The Gut Microbiota of the Greater Horseshoe Bat Confers Rapidly Corresponding Immune Cells in Mice
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
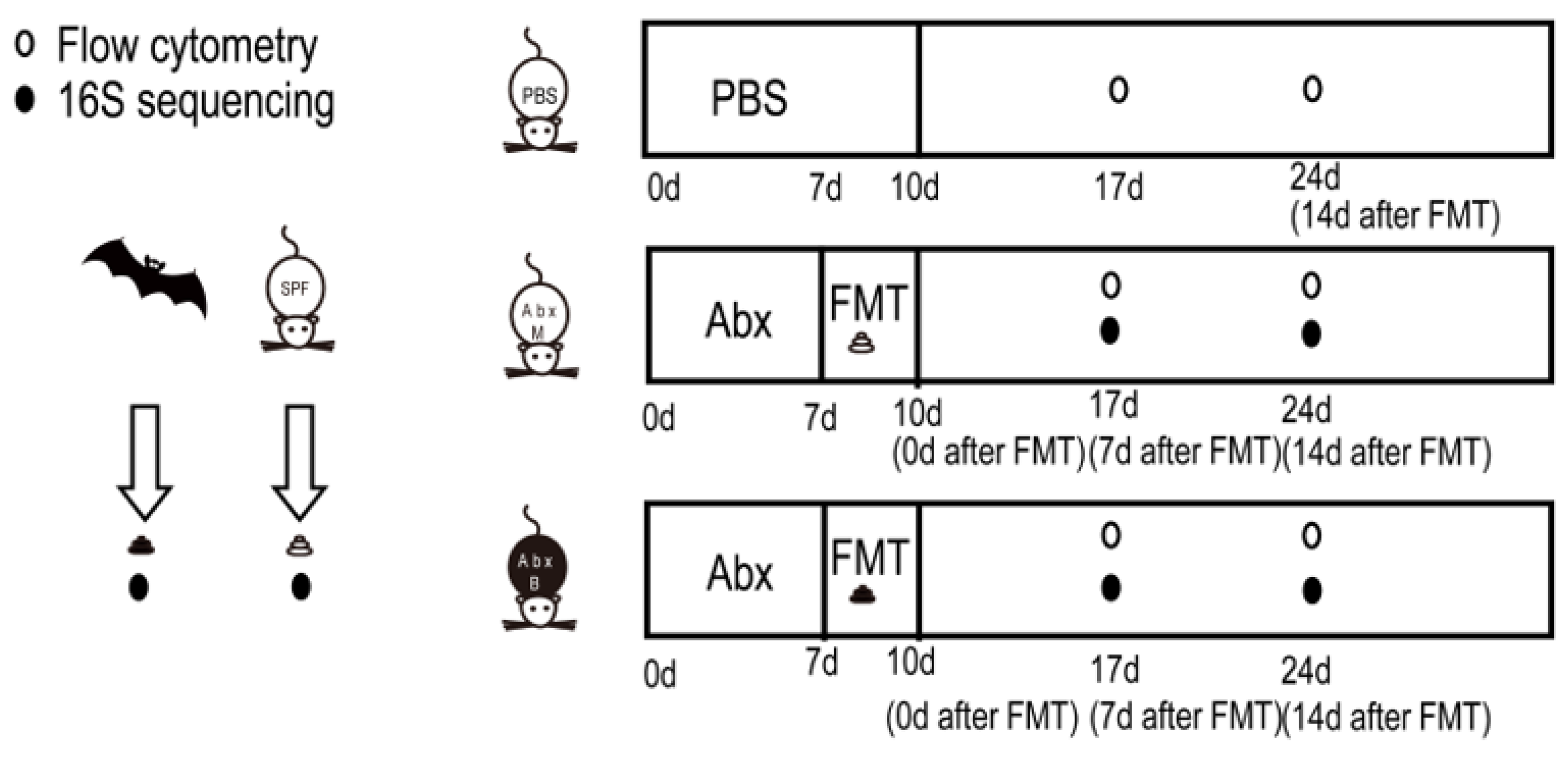
2.1. Study Design, Mice, Collection, and Preservation of Feces
2.2. Fecal Microbiota Transplantation
2.3. 16S rRNA Gene Sequencing
2.4. Flow Cytometry
2.5. Statistical Analyses
3. Results
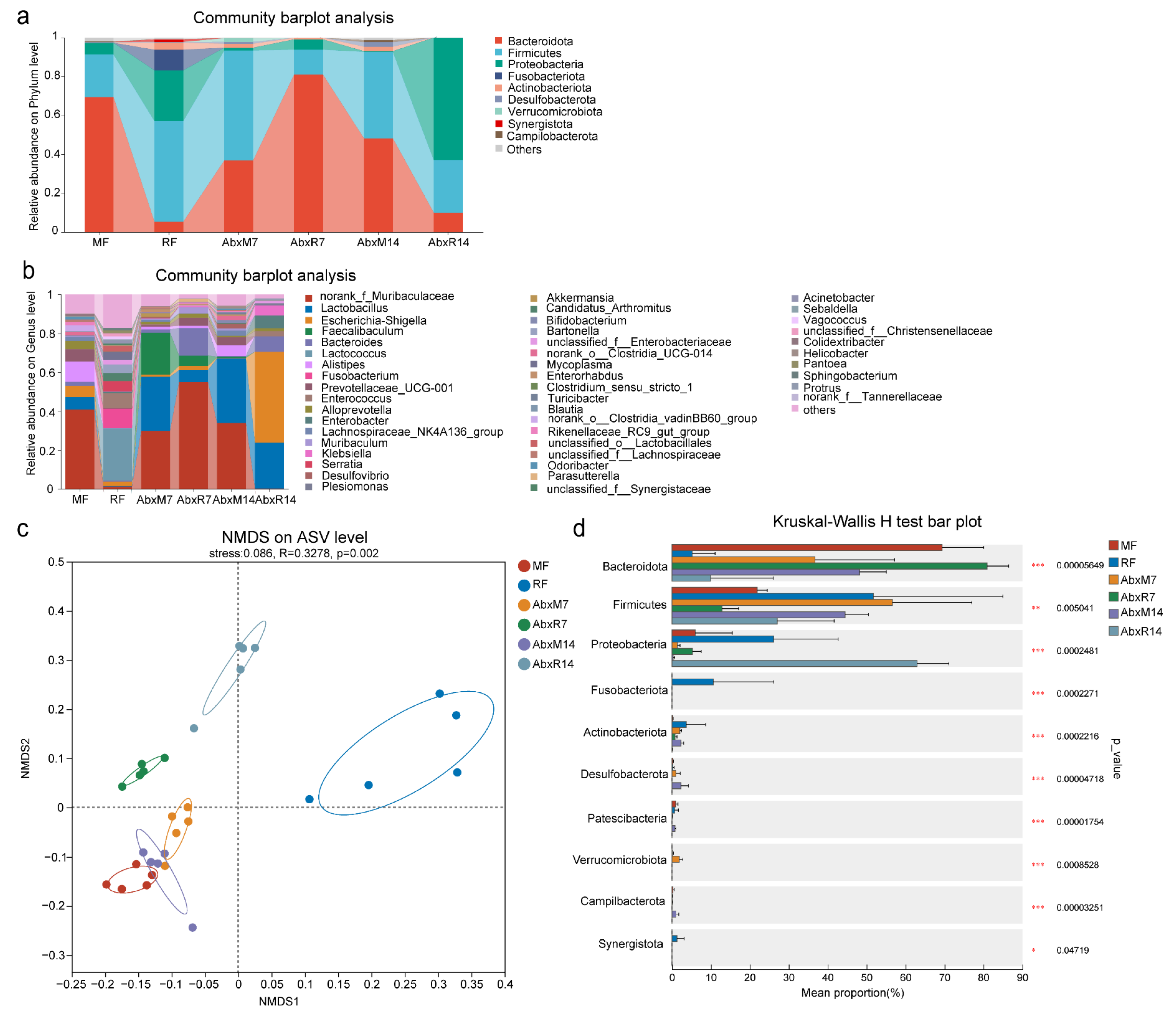
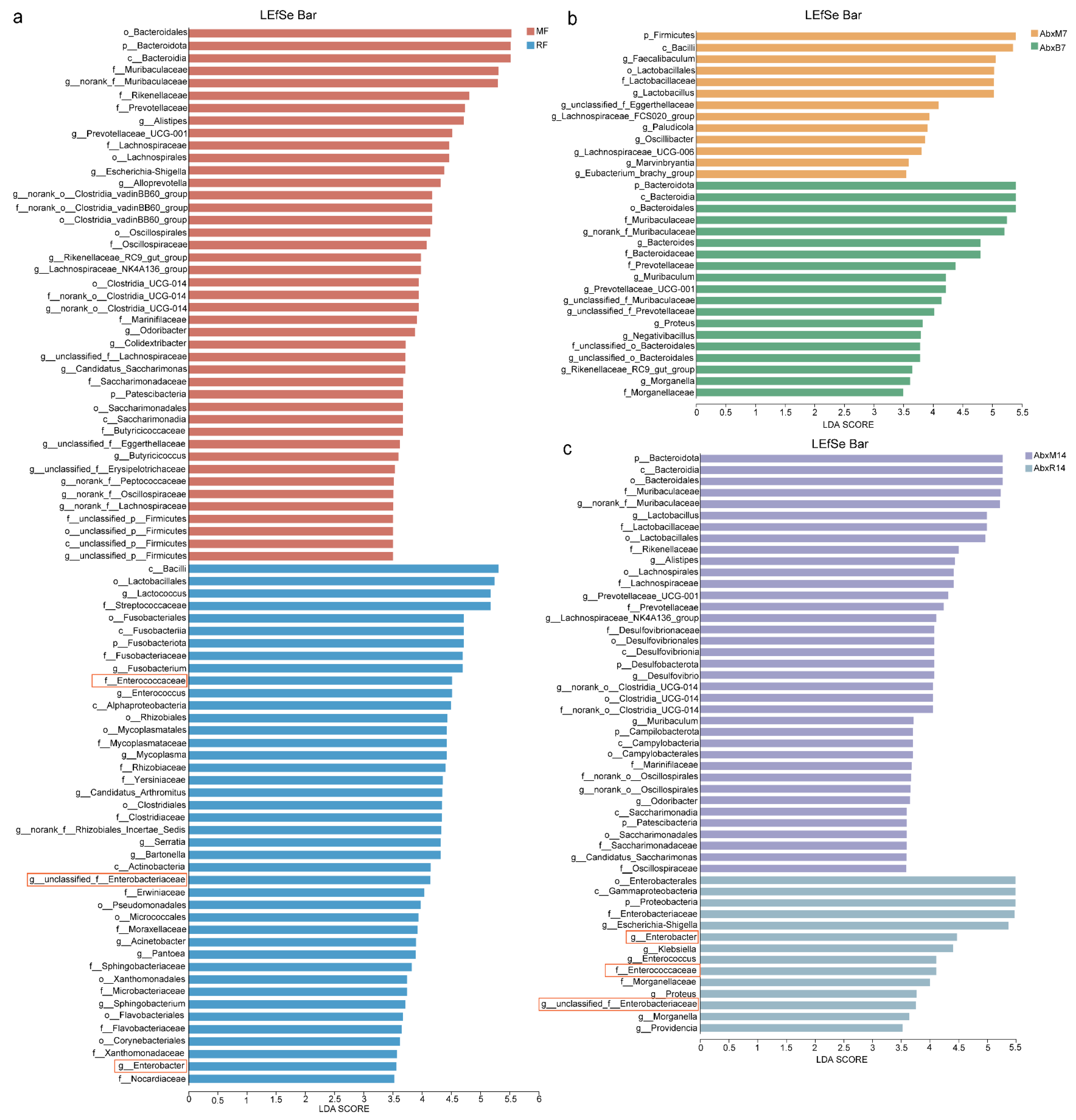
3.1. Microbial Changes in Feces of Mice After FMT
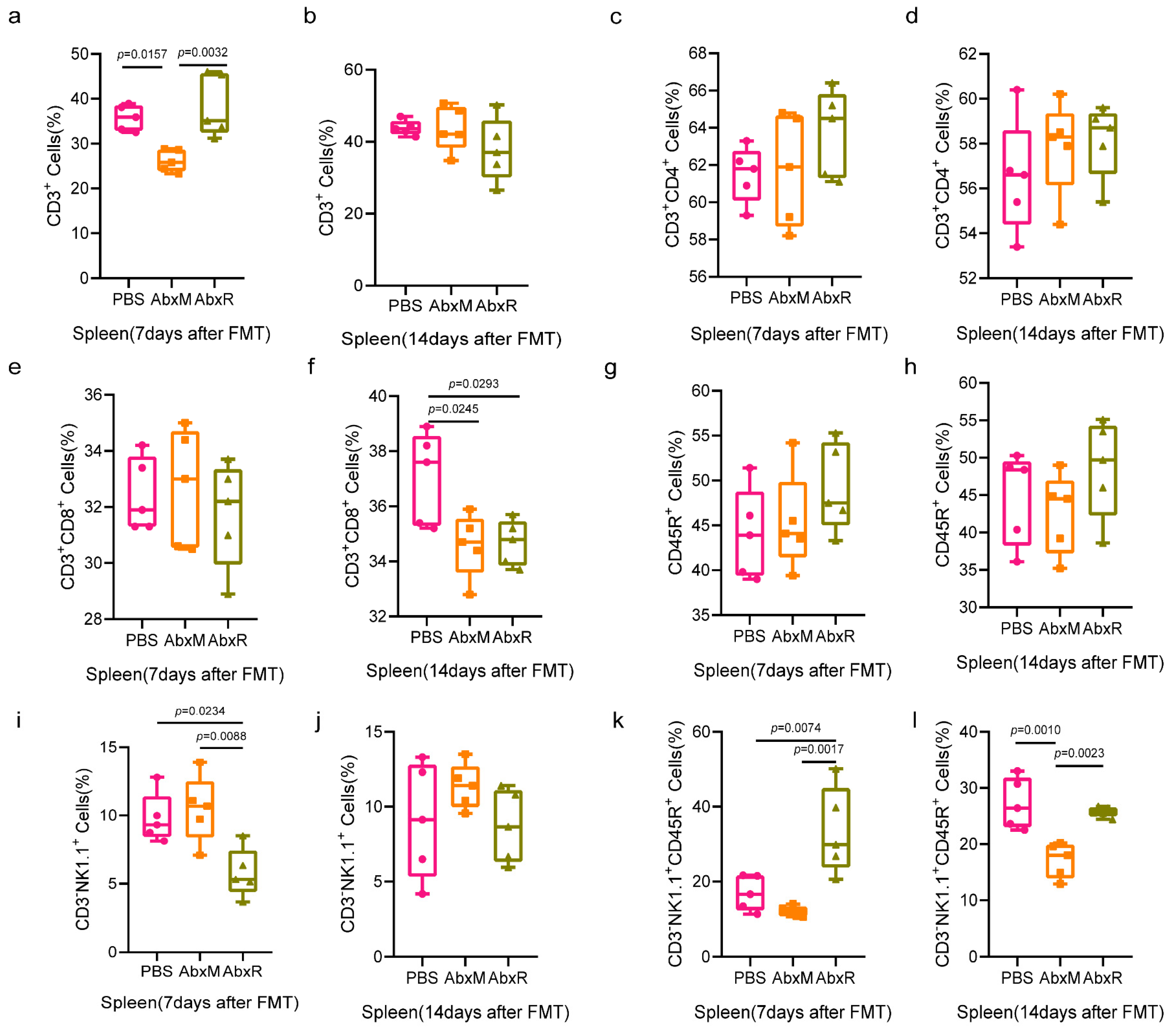
3.2. Immunomodulatory Effect of Bat Gut Microbiota on Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.G.; Liu, Q.L.; Wang, H.F.; Yao, X.S. Severe zoonotic viruses carried by different species of bats and their regional distribution. Clin. Microbiol. Infect. 2024, 30, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Ingala, M.R.; Simmons, N.B.; Perkins, S.L. Bats Are an Untapped System for Understanding Microbiome Evolution in Mammals. mSphere 2018, 3, e00397-18. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020, 18, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, X.; Wang, M.; He, Z.; Li, H.; Wang, J.; Li, Q. Gut microbiota influence immunotherapy responses: Mechanisms and therapeutic strategies. J. Hematol. Oncol. 2022, 15, 47. [Google Scholar] [CrossRef]
- Lee, J.; Song, X.; Hyun, B.; Jeon, C.O.; Hyun, S. Drosophila Gut Immune Pathway Suppresses Host Development-Promoting Effects of Acetic Acid Bacteria. Mol. Cells 2023, 46, 637–653. [Google Scholar] [CrossRef]
