Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals
Simple Summary
Abstract
1. Introduction
2. Analytical Methodologies for Gut Mycobiota
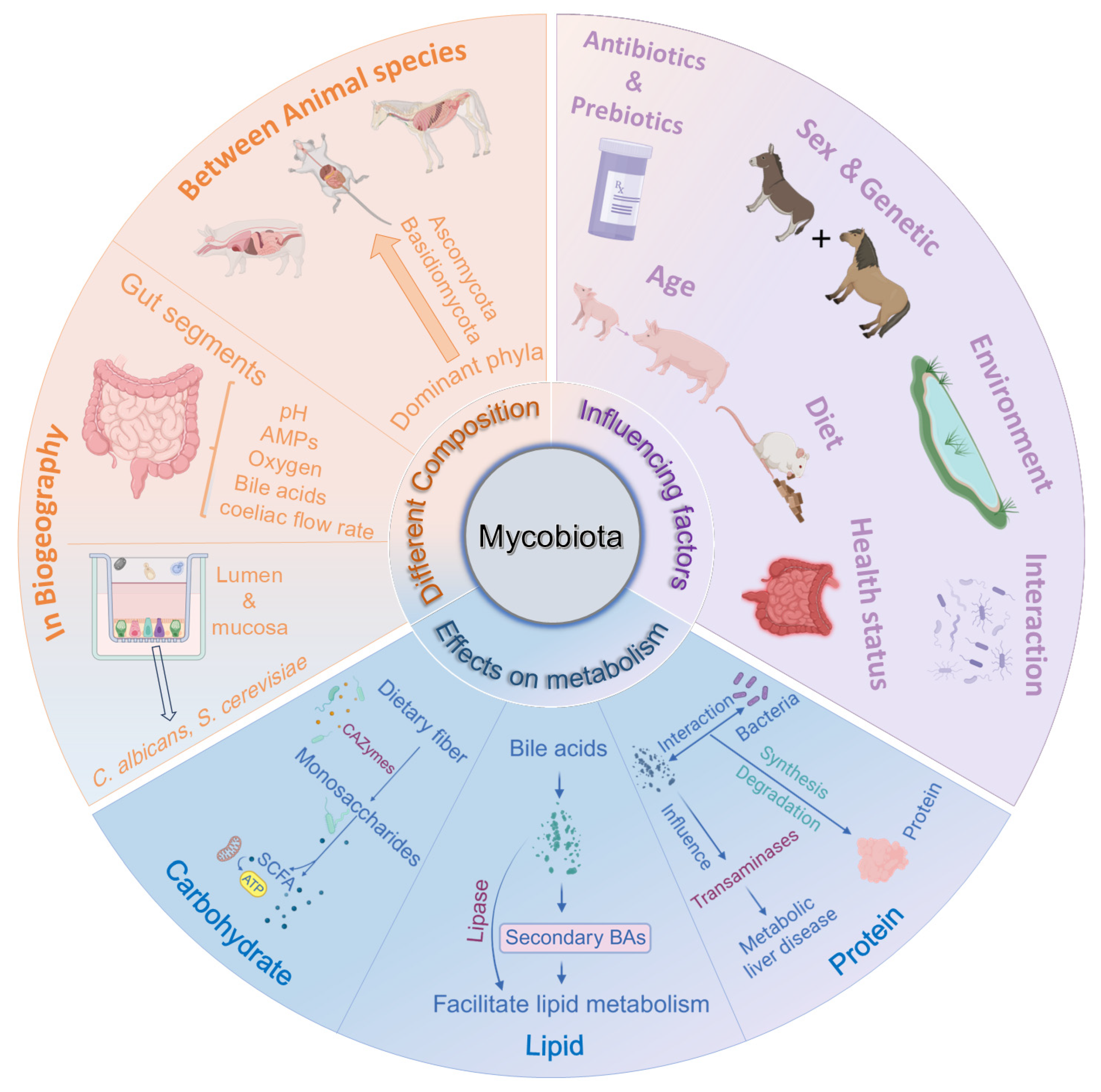
3. Composition and Distribution of Mycobiota in Monogastric Animals
3.1. Composition of Intestinal Fungi in Different Animals
3.1.1. Mouse
3.1.2. Pig
3.1.3. Giant Panda
3.1.4. Dog and Cat
3.1.5. Herbivorous Monogastric Animals
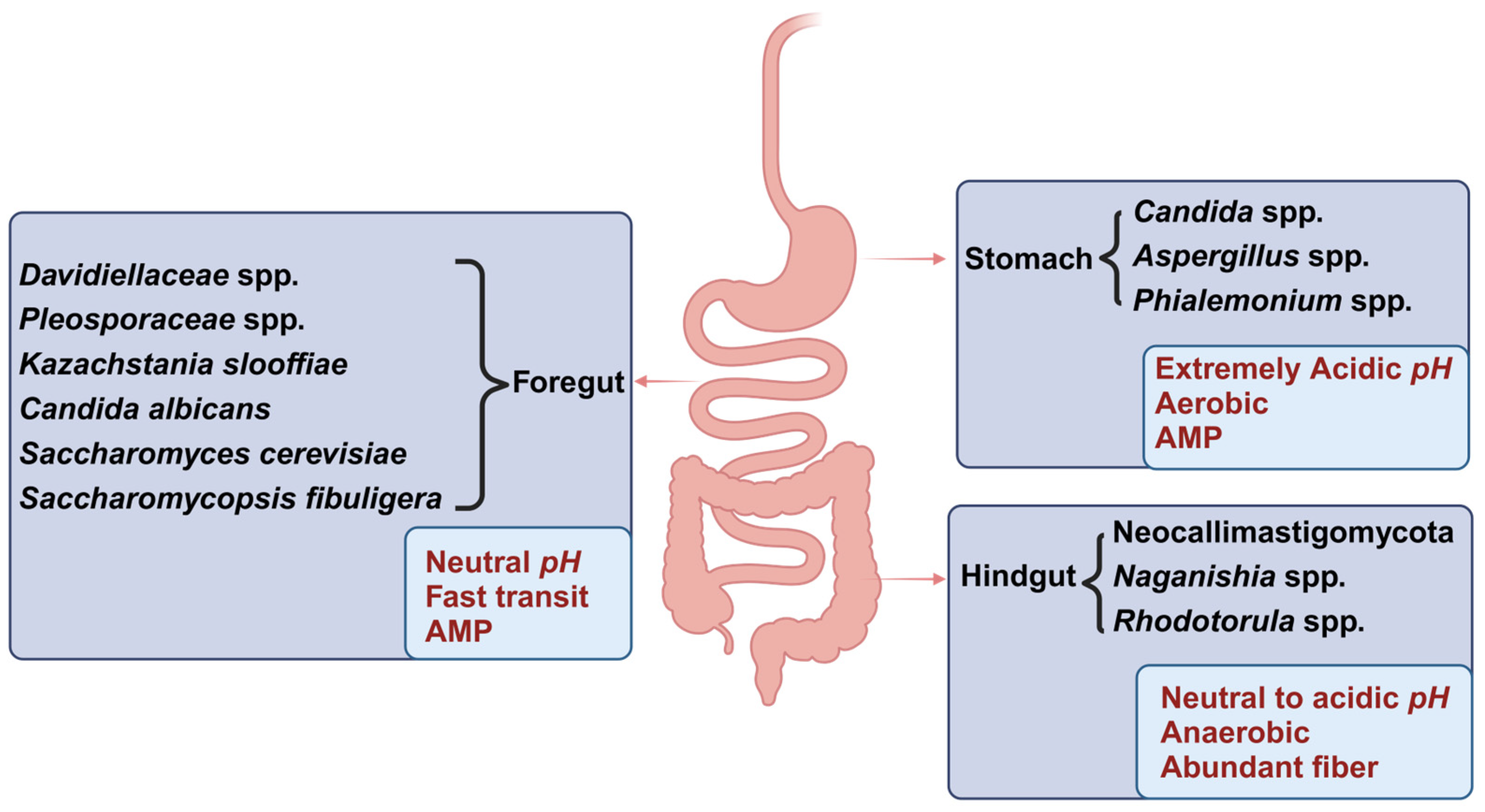
3.2. Distribution of the Mycobiota Across Various Segments of the Gastrointestinal Tract (GIT)
3.3. Distribution of the Mycobiota in Intestinal Lumen and Mucosa
4. Factors Influencing the Composition of the Intestinal Mycobiota
4.1. Age
4.2. Diet
4.3. Health Status
4.4. Utilization of Antibiotics
4.5. Interactions Between Microorganisms
4.5.1. Interactions Between Bacteria and Fungi
4.5.2. Interactions Between Fungi
4.5.3. Interactions Between Mycoviruse and Fungi
4.5.4. Interaction Between Parasite and Fungi
4.6. Environment
4.7. Host Sex and Genetics
4.8. Fungal-Based Probiotics and Prebiotics
5. Effects of Fungi on Metabolism of Nutrients in Monogastric Animals
5.1. Carbohydrate Metabolism
5.2. Lipid Metabolism
5.3. Protein Metabolism
5.4. Vitamins and Minerals
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Charbit-Henrion, F.; Parlato, M.; Malamut, G.; Ruemmele, F.; Cerf-Bensussan, N. Intestinal Immunoregulation: Lessons from Human Mendelian Diseases. Mucosal Immunol. 2021, 14, 1017–1037. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern Recognition Receptors in Health and Diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Cadwell, K. Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology 2020, 160, 1050. [Google Scholar] [CrossRef] [PubMed]
- Aho, V.T.E.; Houser, M.C.; Pereira, P.A.B.; Chang, J.; Rudi, K.; Paulin, L.; Hertzberg, V.; Auvinen, P.; Tansey, M.G.; Scheperjans, F. Relationships of Gut Microbiota, Short-Chain Fatty Acids, Inflammation, and the Gut Barrier in Parkinson’s Disease. Mol. Neurodegener. 2021, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Sun, L.; Gonzalez, F.J. Gut Microbiota-Derived Bile Acids in Intestinal Immunity, Inflammation, and Tumorigenesis. Cell Host Microbe 2022, 30, 289–300. [Google Scholar] [CrossRef]
- Luo, Y.; Ren, W.; Smidt, H.; Wright, A.-D.G.; Yu, B.; Schyns, G.; McCormack, U.M.; Cowieson, A.J.; Yu, J.; He, J.; et al. Dynamic Distribution of Gut Microbiota in Pigs at Different Growth Stages: Composition and Contribution. Microbiol. Spectr. 2022, 10, e00688-21. [Google Scholar] [CrossRef]
- Leonardi, I.; Gao, I.H.; Lin, W.-Y.; Allen, M.; Li, X.V.; Fiers, W.D.; De Celie, M.B.; Putzel, G.G.; Yantiss, R.K.; Johncilla, M.; et al. Mucosal Fungi Promote Gut Barrier Function and Social Behavior via Type 17 Immunity. Cell 2022, 185, 831–846. [Google Scholar] [CrossRef]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yu, B.; He, J.; Zheng, P.; Mao, X.; Yu, J.; Luo, J.; Tian, G.; Huang, Z.; et al. Fungi in Gastrointestinal Tracts of Human and Mice: From Community to Functions. Microb. Ecol. 2018, 75, 821–829. [Google Scholar] [CrossRef]
- Doron, I.; Leonardi, I.; Li, X.V.; Fiers, W.D.; Semon, A.; Bialt-DeCelie, M.; Migaud, M.; Gao, I.H.; Lin, W.-Y.; Kusakabe, T.; et al. Human Gut Mycobiota Tune Immunity via CARD9-Dependent Induction of Anti-Fungal IgG Antibodies. Cell 2021, 184, 1017. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Prenafeta-Boldú, F.; Zingaretti, L.M.; Gonzalez-Rodriguez, O.; Dalmau, A.; Quintanilla, R.; Ballester, M. Gut Eukaryotic Communities in Pigs: Diversity, Composition and Host Genetics Contribution. Anim. Microbiome 2020, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.S. Analysis of the Gut Microbiome in Dogs and Cats. Vet. Clin. Pathol. 2022, 50, 6–17. [Google Scholar] [CrossRef]
- Scupham, A.J.; Presley, L.L.; Wei, B.; Bent, E.; Griffith, N.; McPherson, M.; Zhu, F.; Oluwadara, O.; Rao, N.; Braun, J.; et al. Abundant and Diverse Fungal Microbiota in the Murine Intestine. Appl. Environ. Microbiol. 2006, 72, 793–801. [Google Scholar] [CrossRef]
- Wei, G. Insights into Gut Fungi in Pigs: A Comprehensive Review. J. Anim. Physiol. Anim. Nutr. 2024, 109, 96–112. [Google Scholar] [CrossRef]
- Zhao, Y.; Ren, X.; Wu, H.; Hu, H.; Cheng, C.; Du, M.; Huang, Y.; Zhao, X.; Wang, L.; Yi, L.; et al. Diversity and Functional Prediction of Fungal Communities in Different Segments of Mongolian Horse Gastrointestinal Tracts. BMC Microbiol. 2023, 23, 253. [Google Scholar] [CrossRef] [PubMed]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; Da Costa, G.; et al. Enterobacteriaceae Are Essential for the Modulation of Colitis Severity by Fungi. Microbiome 2018, 6, 152. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal Microbiota Dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Li, X.V.; Leonardi, I.; Putzel, G.G.; Semon, A.; Fiers, W.D.; Kusakabe, T.; Lin, W.-Y.; Gao, I.H.; Doron, I.; Gutierrez-Guerrero, A.; et al. Immune Regulation by Fungal Strain Diversity in Inflammatory Bowel Disease. Nature 2022, 603, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Chehoud, C.; Albenberg, L.G.; Judge, C.; Hoffmann, C.; Grunberg, S.; Bittinger, K.; Baldassano, R.N.; Lewis, J.D.; Bushman, F.D.; Wu, G.D. Fungal Signature in the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 1948–1956. [Google Scholar] [CrossRef]
- Lan, Y.; Li, Y.; Yu, G.; Zhang, Z.; Irshad, I. Dynamic Changes of Gut Fungal Community in Horse at Different Health States. Front. Vet. Sci. 2022, 9, 1047412. [Google Scholar] [CrossRef]
- Vecere, G.; Malka, S.; Sands, N.; Lee, M.; Krumbeck, J.A. Assessment of the Fecal Microbiome of Healthy Rabbits (Oryctolagus Cuniculus Domesticus) Compared with Rabbits with Gastrointestinal Disease Using next-Generation DNA Sequencing. Am. J. Vet. Res. 2024, 86, 193. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, K.; Sun, L.; Cheng, B.; Qiao, S.; Dai, H.; Shi, W.; Ma, J.; Liu, H. Therapeutic Manipulation of Gut Microbiota by Polysaccharides of Wolfiporia Cocos Reveals the Contribution of the Gut Fungi-Induced PGE2 to Alcoholic Hepatic Steatosis. Gut Microbes 2020, 12, 1830693. [Google Scholar] [CrossRef]
- Mims, T.S.; Abdallah, Q.A.; Stewart, J.D.; Watts, S.P.; White, C.T.; Rousselle, T.V.; Gosain, A.; Bajwa, A.; Han, J.C.; Willis, K.A.; et al. The Gut Mycobiome of Healthy Mice Is Shaped by the Environment and Correlates with Metabolic Outcomes in Response to Diet. Commun. Biol. 2021, 4, 281. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List; Bolchacova, E.; et al. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H. ITS1 versus ITS2 as DNA Metabarcodes for Fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Netherway, T.; Frioux, C.; Ferretti, P.; Coelho, L.P.; Geisen, S.; Bork, P.; Hildebrand, F. Metagenomic Assessment of the Global Diversity and Distribution of Bacteria and Fungi. Environ. Microbiol. 2021, 23, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Aoki, T.; Ariyawansa, H.A.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Fungal Taxonomy and Sequence-Based Nomenclature. Nat. Microbiol. 2021, 6, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Saarenpää, S.; Shalev, O.; Ashkenazy, H.; Carlos, V.; Lundberg, D.S.; Weigel, D.; Giacomello, S. Spatial Metatranscriptomics Resolves Host–Bacteria–Fungi Interactomes. Nat. Biotechnol. 2024, 42, 1384–1393. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Zinger, L.; Nilsson, R.H.; Kennedy, P.G.; Yang, T.; Anslan, S.; Mikryukov, V. Best Practices in Metabarcoding of Fungi: From Experimental Design to Results. Mol. Ecol. 2022, 31, 2769–2795. [Google Scholar] [CrossRef]
