2003, 23, 1075–1084. [Google Scholar] [CrossRef]
- Cao, L.; Lin, L.; Li, H.; Pan, X.; Zhou, J.; Yang, B.; Kang, Y.; Qian, A.; Yao, J.; Shan, X. Role of Ras-related C3 botulinum toxin substrate (Rac2) in northern snakehead (Channa argus) resistance to Aeromonas veronii infection. Aquacult. Res. 2022, 53, 4266–4275. [Google Scholar] [CrossRef]
- Laing, K.J.; Purcell, M.K.; Winton, J.R.; Hansen, J.D. A genomic view of the NOD-like receptor family in teleost fish: Identification of a novel NLR subfamily in zebrafish. BMC Evol. Biol. 2008, 8, 42. [Google Scholar] [CrossRef]
- Lessieur, E.M.; Joseph, F.; Gaivin, R.J.; Ping, S.; Perkins, B.D. The Ciliopathy Gene ahi1 Is Required for Zebrafish Cone Photoreceptor Outer Segment Morphogenesis and Survival. Investig. Ophthalmol. Visual Sci. 2017, 58, 448–460. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Liu, C.; Jamsen, J.; Wu, Z.; Wang, Y.; Chen, J.; Zheng, L.; Shen, B. The DNase domain-containing protein TATDN1 plays an important role in chromosomal segregation and cell cycle progression during zebrafish eye development. Cell Cycle 2012, 11, 4626–4632. [Google Scholar] [CrossRef]
- Araya, N.; Arimura, H.; Kawahara, K.-I.; Yagishita, N.; Ishida, J.; Fujii, R.; Aratani, S.; Fujita, H.; Sato, T.; Yamano, Y. Role of Kenae/CCDC125 in cell motility through the deregulation of RhoGTPase. Int. J. Mol. Med. 2009, 24, 605–611. [Google Scholar] [CrossRef]
- Wang, X.; Li, T.; Sukrithan, V.; Ratan, A.; McCarter, M.; Carpten, J.D.; Colman, H.; Ikeguchi, A.; Puzanov, I.; Arnold, S.M. Incorporating long non-coding RNA (lncRNA) into genome-wide biomarker screening for prognostic gene signatures of immunotherapy outcomes. J. Clin. Oncol. 2023, 41, 16. [Google Scholar] [CrossRef]
- Zhang, B.; Li, J.; Ma, Y.; Dong, F.; Tao, Z.; Shen, J.; Tian, Y.; Shi, F.; Lu, L. Molecular cloning and sequence analysis of duck BLVRA gene. CABI Databases 2010, 32, 14–18. [Google Scholar] [CrossRef]
- Johnson, A.C.; Wu, W.; Showmaker, K.C.; Lindsey, M.L.; Garrett, M.R. Physiological Omics Identifies Mechanisms that Attenuate Renal Injury and Blood Pressure in Dahl salt-sensitive Arhgef11−/− Rats. FASEB J. 2019, 33, 571.1. [Google Scholar] [CrossRef]
- Tu, J.; Huang, C.; Huang, Y.; Wang, H. Molecular cloning and expression analysis of jphs in Megalobrama amblycephala. J. Huazhong Agric. Univ. 2020, 39, 126–135. [Google Scholar] [CrossRef]