- Ronan, V.; Yeasin, R.; Claud, E.C. Childhood Development and the Microbiome-The Intestinal Microbiota in Maintenance of Health and Development of Disease During Childhood Development. Gastroenterology 2021, 160, 495–506. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zheng, W.; Zhao, G.; Zhang, H.; Wang, X.; Guo, Y.; Qin, C.; Shi, Y. Peripheral Lymphoid Volume Expansion and Maintenance Are Controlled by Gut Microbiota via RALDH+ Dendritic Cells. Immunity 2016, 44, 330–342. [Google Scholar] [CrossRef]
- Baker, M.L.; Schountz, T.; Wang, L.F. Antiviral immune responses of bats: A review. Zoonoses Public Health 2013, 60, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.F. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Cowled, C.; Shi, Z.L.; Huang, Z.Y.; Bishop-Lilly, K.A.; Fang, X.D.; Wynne, J.W.; Xiong, Z.Q.; Baker, M.L.; Zhao, W.; et al. Comparative Analysis of Bat Genomes Provides Insight into the Evolution of Flight and Immunity. Science 2013, 339, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Papenfuss, A.T.; Baker, M.L.; Feng, Z.P.; Tachedjian, M.; Crameri, G.; Cowled, C.; Ng, J.; Janardhana, V.; Field, H.E.; Wang, L.F. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genom. 2012, 13, 261. [Google Scholar] [CrossRef]
- Zhou, P.; Chionh, Y.T.; Irac, S.E.; Ahn, M.; Jia Ng, J.H.; Fossum, E.; Bogen, B.; Ginhoux, F.; Irving, A.T.; Dutertre, C.A.; et al. Unlocking bat immunology: Establishment of Pteropus alecto bone marrow-derived dendritic cells and macrophages. Sci. Rep. 2016, 6, 38597. [Google Scholar] [CrossRef]
- Martinez Gomez, J.M.; Periasamy, P.; Dutertre, C.A.; Irving, A.T.; Ng, J.H.; Crameri, G.; Baker, M.L.; Ginhoux, F.; Wang, L.F.; Alonso, S. Phenotypic and functional characterization of the major lymphocyte populations in the fruit-eating bat Pteropus alecto. Sci. Rep. 2016, 6, 37796. [Google Scholar] [CrossRef]