- Yan, Q.; Li, S.; Yan, Q.; Huo, X.; Wang, C.; Wang, X.; Sun, Y.; Zhao, W.; Yu, Z.; Zhang, Y.; et al. A Genomic Compendium of Cultivated Human Gut Fungi Characterizes the Gut Mycobiome and Its Relevance to Common Diseases. Cell 2024, 187, 2969–2989.e24. [Google Scholar] [CrossRef]
- Badotti, F.; De Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Góes-Neto, A. Effectiveness of ITS and Sub-Regions as DNA Barcode Markers for the Identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 17, 42. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N.; et al. Environmental DNA Metabarcoding: Transforming How We Survey Animal and Plant Communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Kelly, R.P. Making Environmental DNA Count. Mol. Ecol. Resour. 2016, 16, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous Identification of Fungi: Where Do We Stand and How Accurate and Precise Is Fungal DNA Barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef]
- Mu, P.; Li, W.; Tran, L.-S.P.; Li, X. SmT/SHM-Seq: Simultaneously Capturing Spatial Transcriptome and Microbiome Information in Plants. Trends Plant Sci. 2024, 29, 1277–1278. [Google Scholar] [CrossRef]
- Hirayama, T.; Miyazaki, T.; Ito, Y.; Wakayama, M.; Shibuya, K.; Yamashita, K.; Takazono, T.; Saijo, T.; Shimamura, S.; Yamamoto, K.; et al. Virulence Assessment of Six Major Pathogenic Candida Species in the Mouse Model of Invasive Candidiasis Caused by Fungal Translocation. Sci. Rep. 2020, 10, 3814. [Google Scholar] [CrossRef]
- Yang, S.; Gao, X.; Meng, J.; Zhang, A.; Zhou, Y.; Long, M.; Li, B.; Deng, W.; Jin, L.; Zhao, S.; et al. Metagenomic Analysis of Bacteria, Fungi, Bacteriophages, and Helminths in the Gut of Giant Pandas. Front. Microbiol. 2018, 9, 1717. [Google Scholar] [CrossRef] [PubMed]
- Handl, S.; Dowd, S.E.; Garcia-Mazcorro, J.F.; Steiner, J.M.; Suchodolski, J.S. Massive Parallel 16S rRNA Gene Pyrosequencing Reveals Highly Diverse Fecal Bacterial and Fungal Communities in Healthy Dogs and Cats: Fecal Microbiota in Dogs and Cats Using Pyrosequencing. FEMS Microbiol. Ecol. 2011, 76, 301–310. [Google Scholar] [CrossRef]
- da Silva, J.F.; Bolsoni, J.A.; da Costa, R.M.; Alves, J.V.; Bressan, A.F.M.; Silva, L.E.V.; Costa, T.J.; Oliveira, A.E.R.; Manzato, C.P.; Aguiar, C.A.; et al. Aryl Hydrocarbon Receptor (AhR) Activation Contributes to High-Fat Diet-Induced Vascular Dysfunction. Br. J. Pharmacol. 2022, 179, 2938–2952. [Google Scholar] [CrossRef]
- Du Preez, L.L.; Van Der Walt, E.; Valverde, A.; Rothmann, C.; Neser, F.W.C.; Cason, E.D. A Metagenomic Survey of the Fecal Microbiome of the African Savanna Elephant (Loxodonta africana). Anim. Genet. 2024, 55, 621–643. [Google Scholar] [CrossRef]
- Sun, X.; Sitters, J.; Ruytinx, J.; Wassen, M.J.; Olde Venterink, H. Microbial Community Composition in the Dung of Five Sympatric European Herbivore Species. Ecol. Evol. 2024, 14, e11071. [Google Scholar] [CrossRef]
- Bradshaw, A.J.; Autumn, K.C.; Rickart, E.A. On the Origin of Feces: Fungal Diversity, Distribution, and Conservation Implications from Feces of Small Mammals. Environ. DNA 2022, 4, 608–626. [Google Scholar] [CrossRef]
- Li, J.; Luo, Y.; Chen, D.; Yu, B.; He, J.; Huang, Z.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; et al. The fungal community and its interaction with the concentration of short-chain fatty acids in the caecum and colon of weaned piglets. J. Anim. Physiol. Anim. Nutr. 2020, 104, 616–628. [Google Scholar] [CrossRef]
- Hu, R.; Li, S.; Diao, H.; Huang, C.; Yan, J.; Wei, X.; Zhou, M.; He, P.; Wang, T.; Fu, H.; et al. The Interaction between Dietary Fiber and Gut Microbiota, and Its Effect on Pig Intestinal Health. Front. Immunol. 2023, 14, 1095740. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhou, M.; Song, Z.; Deng, Y.; Xia, S.; Li, Y.; Huang, X.; Xiao, D.; Yin, Y.; Yin, J. Clec7a Drives Gut Fungus-Mediated Host Lipid Deposition. Microbiome 2023, 11, 264. [Google Scholar] [CrossRef] [PubMed]
- Gupta, Y.; Ernst, A.L.; Vorobyev, A.; Beltsiou, F.; Zillikens, D.; Bieber, K.; Sanna-Cherchi, S.; Christiano, A.M.; Sadik, C.D.; Ludwig, R.J.; et al. Impact of Diet and Host Genetics on the Murine Intestinal Mycobiome. Nat. Commun. 2023, 14, 834. [Google Scholar] [CrossRef]
- Hu, J.; Nie, Y.; Chen, J.; Zhang, Y.; Wang, Z.; Fan, Q.; Yan, X. Gradual Changes of Gut Microbiota in Weaned Miniature Piglets. Front. Microbiol. 2016, 7, 1727. [Google Scholar] [CrossRef]
- Heisel, T.; Montassier, E.; Johnson, A.; Al-Ghalith, G.; Lin, Y.-W.; Wei, L.-N.; Knights, D.; Gale, C.A. High-Fat Diet Changes Fungal Microbiomes and Interkingdom Relationships in the Murine Gut. mSphere 2017, 2, e00351-17. [Google Scholar] [CrossRef]
- Wang, T.; Liu, J.; Luo, Y.; Yu, B.; Kong, X.; Zheng, P.; Huang, Z.; Mao, X.; Yu, J.; Luo, J.; et al. Combined Effects of Host Genetics and Diet on Porcine Intestinal Fungi and Their Pathogenic Genes. Front. Microbiol. 2023, 14, 1192288. [Google Scholar] [CrossRef]
- Kong, Q.; Liu, S.; Li, A.; Wang, Y.; Zhang, L.; Iqbal, M.; Jamil, T.; Shang, Z.; Suo, L.; Li, J. Characterization of Fungal Microbial Diversity in Healthy and Diarrheal Tibetan Piglets. BMC Microbiol. 2021, 21, 204. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yu, B.; He, J.; Huang, Z.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; Tian, G.; et al. The Fungal Community and Its Interaction with the Concentration of Short-Chain Fatty Acids in the Faeces of Chenghua, Yorkshire and Tibetan Pigs. Microb. Biotechnol. 2020, 13, 509–521. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yu, B.; He, J.; Huang, Z.; Zheng, P.; Mao, X.; Li, H.; Yu, J.; Luo, J.; et al. Batch and Sampling Time Exert a Larger Influence on the Fungal Community than Gastrointestinal Location in Model Animals: A Meaningful Case Study. Front. Nutr. 2022, 9, 1021215. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, J.; Hou, Q.; Xu, X.; Ren, J.; Ma, L.; Yan, X. Core-Predominant Gut Fungus Kazachstania Slooffiae Promotes Intestinal Epithelial Glycolysis via Lysine Desuccinylation in Pigs. Microbiome 2023, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Huang, Y.; Yang, S.; Wu, D.; Li, C.; Deng, W.; Zhao, K.; He, Y.; Li, B.; Zhang, G.; et al. Diet, Habitat Environment and Lifestyle Conversion Affect the Gut Microbiomes of Giant Pandas. Sci. Total Environ. 2021, 770, 145316. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhan, M.; Pei, E. Succession of Intestinal Microbial Structure of Giant Pandas (Ailuropoda Melanoleuca) during Different Developmental Stages and Its Correlation with Cellulase Activity. Animals 2021, 11, 2358. [Google Scholar] [CrossRef]