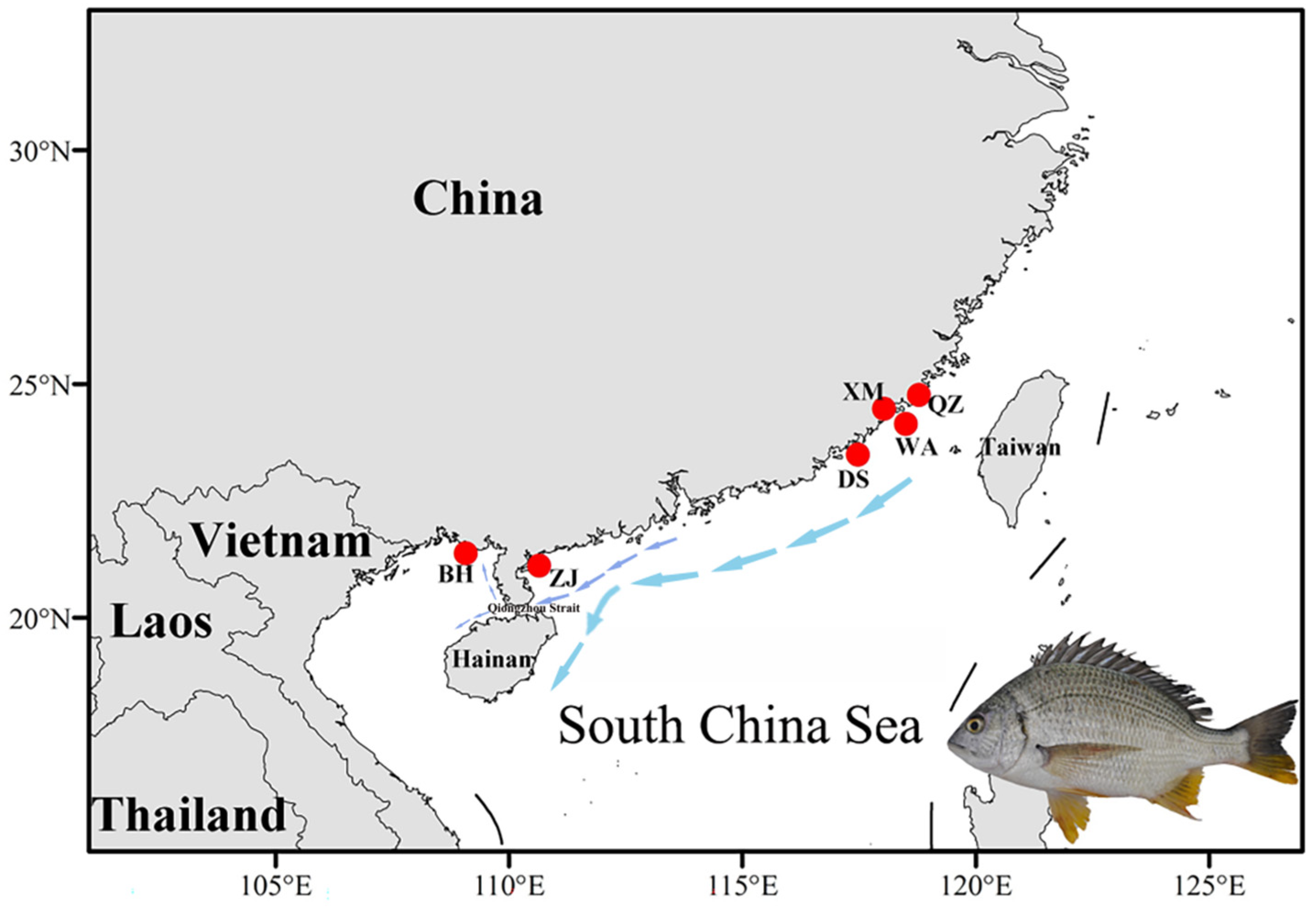

| Sampling Sites | Sampling Coordinates | Number of Samples |
|---|---|---|
| Beihai (BH) | 21°27′23.2″ N; 108°51′59.3″ E | 10 |
| Zhanjiang (ZJ) | 21°09′35.5″ N; 110°44′02.1″ E | 10 |
| Dongshan (DS) | 23°37′48.1″ N; 117°21′01.2″ E | 10 |
| Wuyu (WA) | 24°20′11.7″ N; 118°08′22.5″ E | 10 |
| Xiamen (XM) | 24°25′35.5″ N; 118°04′49.9″ E | 10 |
| Quanzhou (QZ) | 24°49′41.8″ N; 118°43′56.5″ E | 10 |
| Beihai | Dongshan | Quanzhou | Wuyu | Xiamen | Zhanjiang | |
|---|---|---|---|---|---|---|
| πθ | 3.155 × 10−3 | 3.042 × 10−3 | 3.058 × 10−3 | 3.119 × 10−3 | 3.096 × 10−3 | 3.102 × 10−3 |
| Beihai (BH) | Dongshan (DS) | Quanzhou (QZ) | Wuyu (WA) | Zhanjiang (ZJ) | Xiamen (XM) | |
|---|---|---|---|---|---|---|
| BH | ||||||
| DS | −7.971 × 10−4 | |||||
| QZ | −9.355 × 10−4 | −1.572 × 10−3 | ||||
| WA | −8.193 × 10−4 | −1.044 × 10−3 | −1.336 × 10−3 | |||
| ZJ | −4.121 × 10−4 | −8.160 × 10−4 | −1.016 × 10−3 | −7.174 × 10−4 | ||
| XM | −1.016 × 10−3 | −1.420 × 10−3 | −1.670 × 10−3 | −4.395 × 10−3 | −1.034 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yang, J.; Fang, Y.; Zhang, R.; Cai, Z.; Shan, B.; Miao, X.; Lin, L.; Song, P.; Zhang, J. Genomic Insights into the Population Genetics and Adaptive Evolution of Yellow Seabream (Acanthopagrus latus) with Whole-Genome Resequencing. Animals 2025, 15, 745. https://doi.org/10.3390/ani15050745
Li Y, Yang J, Fang Y, Zhang R, Cai Z, Shan B, Miao X, Lin L, Song P, Zhang J. Genomic Insights into the Population Genetics and Adaptive Evolution of Yellow Seabream (Acanthopagrus latus) with Whole-Genome Resequencing. Animals. 2025; 15(5):745. https://doi.org/10.3390/ani15050745
Chicago/Turabian StyleLi, Yuan, Jingyu Yang, Yan Fang, Ran Zhang, Zizi Cai, Binbin Shan, Xing Miao, Longshan Lin, Puqing Song, and Jing Zhang. 2025. "Genomic Insights into the Population Genetics and Adaptive Evolution of Yellow Seabream (Acanthopagrus latus) with Whole-Genome Resequencing" Animals 15, no. 5: 745. https://doi.org/10.3390/ani15050745
APA StyleLi, Y., Yang, J., Fang, Y., Zhang, R., Cai, Z., Shan, B., Miao, X., Lin, L., Song, P., & Zhang, J. (2025). Genomic Insights into the Population Genetics and Adaptive Evolution of Yellow Seabream (Acanthopagrus latus) with Whole-Genome Resequencing. Animals, 15(5), 745. https://doi.org/10.3390/ani15050745