- Luo, J.; Liang, S.; Jin, F. Gut microbiota in antiviral strategy from bats to humans: A missing link in COVID-19. Sci. China Life Sci. 2021, 64, 942–956. [Google Scholar] [CrossRef]
- Jones, D.N.; Ravelomanantsoa, N.A.F.; Yeoman, C.J.; Plowright, R.K.; Brook, C.E. Do gastrointestinal microbiomes play a role in bats’ unique viral hosting capacity? Trends Microbiol. 2022, 30, 632–642. [Google Scholar] [CrossRef]
- Gong, L.; Liu, B.; Wu, H.; Feng, J.; Jiang, T. Seasonal Dietary Shifts Alter the Gut Microbiota of Avivorous Bats: Implication for Adaptation to Energy Harvest and Nutritional Utilization. mSphere 2021, 6, e0046721. [Google Scholar] [CrossRef]
- Xiao, G.; Liu, S.; Xiao, Y.; Zhu, Y.; Zhao, H.; Li, A.; Li, Z.; Feng, J. Seasonal Changes in Gut Microbiota Diversity and Composition in the Greater Horseshoe Bat. Front. Microbiol. 2019, 10, 2247. [Google Scholar] [CrossRef]
- Fleischer, R.; Jones, C.; Ledezma-Campos, P.; Czirjak, G.A.; Sommer, S.; Gillespie, T.R.; Vicente-Santos, A. Gut microbial shifts in vampire bats linked to immunity due to changed diet in human disturbed landscapes. Sci. Total Environ. 2024, 907, 167815. [Google Scholar] [CrossRef] [PubMed]
- Berman, T.S.; Weinberg, M.; Moreno, K.R.; Czirjak, G.A.; Yovel, Y. In sickness and in health: The dynamics of the fruit bat gut microbiota under a bacterial antigen challenge and its association with the immune response. Front. Immunol. 2023, 14, 1152107. [Google Scholar] [CrossRef]
- Liu, B.; Chen, X.; Zhou, L.; Li, J.; Wang, D.; Yang, W.; Wu, H.; Yao, J.; Yang, G.; Wang, C.; et al. The gut microbiota of bats confers tolerance to influenza virus (H1N1) infection in mice. Transbound. Emerg. Dis. 2022, 69, e1469–e1487. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, T.; Debedat, J.; Marquet, F.; Da-Cunha, C.; Ichou, F.; Guerre-Millo, M.; Kapel, N.; Aron-Wisnewsky, J.; Clement, K. Comparative Evaluation of Microbiota Engraftment Following Fecal Microbiota Transfer in Mice Models: Age, Kinetic and Microbial Status Matter. Front. Microbiol. 2018, 9, 3289. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, D.P.; Labrenz, M.; Jurgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Shi, C.W.; Cheng, M.Y.; Yang, X.; Lu, Y.Y.; Yin, H.D.; Zeng, Y.; Wang, R.Y.; Jiang, Y.L.; Yang, W.T.; Wang, J.Z.; et al. Probiotic Lactobacillus rhamnosus GG Promotes Mouse Gut Microbiota Diversity and T Cell Differentiation. Front. Microbiol. 2020, 11, 607735. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Sun, Z.; Zeng, B.; Huang, S.; Zhao, J.; Zhang, Y.; Su, X.; Xu, J.; Wei, H.; Zhang, H. Cow-to-mouse fecal transplantations suggest intestinal microbiome as one cause of mastitis. Microbiome 2018, 6, 200. [Google Scholar] [CrossRef] [PubMed]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J.; et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 2017, 171, 1015–1028.e13. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Stahlman, M.; Ilkayeva, O.; Arnemo, J.M.; Kindberg, J.; Josefsson, J.; Newgard, C.B.; Frobert, O.; Backhed, F. The Gut Microbiota Modulates Energy Metabolism in the Hibernating Brown Bear Ursus arctos. Cell Rep. 2016, 14, 1655–1661. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Song, S.J.; Sanders, J.G.; Delsuc, F.; Metcalf, J.; Amato, K.; Taylor, M.W.; Mazel, F.; Lutz, H.L.; Winker, K.; Graves, G.R.; et al. Comparative Analyses of Vertebrate Gut Microbiomes Reveal Convergence between Birds and Bats. mBio 2020, 11, e02901-19. [Google Scholar] [CrossRef]
- Brook, C.E.; Dobson, A.P. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef]