- Farouq, A.A.; Abdullah, D.K.; Hooi-Ling, F.; Abdullah, N. Isolation and Characterization of Coprophilous Cellulolytic Fungi from Asian Elephant (Elephas Maximus) Dung. J. Biol. 2012, 2, 44–51. [Google Scholar]
- Foster, M.L.; Dowd, S.E.; Stephenson, C.; Steiner, J.M.; Suchodolski, J.S. Characterization of the Fungal Microbiome (Mycobiome) in Fecal Samples from Dogs. Vet. Med. Int. 2013, 2013, 658373. [Google Scholar] [CrossRef]
- Tay, D.D.; Siew, S.W.; Shamzir Kamal, S.; Razali, M.N.; Ahmad, H.F. ITS1 Amplicon Sequencing of Feline Gut Mycobiome of Malaysian Local Breeds Using Nanopore Flongle. Arch. Microbiol. 2022, 204, 314. [Google Scholar] [CrossRef]
- Van Tilburg Bernardes, E.; Pettersen, V.K.; Gutierrez, M.W.; Laforest-Lapointe, I.; Jendzjowsky, N.G.; Cavin, J.-B.; Vicentini, F.A.; Keenan, C.M.; Ramay, H.R.; Samara, J.; et al. Intestinal Fungi Are Causally Implicated in Microbiome Assembly and Immune Development in Mice. Nat. Commun. 2020, 11, 2577. [Google Scholar] [CrossRef]
- Li, J.Q.; Li, J.L.; Xie, Y.H.; Wang, Y.; Shen, X.N.; Qian, Y.; Han, J.X.; Chen, Y.X.; Fang, J. Saccharomyces cerevisiae May Serve as a Probiotic in Colorectal Cancer by Promoting Cancer Cell Apoptosis. J. Digest. Dis. 2020, 21, 571–582. [Google Scholar] [CrossRef]
- Luo, Y.; Li, J.; Zhou, H.; Yu, B.; He, J.; Wu, A.; Huang, Z.; Zheng, P.; Mao, X.; Yu, J.; et al. The Nutritional Significance of Intestinal Fungi: Alteration of Dietary Carbohydrate Composition Triggers Colonic Fungal Community Shifts in a Pig Model. Appl. Environ. Microbiol. 2021, 87, e00038. [Google Scholar] [CrossRef]
- Prisnee, T.L.; Rahman, R.; Fouhse, J.M.; Van Kessel, A.G.; Brook, R.K.; Willing, B.P. Tracking the Fecal Mycobiome through the Lifespan of Production Pigs and a Comparison to the Feral Pig. Appl. Environ. Microbiol. 2023, 89, e00977-23. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.; Wang, A.; Yao, Y.; Zhou, Y.; Zhang, S.; Fu, X.; Zhou, J.; Pei, E.; Wang, L. An Amateur Gut Microbial Configuration Formed in Giant Panda for Striving to Digest Cellulose in Bamboo: Systematic Evidence from Intestinal Digestive Enzymes, Functional Genes and Microbial Structures. Front. Microbiol. 2022, 13, 926515. [Google Scholar] [CrossRef]
- Jin, L.; Wu, H.; Li, G.; Yang, S.; Wei, R.; Huang, Y.; Penttinen, P.; Deng, W.; Chen, J.; Han, X.; et al. Gastrointestinal Microbiome, Resistance Genes, and Risk Assessment of Heavy Metals in Wild Giant Pandas. Sci. Total Environ. 2023, 899, 165671. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.E.; Schennink, A.; Burden, F.; Long, S.; Van Doorn, D.A.; Pellikaan, W.F.; Dijkstra, J.; Saccenti, E.; Smidt, H. Domesticated Equine Species and Their Derived Hybrids Differ in Their Fecal Microbiota. Anim. Microbiome 2020, 2, 8. [Google Scholar] [CrossRef]
- Edwards, J.E.; Shetty, S.A.; Van Den Berg, P.; Burden, F.; Van Doorn, D.A.; Pellikaan, W.F.; Dijkstra, J.; Smidt, H. Multi-Kingdom Characterization of the Core Equine Fecal Microbiota Based on Multiple Equine (Sub) Species. Anim. Microbiome 2020, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, B.; Gao, X.; Shi, X.; Wang, X.; Wang, T.; Wang, Y.; Liu, G.; Wang, C. Dynamic Changes in Fecal Microbiota in Donkey Foals during Weaning: From Pre-Weaning to Post-Weaning. Front. Microbiol. 2023, 14, 1105330. [Google Scholar] [CrossRef]
- Tropini, C. How the Physical Environment Shapes the Microbiota. mSystems 2021, 6, e00675. [Google Scholar] [CrossRef]
- Ng, K.M.; Pannu, S.; Liu, S.; Burckhardt, J.C.; Hughes, T.; Van Treuren, W.; Nguyen, J.; Naqvi, K.; Nguyen, B.; Clayton, C.A.; et al. Single-Strain Behavior Predicts Responses to Environmental pH and Osmolality in the Gut Microbiota. mBio 2023, 14, e00753-23. [Google Scholar] [CrossRef]
- Lkhagva, E.; Chung, H.-J.; Hong, J.; Tang, W.H.W.; Lee, S.-I.; Hong, S.-T.; Lee, S. The Regional Diversity of Gut Microbiome along the GI Tract of Male C57BL/6 Mice. BMC Microbiol. 2021, 21, 44. [Google Scholar] [CrossRef]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut Microbiome and Health: Mechanistic Insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Hooda, S.; Metzler-Zebeli, B.U.; Vasanthan, T.; Zijlstra, R.T. Effects of Viscosity and Fermentability of Dietary Fibre on Nutrient Digestibility and Digesta Characteristics in Ileal-Cannulated Grower Pigs. Br. J. Nutr. 2011, 106, 664–674. [Google Scholar] [CrossRef]
- Thangamani, S.; Monasky, R.; Lee, J.K.; Antharam, V.; HogenEsch, H.; Hazbun, T.R.; Jin, Y.; Gu, H.; Guo, G.L. Bile Acid Regulates the Colonization and Dissemination of Candida Albicans from the Gastrointestinal Tract by Controlling Host Defense System and Microbiota. JoF 2021, 7, 1030. [Google Scholar] [CrossRef] [PubMed]
- Pierre, J.F.; Peters, B.M.; La Torre, D.; Sidebottom, A.M.; Tao, Y.; Zhu, X.; Cham, C.M.; Wang, L.; Kambal, A.; Harris, K.G.; et al. Peptide YY: A Paneth Cell Antimicrobial Peptide That Maintains Candida Gut Commensalism. Science 2023, 381, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Arfken, A.M.; Frey, J.F.; Ramsay, T.G.; Summers, K.L. Yeasts of Burden: Exploring the Mycobiome–Bacteriome of the Piglet GI Tract. Front. Microbiol. 2019, 10, 2286. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The Role of the Gut Microbiota in Nutrition and Health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Kiela, P.R.; Ghishan, F.K. Physiology of Intestinal Absorption and Secretion. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Guan, G.; Li, S.; Bing, J.; Liu, L.; Tao, L. The Rfg1 and Bcr1 Transcription Factors Regulate Acidic pH–Induced Filamentous Growth in Candida albicans. Microbiol. Spectr. 2023, 11, e01789-23. [Google Scholar] [CrossRef]
- Von Rosenvinge, E.C.; Song, Y.; White, J.R.; Maddox, C.; Blanchard, T.; Fricke, W.F. Immune Status, Antibiotic Medication and pH Are Associated with Changes in the Stomach Fluid Microbiota. ISME J. 2013, 7, 1354–1366. [Google Scholar] [CrossRef]
- McCallum, G.; Tropini, C. The Gut Microbiota and Its Biogeography. Nat. Rev. Microbiol. 2024, 22, 105–118. [Google Scholar] [CrossRef]
- Askew, C.; Sellam, A.; Epp, E.; Hogues, H.; Mullick, A.; Nantel, A.; Whiteway, M. Transcriptional Regulation of Carbohydrate Metabolism in the Human Pathogen Candida Albicans. PLoS Pathog. 2009, 5, e1000612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Ai, Y.-H.; Xu, Y.; Yu, X.-W. High-Level Expression of Aspergillus Niger Lipase in Pichia Pastoris: Characterization and Gastric Digestion in Vitro. Food Chem. 2019, 274, 305–313. [Google Scholar] [CrossRef]
- Barig, S.; Alisch, R.; Nieland, S.; Wuttke, A.; Gräser, Y.; Huddar, M.; Schnitzlein, K.; Stahmann, K. Monoseptic Growth of Fungal Lipase Producers under Minimized Sterile Conditions: Cultivation of Phialemonium curvatum in 350 L Scale. Eng. Life Sci. 2011, 11, 387–394. [Google Scholar] [CrossRef]
- Petri, D.; Hill, J.E.; Van Kessel, A.G. Microbial Succession in the Gastrointestinal Tract (GIT) of the Preweaned Pig. Livest. Sci. 2010, 133, 107–109. [Google Scholar] [CrossRef]
- Urubschurov, V.; Büsing, K.; Souffrant, W.-B.; Schauer, N.; Zeyner, A. Porcine Intestinal Yeast Species, Kazachstania Slooffiae, a New Potential Protein Source with Favourable Amino Acid Composition for Animals. J. Anim. Physiol. Anim. Nutr. 2018, 102, e892–e901. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, F.; Yang, X.; Wu, N.; Jiang, W.; Li, X.; Li, X.; Liu, Y. Changes in the Composition of Intestinal Fungi and Their Role in Mice with Dextran Sulfate Sodium-Induced Colitis. Sci. Rep. 2015, 5, 10416. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Chandra, J.; Retuerto, M.; Sikaroodi, M.; Brown, R.E.; Jurevic, R.; Salata, R.A.; Lederman, M.M.; Gillevet, P.M.; Ghannoum, M.A. Oral Mycobiome Analysis of HIV-Infected Patients: Identification of Pichia as an Antagonist of Opportunistic Fungi. PLoS Pathog. 2014, 10, e1003996. [Google Scholar] [CrossRef]
- Groestlinger, J.; Spindler, V.; Pahlke, G.; Rychlik, M.; Del Favero, G.; Marko, D. Alternaria alternata Mycotoxins Activate the Aryl Hydrocarbon Receptor and Nrf2-ARE Pathway to Alter the Structure and Immune Response of Colon Epithelial Cells. Chem. Res. Toxicol. 2022, 35, 731–749. [Google Scholar] [CrossRef]
- Solomon, K.V.; Haitjema, C.H.; Henske, J.K.; Gilmore, S.P.; Borges-Rivera, D.; Lipzen, A.; Brewer, H.M.; Purvine, S.O.; Wright, A.T.; Theodorou, M.K.; et al. Early-Branching Gut Fungi Possess a Large, Comprehensive Array of Biomass-Degrading Enzymes. Science 2016, 351, 1192–1195. [Google Scholar] [CrossRef]
- Manici, L.M.; Caputo, F.; De Sabata, D.; Fornasier, F. The Enzyme Patterns of Ascomycota and Basidiomycota Fungi Reveal Their Different Functions in Soil. Appl. Soil Ecol. 2024, 196, 105323. [Google Scholar] [CrossRef]
- Sathiyamoorthi, E.; Dikshit, P.K.; Kumar, P.; Kim, B.S. Co-fermentation of Agricultural and Industrial Waste by Naganishia albida for Microbial Lipid Production in Fed-batch Fermentation. J. Chem. Tech. Biotechnol. 2020, 95, 813–821. [Google Scholar] [CrossRef]
- Yen, H.-W.; Chang, J.-T. Growth of Oleaginous Rhodotorula glutinis in an Internal-Loop Airlift Bioreactor by Using Lignocellulosic Biomass Hydrolysate as the Carbon Source. J. Biosci. Bioeng. 2015, 119, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.-J.; Li, X.-C.; Liu, J.; Zhang, X.-T.; Xin, Z.-Z.; Jiang, W.-W.; Zhang, J.-Y. Efficient Sugar Utilization and High Tolerance to Inhibitors Enable Rhodotorula toruloides C23 to Robustly Produce Lipid and Carotenoid from Lignocellulosic Feedstock. Bioresour. Technol. 2024, 407, 131146. [Google Scholar] [CrossRef] [PubMed]
- Sargeant, L.A.; Mardell, M.; Saad-Allah, K.M.; Hussein, A.H.; Whiffin, F.; Santomauro, F.; Chuck, C.J. Production of Lipid from Depolymerised Lignocellulose Using the Biocontrol Yeast, Rhodotorula minuta: The Fatty Acid Profile Remains Stable Irrespective of Environmental Conditions. Eur. J. Lipid. Sci. Technol. 2016, 118, 777–787. [Google Scholar] [CrossRef]
- Stanforth, K.J.; Wilcox, M.D.; Chater, P.I.; Brownlee, I.A.; Zakhour, M.I.; Banecki, K.M.R.M.; Pearson, J.P. Pepsin Properties, Structure, and Its Accurate Measurement: A Narrative Review. Ann. Esophagus 2022, 5, 31. [Google Scholar] [CrossRef]
- Verma, A.H.; Richardson, J.P.; Zhou, C.; Coleman, B.M.; Moyes, D.L.; Ho, J.; Huppler, A.R.; Ramani, K.; McGeachy, M.J.; Mufazalov, I.A.; et al. Oral Epithelial Cells Orchestrate Innate Type 17 Responses to Candida albicans through the Virulence Factor Candidalysin. Sci. Immunol. 2017, 2, eaam8834. [Google Scholar] [CrossRef]
- Doron, I.; Mesko, M.; Li, X.V.; Kusakabe, T.; Leonardi, I.; Shaw, D.G.; Fiers, W.D.; Lin, W.-Y.; Bialt-DeCelie, M.; Román, E.; et al. Mycobiota-Induced IgA Antibodies Regulate Fungal Commensalism in the Gut and Are Dysregulated in Crohn’s Disease. Nat. Microbiol. 2021, 6, 1493–1504. [Google Scholar] [CrossRef]
- Wlazło, Ł.; Kowalska, D.; Bielański, P.; Chmielowiec-Korzeniowska, A.; Ossowski, M.; Łukaszewicz, M.; Czech, A.; Nowakowicz-Dębek, B. Effect of Fermented Rapeseed Meal on the Gastrointestinal Microbiota and Immune Status of Rabbit (Oryctolagus Cuniculus). Animals 2021, 11, 716. [Google Scholar] [CrossRef]
- van der Merwe, M.; Sharma, S.; Caldwell, J.L.; Smith, N.J.; Gomes, C.K.; Bloomer, R.J.; Buddington, R.K.; Pierre, J.F. Time of Feeding Alters Obesity-Associated Parameters and Gut Bacterial Communities, but Not Fungal Populations, in C57BL/6 Male Mice. Curr. Dev. Nutr. 2020, 4, nzz145. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Mao, B.; Yang, Q.; Zhao, J.; Gu, Z.; Zhang, H.; Chen, Y.Q.; Chen, W. Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci. Rep. 2017, 7, 45840. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, L.; Lei, Y.L.; Ren, B.; Zhou, X. The Interactions Between Candida Albicans and Mucosal Immunity. Front. Microbiol. 2021, 12, 652725. [Google Scholar] [CrossRef]
- Eckstein, M.-T.; Moreno-Velásquez, S.D.; Pérez, J.C. Gut Bacteria Shape Intestinal Microhabitats Occupied by the Fungus Candida Albicans. Curr. Biol. 2020, 30, 4799–4807.e4. [Google Scholar] [CrossRef]
- Boutin, R.C.; Petersen, C.; Woodward, S.E.; Serapio-Palacios, A.; Bozorgmehr, T.; Loo, R.; Chalanuchpong, A.; Cirstea, M.; Lo, B.; Huus, K.E.; et al. Bacterial–Fungal Interactions in the Neonatal Gut Influence Asthma Outcomes Later in Life. eLife 2021, 10, e67740. [Google Scholar] [CrossRef] [PubMed]
- Auchtung, T.A.; Stewart, C.J.; Smith, D.P.; Triplett, E.W.; Agardh, D.; Hagopian, W.A.; Ziegler, A.G.; Rewers, M.J.; She, J.-X.; Toppari, J.; et al. Temporal Changes in Gastrointestinal Fungi and the Risk of Autoimmunity during Early Childhood: The TEDDY Study. Nat. Commun. 2022, 13, 3151. [Google Scholar] [CrossRef]
- Arfken, A.M.; Frey, J.F.; Summers, K.L. Temporal Dynamics of the Gut Bacteriome and Mycobiome in the Weanling Pig. Microorganisms 2020, 8, 868. [Google Scholar]
- Spatz, M.; Da Costa, G.; Ventin-Holmberg, R.; Planchais, J.; Michaudel, C.; Wang, Y.; Danne, C.; Lapiere, A.; Michel, M.-L.; Kolho, K.-L.; et al. Antibiotic Treatment Using Amoxicillin-Clavulanic Acid Impairs Gut Mycobiota Development through Modification of the Bacterial Ecosystem. Microbiome 2023, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Nel Van Zyl, K.; Whitelaw, A.C.; Hesseling, A.C.; Seddon, J.A.; Demers, A.-M.; Newton-Foot, M. Fungal Diversity in the Gut Microbiome of Young South African Children. BMC Microbiol. 2022, 22, 201. [Google Scholar] [CrossRef]
- Al Bataineh, M.T.; Dash, N.R.; Bel Lassen, P.; Banimfreg, B.H.; Nada, A.M.; Belda, E.; Clément, K. Revealing Links between Gut Microbiome and Its Fungal Community in Type 2 Diabetes Mellitus among Emirati Subjects: A Pilot Study. Sci. Rep. 2020, 10, 9624. [Google Scholar] [CrossRef] [PubMed]
- Summers, K.L.; Frey, J.F.; Ramsay, T.G.; Arfken, A.M. The Piglet Mycobiome during the Weaning Transition: A Pilot Study1. J. Anim. Sci. 2019, 97, 2889–2900. [Google Scholar] [CrossRef]
- Zhan, M.; Wang, L.; Xie, C.; Fu, X.; Zhang, S.; Wang, A.; Zhou, Y.; Xu, C.; Zhang, H. Succession of Gut Microbial Structure in Twin Giant Pandas During the Dietary Change Stage and Its Role in Polysaccharide Metabolism. Front. Microbiol. 2020, 11, 551038. [Google Scholar] [CrossRef]