- Brook, C.E.; Boots, M.; Chandran, K.; Dobson, A.P.; Drosten, C.; Graham, A.L.; Grenfell, B.T.; Müller, M.A.; Ng, M.; Wang, L.F.; et al. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 2020, 9, e48401. [Google Scholar] [CrossRef]
- Santillán, D.D.M.; Lama, T.M.; Gutierrez, Y.T.G.; Brown, A.M.; Donat, P.; Zhao, H.; Rossiterao, S.J.; Yohe, L.R.; Potter, J.H.; Teeling, E.C.; et al. Large-scale genome sampling reveals unique immunity and metabolic adaptations in bats (vol 30, pg 6449, 2021). Mol. Ecol. 2022, 31, 5124. [Google Scholar] [CrossRef]
- Guito, J.C.; Prescott, J.B.; Arnold, C.E.; Amman, B.R.; Schuh, A.J.; Spengler, J.R.; Sealy, T.K.; Harmon, J.R.; Coleman-McCray, J.D.; Kulcsar, K.A.; et al. Asymptomatic Infection of Marburg Virus Reservoir Bats Is Explained by a Strategy of Immunoprotective Disease Tolerance. Curr. Biol. 2021, 31, 257–270.e5. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.H. Epidermal stem cells: Interplay with the skin microenvironment during wound healing. Mol. Cells 2024, 47, 100138. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Lim, W.; Kim, M.; Jang, H.; Park, S.J.; Song, G.; Park, S. Female reproductive disease, endometriosis: From inflammation to infertility. Mol. Cells 2025, 48, 100164. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.J.; Yi, Y.; Kang, Y.; Kim, S.J.; Yoon, Y.I.; Tran, P.H.; Kang, T.; Kim, M.K.; Han, J.; Tak, E.; et al. Reduced Ceramides Are Associated with Acute Rejection in Liver Transplant Patients and Skin Graft and Hepatocyte Transplant Mice, Reducing Tolerogenic Dendritic Cells. Mol. Cells 2023, 46, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Ekmekciu, I.; von Klitzing, E.; Fiebiger, U.; Escher, U.; Neumann, C.; Bacher, P.; Scheffold, A.; Kuhl, A.A.; Bereswill, S.; Heimesaat, M.M. Immune Responses to Broad-Spectrum Antibiotic Treatment and Fecal Microbiota Transplantation in Mice. Front. Immunol. 2017, 8, 397. [Google Scholar] [CrossRef]