- Sun, Y.; Zuo, T.; Cheung, C.P.; Gu, W.; Wan, Y.; Zhang, F.; Chen, N.; Zhan, H.; Yeoh, Y.K.; Niu, J.; et al. Population-Level Configurations of Gut Mycobiome Across 6 Ethnicities in Urban and Rural China. Gastroenterology 2021, 160, 272–286. [Google Scholar] [CrossRef]
- Martínez-Romero, E.; Aguirre-Noyola, J.L.; Bustamante-Brito, R.; González-Román, P.; Hernández-Oaxaca, D.; Higareda-Alvear, V.; Montes-Carreto, L.M.; Martínez-Romero, J.C.; Rosenblueth, M.; Servín-Garcidueñas, L.E. We and Herbivores Eat Endophytes. Microb. Biotechnol. 2021, 14, 1282–1299. [Google Scholar] [CrossRef] [PubMed]
- James, S.A.; Phillips, S.; Telatin, A.; Baker, D.; Ansorge, R.; Clarke, P.; Hall, L.J.; Carding, S.R. Preterm Infants Harbour a Rapidly Changing Mycobiota That Includes Candida Pathobionts. J. Fungi 2020, 6, 273. [Google Scholar] [CrossRef] [PubMed]
- Koechli, C.; Campbell, A.N.; Pepe-Ranney, C.; Buckley, D.H. Assessing Fungal Contributions to Cellulose Degradation in Soil by Using High-Throughput Stable Isotope Probing. Soil Biol. Biochem. 2019, 130, 150–158. [Google Scholar] [CrossRef]
- Bai, Y.; Zhou, X.; Li, N.; Zhao, J.; Ye, H.; Zhang, S.; Yang, H.; Pi, Y.; Tao, S.; Han, D.; et al. In Vitro Fermentation Characteristics and Fiber-Degrading Enzyme Kinetics of Cellulose, Arabinoxylan, β-Glucan and Glucomannan by Pig Fecal Microbiota. Microorganisms 2021, 9, 1071. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, D.; Weinstock, A.; Antharam, V.C.; Gu, H.; Jasbi, P.; Shi, X.; Dirks, B.; Krajmalnik-Brown, R.; Maldonado, J.; Guinan, J.; et al. Antibiotic-Induced Gut Metabolome and Microbiome Alterations Increase the Susceptibility to Candida Albicans Colonization in the Gastrointestinal Tract. FEMS Microbiol. Ecol. 2019, 96, fiz187. [Google Scholar] [CrossRef]
- Guinan, J.; Wang, S.; Hazbun, T.R.; Yadav, H.; Thangamani, S. Antibiotic-Induced Decreases in the Levels of Microbial-Derived Short-Chain Fatty Acids Correlate with Increased Gastrointestinal Colonization of Candida Albicans. Sci. Rep. 2019, 9, 8872. [Google Scholar] [CrossRef]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Miramón, P.; Lorenz, M.C. The SPS Amino Acid Sensor Mediates Nutrient Acquisition and Immune Evasion in Candida Albicans: Nutrient Sensing and Immune Evasion in C. Albicans. Cell. Microbiol. 2016, 18, 1611–1624. [Google Scholar] [CrossRef]
- Patra, A.K.; Kar, I. Heat Stress on Microbiota Composition, Barrier Integrity, and Nutrient Transport in Gut, Production Performance, and Its Amelioration in Farm Animals. J. Anim. Sci. Technol. 2021, 63, 211–247. [Google Scholar] [CrossRef]
- Iliev, I.D.; Leonardi, I. Fungal Dysbiosis: Immunity and Interactions at Mucosal Barriers. Nat. Rev. Immunol. 2017, 17, 635–646. [Google Scholar] [CrossRef]
- Martini, G.R.; Tikhonova, E.; Rosati, E.; DeCelie, M.B.; Sievers, L.K.; Tran, F.; Lessing, M.; Bergfeld, A.; Hinz, S.; Nikolaus, S.; et al. Selection of Cross-Reactive T Cells by Commensal and Food-Derived Yeasts Drives Cytotoxic TH1 Cell Responses in Crohn’s Disease. Nat. Med. 2023, 29, 2602–2614. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, H.; Li, K.-D.; Wang, Y.-Y.; Huang, R.-G.; Du, Y.-J.; Jin, X.; Zhang, Q.-R.; Li, X.-B.; Li, B.-Z. Intestinal Fungi and Systemic Autoimmune Diseases. Autoimmun. Rev. 2023, 22, 103234. [Google Scholar] [CrossRef] [PubMed]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377. [Google Scholar] [CrossRef]
- Bacher, P.; Hohnstein, T.; Beerbaum, E.; Röcker, M.; Blango, M.G.; Kaufmann, S.; Röhmel, J.; Eschenhagen, P.; Grehn, C.; Seidel, K.; et al. Human Anti-Fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida Albicans. Cell 2019, 176, 1340–1355. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin Is a Fungal Peptide Toxin Critical for Mucosal Infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef]
- Leonardi, I.; Li, X.; Semon, A.; Li, D.; Doron, I.; Putzel, G.; Bar, A.; Prieto, D.; Rescigno, M.; McGovern, D.P.B.; et al. CX3CR1+ Mononuclear Phagocytes Control Immunity to Intestinal Fungi. Science 2018, 359, 232–236. [Google Scholar] [CrossRef]
- Richardson, J.P.; Willems, H.M.E.; Moyes, D.L.; Shoaie, S.; Barker, K.S.; Tan, S.L.; Palmer, G.E.; Hube, B.; Naglik, J.R.; Peters, B.M. Candidalysin Drives Epithelial Signaling, Neutrophil Recruitment, and Immunopathology at the Vaginal Mucosa. Infect. Immun. 2018, 86, e00645-17. [Google Scholar] [CrossRef]
- Lv, Q.-Z.; Li, D.-D.; Han, H.; Yang, Y.-H.; Duan, J.-L.; Ma, H.-H.; Yu, Y.; Chen, J.-Y.; Jiang, Y.-Y.; Jia, X.-M. Priming with FLO8-Deficient Candida Albicans Induces Th1-Biased Protective Immunity against Lethal Polymicrobial Sepsis. Cell Mol. Immunol. 2021, 18, 2010–2023. [Google Scholar] [CrossRef]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida Albicans Cell-Type Switching and Functional Plasticity in the Mammalian Host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef]
- Huo, X.; Li, D.; Wu, F.; Li, S.; Qiao, Y.; Wang, C.; Wang, Y.; Zhou, C.; Sun, L.; Luan, Z.; et al. Cultivated Human Intestinal Fungus Candida Metapsilosis M2006B Attenuates Colitis by Secreting Acyclic Sesquiterpenoids as FXR Agonists. Gut 2022, 71, 2205–2217. [Google Scholar] [CrossRef]
- Standaert–Vitse, A.; Jouault, T.; Vandewalle, P.; Mille, C.; Seddik, M.; Sendid, B.; Mallet, J.; Colombel, J.; Poulain, D. Candida Albicans Is an Immunogen for Anti–Saccharomyces Cerevisiae Antibody Markers of Crohn’s Disease. Gastroenterology 2006, 130, 1764–1775. [Google Scholar] [CrossRef] [PubMed]
- Kasper, L.; König, A.; Koenig, P.-A.; Gresnigt, M.S.; Westman, J.; Drummond, R.A.; Lionakis, M.S.; Groß, O.; Ruland, J.; Naglik, J.R.; et al. The Fungal Peptide Toxin Candidalysin Activates the NLRP3 Inflammasome and Causes Cytolysis in Mononuclear Phagocytes. Nat. Commun. 2018, 9, 4260. [Google Scholar] [CrossRef]
- Panpetch, W.; Sawaswong, V.; Chanchaem, P.; Ondee, T.; Dang, C.P.; Payungporn, S.; Tumwasorn, S.; Leelahavanichkul, A. Candida Administration Worsens Cecal Ligation and Puncture-Induced Sepsis in Obese Mice Through Gut Dysbiosis Enhanced Systemic Inflammation, Impact of Pathogen-Associated Molecules From Gut Translocation and Saturated Fatty Acid. Front. Immunol. 2020, 11, 561652. [Google Scholar] [CrossRef]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary Gut Microbial Metabolites, Short-Chain Fatty Acids, and Host Metabolic Regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef]
- Sun, S.; Sun, L.; Wang, K.; Qiao, S.; Zhao, X.; Hu, X.; Chen, W.; Zhang, S.; Li, H.; Dai, H.; et al. The Gut Commensal Fungus, Candida Parapsilosis, Promotes High Fat-Diet Induced Obesity in Mice. Commun. Biol. 2021, 4, 1220. [Google Scholar] [CrossRef]
- Gryaznova, M.; Smirnova, Y.; Burakova, I.; Morozova, P.; Nesterova, E.; Gladkikh, M.; Mikhaylov, E.; Syromyatnikov, M. Characteristics of the Fecal Microbiome of Piglets with Diarrhea Identified Using Shotgun Metagenomics Sequencing. Animals 2023, 13, 2303. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular Mechanisms of Antibiotic Resistance Revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Seelbinder, B.; Chen, J.; Brunke, S.; Vazquez-Uribe, R.; Santhaman, R.; Meyer, A.-C.; De Oliveira Lino, F.S.; Chan, K.-F.; Loos, D.; Imamovic, L.; et al. Antibiotics Create a Shift from Mutualism to Competition in Human Gut Communities with a Longer-Lasting Impact on Fungi than Bacteria. Microbiome 2020, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Drummond, R.A.; Desai, J.V.; Ricotta, E.E.; Swamydas, M.; Deming, C.; Conlan, S.; Quinones, M.; Matei-Rascu, V.; Sherif, L.; Lecky, D.; et al. Long-Term Antibiotic Exposure Promotes Mortality after Systemic Fungal Infection by Driving Lymphocyte Dysfunction and Systemic Escape of Commensal Bacteria. Cell Host Microbe 2022, 30, 1020–1033. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, Y.; Yuan, Y.; Xie, Y. A Systematic Review on Antibiotics Misuse in Livestock and Aquaculture and Regulation Implications in China. Sci. Total Environ. 2021, 798, 149205. [Google Scholar] [CrossRef]
- Dewulf, J.; Joosten, P.; Chantziaras, I.; Bernaerdt, E.; Vanderhaeghen, W.; Postma, M.; Maes, D. Antibiotic Use in European Pig Production: Less Is More. Antibiotics 2022, 11, 1493. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.A.; Arenas, N.E.; Luiza, V.L.; Bermudez, J.A.Z.; Clarke, S.E. Regulations on the Use of Antibiotics in Livestock Production in South America: A Comparative Literature Analysis. Antibiotics 2023, 12, 1303. [Google Scholar] [CrossRef] [PubMed]
- Ventin-Holmberg, R.; Saqib, S.; Korpela, K.; Nikkonen, A.; Peltola, V.; Salonen, A.; De Vos, W.M.; Kolho, K.-L. The Effect of Antibiotics on the Infant Gut Fungal Microbiota. JoF 2022, 8, 328. [Google Scholar] [CrossRef]
- Dollive, S.; Chen, Y.-Y.; Grunberg, S.; Bittinger, K.; Hoffmann, C.; Vandivier, L.; Cuff, C.; Lewis, J.D.; Wu, G.D.; Bushman, F.D. Fungi of the Murine Gut: Episodic Variation and Proliferation during Antibiotic Treatment. PLoS ONE 2013, 8, e71806. [Google Scholar] [CrossRef] [PubMed]
- Hedin, K.A.; Rees, V.E.; Zhang, H.; Kruse, V.; Vazquez-Uribe, R.; Sommer, M.O.A. Effects of Broad-Spectrum Antibiotics on the Colonisation of Probiotic Yeast Saccharomyces Boulardii in the Murine Gastrointestinal Tract. Sci. Rep. 2022, 12, 8862. [Google Scholar] [CrossRef]
- Tan, C.T.; Xu, X.; Qiao, Y.; Wang, Y. A Peptidoglycan Storm Caused by β-Lactam Antibiotic’s Action on Host Microbiota Drives Candida Albicans Infection. Nat. Commun. 2021, 12, 2560. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Y.; Tu, J.; Yang, W.; Liu, N.; Wang, W.; Sheng, C. Discovery of BRD4–HDAC Dual Inhibitors with Improved Fungal Selectivity and Potent Synergistic Antifungal Activity against Fluconazole-Resistant Candida Albicans. J. Med. Chem. 2023, 66, 5950–5964. [Google Scholar] [CrossRef]
- Richard, M.L.; Sokol, H. The Gut Mycobiota: Insights into Analysis, Environmental Interactions and Role in Gastrointestinal Diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef]
- Zohair, M.M.; Dongmei, W.; Shimizu, K. Metabolic Picture of Microbial Interaction: Chemical Crosstalk during Co-Cultivation between Three Dominant Genera of Bacteria and Fungi in Medicinal Plants Rhizosphere. Metabolomics 2024, 20, 75. [Google Scholar] [CrossRef]
- Mirhakkak, M.H.; Schäuble, S.; Klassert, T.E.; Brunke, S.; Brandt, P.; Loos, D.; Uribe, R.V.; Senne De Oliveira Lino, F.; Ni, Y.; Vylkova, S.; et al. Metabolic Modeling Predicts Specific Gut Bacteria as Key Determinants for Candida albicans Colonization Levels. ISME J. 2021, 15, 1257–1270. [Google Scholar] [CrossRef]
- Charlet, R.; Pruvost, Y.; Tumba, G.; Istel, F.; Poulain, D.; Kuchler, K.; Sendid, B.; Jawhara, S. Remodeling of the Candida Glabrata Cell Wall in the Gastrointestinal Tract Affects the Gut Microbiota and the Immune Response. Sci. Rep. 2018, 8, 3316. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-N.; Jiao, N.; Tan, J.-C.; Wang, Z.; Wu, D.; Wang, A.-J.; Chen, J.; Tao, L.; Zhou, C.; Fang, W.; et al. Multi-Kingdom Microbiota Analyses Identifybacterial–Fungal Interactions and Biomarkers of Colorectal Cancer Acrosscohorts. Nat. Microbiol. 2022, 7, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G. Bacteriophage Therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 2019, 25, 803–814. [Google Scholar] [CrossRef]
- Pei, Z.; Sadiq, F.A.; Han, X.; Zhao, J.; Zhang, H.; Ross, R.P.; Lu, W.; Chen, W. Comprehensive Scanning of Prophages in Lactobacillus: Distribution, Diversity, Antibiotic Resistance Genes, and Linkages with CRISPR-Cas Systems. mSystems 2021, 6, e01211-20. [Google Scholar] [CrossRef]
- Mottawea, W.; Duceppe, M.-O.; Dupras, A.A.; Usongo, V.; Jeukens, J.; Freschi, L.; Emond-Rheault, J.-G.; Hamel, J.; Kukavica-Ibrulj, I.; Boyle, B.; et al. Salmonella Enterica Prophage Sequence Profiles Reflect Genome Diversity and Can Be Used for High Discrimination Subtyping. Front. Microbiol. 2018, 9, 836. [Google Scholar] [CrossRef]
- Rezaei Javan, R.; Ramos-Sevillano, E.; Akter, A.; Brown, J.; Brueggemann, A.B. Prophages and Satellite Prophages Are Widespread in Streptococcus and May Play a Role in Pneumococcal Pathogenesis. Nat. Commun. 2019, 10, 4852. [Google Scholar] [CrossRef]
- Sekeresova Kralova, J.; Donic, C.; Dassa, B.; Livyatan, I.; Jansen, P.M.; Ben-Dor, S.; Fidel, L.; Trzebanski, S.; Narunsky-Haziza, L.; Asraf, O.; et al. Competitive Fungal Commensalism Mitigates Candidiasis Pathology. J. Exp. Med. 2024, 221, e20231686. [Google Scholar] [CrossRef]
- Kotta-Loizou, I. Mycoviruses and Their Role in Fungal Pathogenesis. Curr. Opin. Microbiol. 2021, 63, 10–18. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus Years of Fungal Viruses. Virology 2015, 479–480, 356–368. [Google Scholar] [CrossRef]
- Lemus-Minor, C.G.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Fusarium Oxysporum f. Sp. Dianthi Virus 1 Accumulation Is Correlated with Changes in Virulence and Other Phenotypic Traits of Its Fungal Host. Phytopathology 2018, 108, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Crucitti, D.; Chiapello, M.; Oliva, D.; Forgia, M.; Turina, M.; Carimi, F.; La Bella, F.; Pacifico, D. Identification and Molecular Characterization of Novel Mycoviruses in Saccharomyces and Non-Saccharomyces Yeasts of Oenological Interest. Viruses 2021, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Taggart, N.T.; Crabtree, A.M.; Creagh, J.W.; Bizarria, R.; Li, S.; De La Higuera, I.; Barnes, J.E.; Shipley, M.A.; Boyer, J.M.; Stedman, K.M.; et al. Novel Viruses of the Family Partitiviridae Discovered in Saccharomyces Cerevisiae. PLoS Pathog. 2023, 19, e1011418. [Google Scholar] [CrossRef] [PubMed]
- Nibert, M.L.; Ghabrial, S.A.; Maiss, E.; Lesker, T.; Vainio, E.J.; Jiang, D.; Suzuki, N. Taxonomic Reorganization of Family Partitiviridae and Other Recent Progress in Partitivirus Research. Virus Res. 2014, 188, 128–141. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Van Der Giezen, M. Associations between Gut Microbiota and Common Luminal Intestinal Parasites. Trends Parasitol. 2018, 34, 369–377. [Google Scholar] [CrossRef]
- Aivelo, T.; Norberg, A. Parasite–Microbiota Interactions Potentially Affect Intestinal Communities in Wild Mammals. J. Anim. Ecol. 2018, 87, 438–447. [Google Scholar] [CrossRef]