- Lewis, S.M.; Williams, A.; Eisenbarth, S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019, 4, eaau6085. [Google Scholar] [CrossRef]
- Periasamy, P.; Hutchinson, P.E.; Chen, J.; Bonne, I.; Shahul Hameed, S.S.; Selvam, P.; Hey, Y.Y.; Fink, K.; Irving, A.T.; Dutertre, C.A.; et al. Studies on B Cells in the Fruit-Eating Black Flying Fox (Pteropus alecto). Front. Immunol. 2019, 10, 489. [Google Scholar] [CrossRef]
- Shim, J.A.; Ryu, J.H.; Jo, Y.; Hong, C. The role of gut microbiota in T cell immunity and immune mediated disorders. Int. J. Biol. Sci. 2023, 19, 1178–1191. [Google Scholar] [CrossRef]
- Vivier, E.; Rebuffet, L.; Narni-Mancinelli, E.; Cornen, S.; Igarashi, R.Y.; Fantin, V.R. Natural killer cell therapies. Nature 2024, 626, 727–736. [Google Scholar] [CrossRef]
- Heo, M.J.; Suh, J.H.; Poulsen, K.L.; Ju, C.; Kim, K.H. Updates on the Immune Cell Basis of Hepatic Ischemia-Reperfusion Injury. Mol. Cells 2023, 46, 527–534. [Google Scholar] [CrossRef]
- Pavlovich, S.S.; Lovett, S.P.; Koroleva, G.; Guito, J.C.; Arnold, C.E.; Nagle, E.R.; Kulcsar, K.; Lee, A.; Thibaud-Nissen, F.; Hume, A.J.; et al. The Egyptian Rousette Genome Reveals Unexpected Features of Bat Antiviral Immunity. Cell 2018, 173, 1098–1110.e1018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Tachedjian, M.; Wynne, J.W.; Boyd, V.; Cui, J.; Smith, I.; Cowled, C.; Ng, J.H.; Mok, L.; Michalski, W.P.; et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc. Natl. Acad. Sci. USA 2016, 113, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- Holzer, M.; Schoen, A.; Wulle, J.; Muller, M.A.; Drosten, C.; Marz, M.; Weber, F. Virus- and