- Duarte, A.M.; Jenkins, T.P.; Latrofa, M.S.; Giannelli, A.; Papadopoulos, E.; De Carvalho, L.M.; Nolan, M.J.; Otranto, D.; Cantacessi, C. Helminth Infections and Gut Microbiota—A Feline Perspective. Parasites Vectors 2016, 9, 625. [Google Scholar] [CrossRef]
- Partida-Rodríguez, O.; Serrano-Vázquez, A.; Nieves-Ramírez, M.E.; Moran, P.; Rojas, L.; Portillo, T.; González, E.; Hernández, E.; Finlay, B.B.; Ximenez, C. Human Intestinal Microbiota: Interaction Between Parasites and the Host Immune Response. Arch. Med. Res. 2017, 48, 690–700. [Google Scholar] [CrossRef]
- Grondin, J.A.; Jamal, A.; Mowna, S.; Seto, T.; Khan, W.I. Interaction between Intestinal Parasites and the Gut Microbiota: Implications for the Intestinal Immune Response and Host Defence. Pathogens 2024, 13, 608. [Google Scholar] [CrossRef]
- Leung, J.M.; Graham, A.L.; Knowles, S.C.L. Parasite-Microbiota Interactions With the Vertebrate Gut: Synthesis Through an Ecological Lens. Front. Microbiol. 2018, 9, 843. [Google Scholar] [CrossRef]
- Barelli, C.; Donati, C.; Albanese, D.; Pafčo, B.; Modrý, D.; Rovero, F.; Hauffe, H.C. Interactions between Parasitic Helminths and Gut Microbiota in Wild Tropical Primates from Intact and Fragmented Habitats. Sci. Rep. 2021, 11, 21569. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.A.; Smith, K.A.; Filbey, K.J.; Harcus, Y.; Hewitson, J.P.; Redpath, S.A.; Valdez, Y.; Yebra, M.J.; Finlay, B.B.; Maizels, R.M. Commensal-Pathogen Interactions in the Intestinal Tract: Lactobacilli Promote Infection with, and Are Promoted by, Helminth Parasites. Gut Microbes 2014, 5, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mondal, C.; Mukherjee, T.; Saha, S.; Kundu, A.; Ghosh, S.; Lyndem, L.M. Hymenolepis diminuta Reduce Lactic Acid Bacterial Load and Induce Dysbiosis in the Early Infection of the Probiotic Colonization of Swiss Albino Rat. Microorganisms 2022, 10, 2328. [Google Scholar] [CrossRef] [PubMed]
- Yason, J.A.; Liang, Y.R.; Png, C.W.; Zhang, Y.; Tan, K.S.W. Interactions between a Pathogenic Blastocystis Subtype and Gut Microbiota: In Vitro and in Vivo Studies. Microbiome 2019, 7, 30. [Google Scholar] [CrossRef]
- Guan, W.; Xu, D.; Yang, S.; Zhao, Y.; Xie, Y.; Lin, M.; Liu, Y.; Zheng, Y.; Li, J. Observation of Intestinal Flora Diversity with the Parasites Infection Process in a Nonlethal Malaria Model of BALB/c Mice Induced by Plasmodium Yoelii 17XNL Strain. Decod. Infect. Transm. 2023, 1, 100004. [Google Scholar] [CrossRef]
- Boutin, R.C.T.; Sbihi, H.; McLaughlin, R.J.; Hahn, A.S.; Konwar, K.M.; Loo, R.S.; Dai, D.; Petersen, C.; Brinkman, F.S.L.; Winsor, G.L.; et al. Composition and Associations of the Infant Gut Fungal Microbiota with Environmental Factors and Childhood Allergic Outcomes. mBio 2021, 12, e03396-20. [Google Scholar] [CrossRef]
- Burden, F.; Thiemann, A. Donkeys Are Different. J. Equine Vet. Sci. 2015, 35, 376–382. [Google Scholar] [CrossRef]
- Tang, Q.; Huang, K.; Liu, J.; Shen, D.; Dai, P.; Li, Y.; Li, C. Seasonal Variations of Microbial Assemblage in Fine Particulate Matter from a Nursery Pig House. Sci. Total Environ. 2020, 708, 134921. [Google Scholar] [CrossRef]
- Masunga, G.S.; Andresen, Ø.; Taylor, J.E.; Dhillion, S.S. Elephant Dung Decomposition and Coprophilous Fungi in Two Habitats of Semi-Arid Botswana. Mycol. Res. 2006, 110, 1214–1226. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Buddington, K.K.; Donahoo, J.B.; Buddington, R.K. Dietary Oligofructose and Inulin Protect Mice from Enteric and Systemic Pathogens and Tumor Inducers. J. Nutr. 2002, 132, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Chantanawilas, P.; Pahumunto, N.; Thananimit, S.; Teanpaisan, R. Anticandidal Activity of Various Probiotic Lactobacillus Strains and Their Efficacy Enhanced by Prebiotic Supplementation. Curr. Microbiol. 2024, 81, 271. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, A.; Tester, R.; Al-Ghazzewi, F.; Mcculloch, E.; Connolly, M. Glucomannan Hydrolysate (GMH) Inhibition of Candida Albicans Growth in the Presence of Lactobacillus and Lactococcus Species. Microb. Ecol. Health Dis. 2008, 20, 127–134. [Google Scholar] [CrossRef]
- García-Gamboa, R.; Domínguez-Simi, M.Á.; Gradilla-Hernández, M.S.; Bravo-Madrigal, J.; Moya, A.; González-Avila, M. Antimicrobial and Antibiofilm Effect of Inulin-Type Fructans, Used in Synbiotic Combination with Lactobacillus spp. Against Candida Albicans. Plant Foods Hum. Nutr. 2022, 77, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Lange, L.; Barrett, K.; Pilgaard, B.; Gleason, F.; Tsang, A. Enzymes of Early-Diverging, Zoosporic Fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6885–6902. [Google Scholar] [CrossRef]
- Urubschurov, V.; Herlemann, D.P.R.; Souffrant, W.-B.; Zeyner, A. New Insights into the Role of the Porcine Intestinal Yeast, Kazachstania slooffiae, in Intestinal Environment of Weaned Piglets. FEMS Microbiol. Ecol. 2017, 93, fiw245. [Google Scholar] [CrossRef]
- Vicente, G.; Bautista, L.F.; Rodríguez, R.; Gutiérrez, F.J.; Sádaba, I.; Ruiz-Vázquez, R.M.; Torres-Martínez, S.; Garre, V. Biodiesel Production from Biomass of an Oleaginous Fungus. Biochem. Eng. J. 2009, 48, 22–27. [Google Scholar] [CrossRef]
- Amend, A. From Dandruff to Deep-Sea Vents: Malassezia-like Fungi Are Ecologically Hyper-Diverse. PLoS Pathog. 2014, 10, e1004277. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Trauner, M. Role of Bile Acids and Their Receptors in Gastrointestinal and Hepatic Pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 432–450. [Google Scholar] [CrossRef]
- Kollerov, V.V.; Lobastova, T.G.; Monti, D.; Deshcherevskaya, N.O.; Ferrandi, E.E.; Fronza, G.; Riva, S.; Donova, M.V. Deoxycholic Acid Transformations Catalyzed by Selected Filamentous Fungi. Steroids 2016, 107, 20–29. [Google Scholar] [CrossRef]
- Wiedmeier, R.D.; Arambel, M.J.; Walters, J.L. Effect of Yeast Culture and Aspergillus oryzae Fermentation Extract on Ruminal Characteristics and Nutrient Digestibility. J. Dairy Sci. 1987, 70, 2063–2068. [Google Scholar] [CrossRef]
- Macchione, M.M.; Merheb, C.W.; Gomes, E.; Da Silva, R. Protease Production by Different Thermophilic Fungi. Appl. Biochem. Biotechnol. 2008, 146, 223–230. [Google Scholar] [CrossRef]
- Li, M.; Petteys, B.J.; McClure, J.M.; Valsakumar, V.; Bekiranov, S.; Frank, E.L.; Smith, J.S. Thiamine Biosynthesis in Saccharomyces cerevisiae Is Regulated by the NAD+ -Dependent Histone Deacetylase Hst1. Mol. Cell. Biol. 2010, 30, 3329–3341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paxhia, M.D.; Downs, D.M. SNZ3 Encodes a PLP Synthase Involved in Thiamine Synthesis in Saccharomyces cerevisiae. G3 Genes|Genomes|Genet. 2019, 9, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Magliano, P.; Flipphi, M.; Arpat, B.A.; Delessert, S.; Poirier, Y. Contributions of the Peroxisome and β-Oxidation Cycle to Biotin Synthesis in Fungi. J. Biol. Chem. 2011, 286, 42133–42140. [Google Scholar] [CrossRef] [PubMed]
- Perli, T.; Moonen, D.P.I.; Van Den Broek, M.; Pronk, J.T.; Daran, J.-M. Adaptive Laboratory Evolution and Reverse Engineering of Single-Vitamin Prototrophies in Saccharomyces Cerevisiae. Appl. Environ. Microbiol. 2020, 86, e00388-20. [Google Scholar] [CrossRef]
- Olzhausen, J.; Grigat, M.; Seifert, L.; Ulbricht, T.; Schüller, H.-J. Increased Biosynthesis of Acetyl-CoA in the Yeast Saccharomyces Cerevisiae by Overexpression of a Deregulated Pantothenate Kinase Gene and Engineering of the Coenzyme A Biosynthetic Pathway. Appl. Microbiol. Biotechnol. 2021, 105, 7321–7337. [Google Scholar] [CrossRef]
- James Theoga Raj, C.; Croft, T.; Venkatakrishnan, P.; Groth, B.; Dhugga, G.; Cater, T.; Lin, S.-J. The Copper-Sensing Transcription Factor Mac1, the Histone Deacetylase Hst1, and Nicotinic Acid Regulate de Novo NAD+ Biosynthesis in Budding Yeast. J. Biol. Chem. 2019, 294, 5562–5575. [Google Scholar] [CrossRef]
- Ohashi, K.; Kawai, S.; Murata, K. Secretion of Quinolinic Acid, an Intermediate in the Kynurenine Pathway, for Utilization in NAD+ Biosynthesis in the Yeast Saccharomyces Cerevisiae. Eukaryot Cell 2013, 12, 648–653. [Google Scholar] [CrossRef]
- Shen, B.; Zhou, P.; Jiao, X.; Yao, Z.; Ye, L.; Yu, H. Fermentative Production of Vitamin E Tocotrienols in Saccharomyces Cerevisiae under Cold-Shock-Triggered Temperature Control. Nat. Commun. 2020, 11, 5155. [Google Scholar] [CrossRef]
- Sun, L.; Kwak, S.; Jin, Y.-S. Vitamin A Production by Engineered Saccharomyces cerevisiae from Xylose via Two-Phase in Situ Extraction. ACS Synth. Biol. 2019, 8, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Tarento, T.D.C.; McClure, D.D.; Talbot, A.M.; Regtop, H.L.; Biffin, J.R.; Valtchev, P.; Dehghani, F.; Kavanagh, J.M. A Potential Biotechnological Process for the Sustainable Production of Vitamin K1. Crit. Rev. Biotechnol. 2019, 39, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, L.; Wang, X.; Shah, D.; Song, X.; Kumar, V.; Shakoor, A.; Tripathi, K.; Ramteke, P.W.; Rani, R. Biosynthesis Pathways, Transport Mechanisms and Biotechnological Applications of Fungal Siderophores. JoF 2021, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Happacher, I.; Aguiar, M.; Yap, A.; Decristoforo, C.; Haas, H. Fungal Siderophore Metabolism with a Focus on Aspergillus fumigatus: Impact on Biotic Interactions and Potential Translational Applications. Essays Biochem. 2023, 67, 829–842. [Google Scholar] [CrossRef]
- Wang, X.; Liang, J.; Liu, Z.; Kuang, Y.; Han, L.; Chen, H.; Xie, X.; Hu, W.; Tang, M. Transcriptional Regulation of Metal Metabolism- and Nutrient Absorption-Related Genes in Eucalyptus Grandis by Arbuscular Mycorrhizal Fungi at Different Zinc Concentrations. BMC Plant Biol. 2022, 22, 76. [Google Scholar] [CrossRef]
- Robinson, J.R.; Isikhuemhen, O.S.; Anike, F.N. Fungal–Metal Interactions: A Review of Toxicity and Homeostasis. JoF 2021, 7, 225. [Google Scholar] [CrossRef]
- Barata-Antunes, C.; Alves, R.; Talaia, G.; Casal, M.; Gerós, H.; Mans, R.; Paiva, S. Endocytosis of Nutrient Transporters in Fungi: The ART of Connecting Signaling and Trafficking. Comput. Struct. Biotechnol. J. 2021, 19, 1713–1737. [Google Scholar] [CrossRef]
- Rekha, C.R.; Vijayalakshmi, G. Bioconversion of Isoflavone Glycosides to Aglycones, Mineral Bioavailability and Vitamin B Complex in Fermented Soymilk by Probiotic Bacteria and Yeast: Biotransformation of Isoflavones. J. Appl. Microbiol. 2010, 109, 1198–1208. [Google Scholar] [CrossRef]
- Klukovich, R.; Courchesne, W.E. Functions of Saccharomyces Cerevisiae Ecm27p, a Putative Na+/Ca2+ Exchanger, in Calcium Homeostasis, Carbohydrate Storage and Cell Cycle Reentry from the Quiescent Phase. Microbiol. Res. 2016, 186–187, 81–89. [Google Scholar] [CrossRef]
- Martínez-Pastor, M.T.; Perea-García, A.; Puig, S. Mechanisms of Iron Sensing and Regulation in the Yeast Saccharomyces Cerevisiae. World J. Microbiol. Biotechnol. 2017, 33, 75. [Google Scholar] [CrossRef]
- Poor, C.B.; Wegner, S.V.; Li, H.; Dlouhy, A.C.; Schuermann, J.P.; Sanishvili, R.; Hinshaw, J.R.; Riggs-Gelasco, P.J.; Outten, C.E.; He, C. Molecular Mechanism and Structure of the Saccharomyces cerevisiae Iron Regulator Aft2. Proc. Natl. Acad. Sci. USA 2014, 111, 4043–4048. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gravelat, F.N.; Chiang, L.Y.; Chen, D.; Vanier, G.; Ejzykowicz, D.E.; Ibrahim, A.S.; Nierman, W.C.; Sheppard, D.C.; Filler, S.G. Aspergillus fumigatus AcuM Regulates Both Iron Acquisition and Gluconeogenesis: A. Fumigatus AcuM. Mol. Microbiol. 2010, 78, 1038–1054. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gsaller, F.; Eisendle, M.; Lechner, B.E.; Schrettl, M.; Lindner, H.; Müller, D.; Geley, S.; Haas, H. The Interplay between Vacuolar and Siderophore-Mediated Iron Storage in Aspergillus fumigatus. Metallomics 2012, 4, 1262. [Google Scholar] [CrossRef] [PubMed]
- Manfiolli, A.O.; De Castro, P.A.; Dos Reis, T.F.; Dolan, S.; Doyle, S.; Jones, G.; Riaño Pachón, D.M.; Ulaş, M.; Noble, L.M.; Mattern, D.J.; et al. Aspergillus fumigatus Protein Phosphatase PpzA Is Involved in Iron Assimilation, Secondary Metabolite Production, and Virulence. Cell. Microbiol. 2017, 19, e12770. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S.; Gientka, I.; Bzducha-Wróbel, A. Accumulation and Metabolism of Selenium by Yeast Cells. Appl. Microbiol. Biotechnol. 2015, 99, 5373–5382. [Google Scholar] [CrossRef]
- Citiulo, F.; Jacobsen, I.D.; Miramón, P.; Schild, L.; Brunke, S.; Zipfel, P.; Brock, M.; Hube, B.; Wilson, D. Candida Albicans Scavenges Host Zinc via Pra1 during Endothelial Invasion. PLoS Pathog. 2012, 8, e1002777. [Google Scholar] [CrossRef]
- Łoboda, D.; Rowińska-Żyrek, M. Zinc Binding Sites in Pra1, a Zincophore from Candida Albicans. Dalton Trans. 2017, 46, 13695–13703. [Google Scholar] [CrossRef]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic Response to Iron Deficiency in Saccharomyces Cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef]
- Drutel, A.; Archambeaud, F.; Caron, P. Selenium and the Thyroid Gland: More Good News for Clinicians. Clin. Endocrinol. 2013, 78, 155–164. [Google Scholar] [CrossRef]
| Animals | Relative Abundance | ||
|---|---|---|---|
| Phyla | Genus | Species | |
| Mouse |
| ||
| Pig |
| ||
| Giant Panda | |||
| Equines |
| ||
| Rabbit |
| ||
| Elephant | |||
| Dog |
| ||
| Cat |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, X.; Li, H.; Wu, A.; He, J.; Mao, X.; Dai, Z.; Tian, G.; Cai, J.; Tang, J.; Luo, Y. Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals. Animals 2025, 15, 710. https://doi.org/10.3390/ani15050710
Deng X, Li H, Wu A, He J, Mao X, Dai Z, Tian G, Cai J, Tang J, Luo Y. Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals. Animals. 2025; 15(5):710. https://doi.org/10.3390/ani15050710
Chicago/Turabian StyleDeng, Xiaofeng, Hua Li, Aimin Wu, Jun He, Xiangbing Mao, Zhaolai Dai, Gang Tian, Jingyi Cai, Jiayong Tang, and Yuheng Luo. 2025. "Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals" Animals 15, no. 5: 710. https://doi.org/10.3390/ani15050710
APA StyleDeng, X., Li, H., Wu, A., He, J., Mao, X., Dai, Z., Tian, G., Cai, J., Tang, J., & Luo, Y. (2025). Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals. Animals, 15(5), 710. https://doi.org/10.3390/ani15050710