Interferon Alpha-Induced Transcriptomes of Cells from the Microbat Myotis daubentonii. iScience 2019, 19, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.E.; Hughes, J.; Gu, Q.; Behdenna, A.; Singer, J.B.; Dennis, T.; Orton, R.J.; Varela, M.; Gifford, R.J.; Wilson, S.J.; et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017, 15, e2004086. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sia, W.R.; Tang, L.J.W.; Gamage, A.M.; Chan, W.O.Y.; Zhu, F.; Chia, W.; Kwek, M.S.S.; Kong, P.S.; Lim, B.L.; et al. Application of a bespoke monoclonal antibody panel to characterize immune cell populations in cave nectar bats. Cell Rep. 2024, 43, 114767. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, S.; Huang, X.; Chen, S.; Li, J.; Wu, H.; He, Y.; Zhou, L.; Liu, B.; Feng, J. The Gut Microbiota of the Greater Horseshoe Bat Confers Rapidly Corresponding Immune Cells in Mice. Animals 2025, 15, 685. https://doi.org/10.3390/ani15050685
Luo S, Huang X, Chen S, Li J, Wu H, He Y, Zhou L, Liu B, Feng J. The Gut Microbiota of the Greater Horseshoe Bat Confers Rapidly Corresponding Immune Cells in Mice. Animals. 2025; 15(5):685. https://doi.org/10.3390/ani15050685
Chicago/Turabian StyleLuo, Shan, Xinlei Huang, Siyu Chen, Junyi Li, Hui Wu, Yuhua He, Lei Zhou, Boyu Liu, and Jiang Feng. 2025. "The Gut Microbiota of the Greater Horseshoe Bat Confers Rapidly Corresponding Immune Cells in Mice" Animals 15, no. 5: 685. https://doi.org/10.3390/ani15050685
APA StyleLuo, S., Huang, X., Chen, S., Li, J., Wu, H., He, Y., Zhou, L., Liu, B., & Feng, J. (2025). The Gut Microbiota of the Greater Horseshoe Bat Confers Rapidly Corresponding Immune Cells in Mice. Animals, 15(5), 685. https://doi.org/10.3390/ani15050685




