The Molecular Monitoring of an Invasive Freshwater Fish, Brown Trout (Salmo trutta), Using Real-Time PCR Assay and Environmental Water Samples
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Fish Specimens and Genomic DNA (gDNA) Extraction
2.2. Modification of Primers and Probe
2.3. Environmental Water Sampling and eDNA Extraction
2.4. qPCR Assay
3. Results
3.1. Modified Primers and Probe
3.2. Sensitivity and Specificity Tests
3.3. qPCR Assay of Environmental Water Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elliott, J.M. Quantitative Ecology and the Brown Trout; Oxford University Press: Oxford, UK, 1994. [Google Scholar]
- Klemetsen, A.; Amundsen, P.A.; Dempson, J.B.; Jonsson, B.; Jonsson, N.; O’Connell, M.F.; Mortensen, E. Atlantic salmon Salmo salar L., brown trout Salmo trutta L. and Arctic charr Salvelinus alpinus (L.): A review of aspects of their life histories. Ecol. Freshw. Fish 2003, 12, 1–59. [Google Scholar] [CrossRef]
- Townsend, C.R. Invasion biology and ecological impacts of brown trout Salmo trutta in New Zealand. Biol. Conserv. 1996, 78, 13–22. [Google Scholar] [CrossRef]
- Behnke, R. Trout and Salmon of North America; The Free Press, Simon & Schuster Inc.: New York, NY, USA, 2010. [Google Scholar]
- McDowall, R.M. Impacts of introduced salmonids on native galaxiids in New Zealand upland streams: A new look at an old problem. Trans. Am. Fish. Soc. 2003, 132, 229–238. [Google Scholar] [CrossRef]
- Korsu, K.; Huusko, A.; Muotka, T. Impacts of invasive stream salmonids on native fish: Using meta-analysis to summarize four decades of research. Boreal Environ. Res. 2010, 15, 491–500. [Google Scholar]
- Hasegawa, K. Invasions of rainbow trout and brown trout in Japan: A comparison of invasiveness and impact on native species. Ecol. Freshw. Fish 2020, 29, 419–428. [Google Scholar] [CrossRef]
- Park, C.W.; Yun, Y.J.; Kim, J.W.; Bae, D.Y.; Kim, J.G.; Kim, S.H. An Identification of domestic habitat and settlement of the invasive exotic fish brown trout, Salmo trutta. Korean J. Ichthyol. 2022, 34, 270–276. (In Korean) [Google Scholar]
- Townsend, C.R.; Crowl, T.A. Fragmented population structure in a native New Zealand fish: An effect of introduced brown trout? Oikos 1991, 61, 347–354. [Google Scholar] [CrossRef]
- McHugh, P.; Budy, P. Experimental effects of nonnative brown trout on the individual- and populations-level performance of native Bonneville cutthroat trout. Trans. Am. Fish. Soc. 2006, 135, 1441–1455. [Google Scholar] [CrossRef]
- McIntosh, A.R.; McHugh, P.A.; Dunn, N.R.; Goodman, J.M.; Howard, S.W.; Jellyman, P.G.; O’Brien, L.K.; Nyström, P.; Woodford, D.J. The impact of trout on galaxiid fishes in New Zealand. N. Z. J. Ecol. 2010, 34, 195–206. [Google Scholar]
- Meldgaard, T.; Crivelli, A.J.; Jesensek, D.; Poizat, G.; Rubin, J.F.; Berrebi, P. Hybridization mechanisms between the endangered marble trout (Salmo marmoratus) and the brown trout (Salmo trutta) as revealed by in-stream experiments. Biol. Conserv. 2007, 136, 602–611. [Google Scholar] [CrossRef]
- Castillo, A.G.F.; Ayllon, F.; Moran, P.; Izquierdo, J.I.; Martinez, J.L.; Beall, E.; Garcia-Vazquez, E. Interspecific hybridization and introgression are associated with stock transfers in salmonids. Aquaculture 2008, 278, 31–36. [Google Scholar] [CrossRef]
- Lowe, S.; Browne, M.; Boudjelas, S.; De Poorter, M. 100 of the World’s Worst Invasive Alien Species: A Selection from the Global Invasive Species Database; Invasive Species Specialist Group: Auckland, New Zealand, 2000. [Google Scholar]
- Molony, B. Environmental Requirements and Tolerances of Rainbow Trout (Oncorhynchus mykiss) and Brown Trout (Salmo trutta) with Special Reference to Western Australia: A Review; Department of Fisheries, Government of Western Australia: Perth, Australia, 2001. [Google Scholar]
- Ministry of Environment. Notice on Designation of Ecosystem-disrupting Organisms; Ministry of Environment: Sejong, Republic of Korea, 2021. (In Korean) [Google Scholar]
- National Institute of Ecology. Monitoring of Invasive Alien species in 2023; National Institute of Ecology: Seocheon, Republic of Korea, 2023. (In Korean) [Google Scholar]
- Wilcox, T.M.; Carim, K.J.; McKelvey, K.S.; Young, M.K.; Schwartz, M.K. The dual challenges of generality and specificity when developing environmental DNA markers for species and subspecies of Oncorhynchus. PLoS ONE 2015, 10, e0142008. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.; Carlsson, J.E.L.; Ball, B.; Egan, D.; Kelly-Quinn, M.; Whelan, K.; Carlsson, J. A quantitative PCR-based environmental DNA assay for detecting Atlantic salmon (Salmo salar L.). Aquat. Conserv. 2018, 28, 1238–1243. [Google Scholar] [CrossRef]
- Fernandez, S.; Sandin, M.M.; Beaulieu, P.G.; Clusa, L.; Martinez, J.L.; Ardura, A.; García-Vázquez, E. Environmental DNA for freshwater fish monitoring: Insights for conservation within a protected area. PeerJ 2018, 6, e4486. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, S.W.; Ebert, R.B.; Hesselsøe, M.; Kuntke, F.; Hassingboe, J.; Mortensen, P.B.; Thomsen, P.F.; Sigsgaard, E.E.; Hansen, B.K.; Nielsen, E.E.; et al. Species-specific detection and quantification of environmental DNA from marine fishes in the Baltic Sea. J. Exp. Mar. Biol. Ecol. 2019, 510, 31–45. [Google Scholar] [CrossRef]
- Hernandez, C.; Bougas, B.; Perreault-Payette, A.; Simard, A.; Côté, G.; Bernatchez, L. 60 specific eDNA qPCR assays to detect invasive, threatened, and exploited freshwater vertebrates and invertebrates in Eastern Canada. Environ. DNA 2020, 2, 373–386. [Google Scholar] [CrossRef]
- Kim, K.-Y.; Heo, J.S.; Moon, S.Y.; Kim, K.-S.; Choi, J.-H.; Yoo, J.-T. Preliminary application of molecular monitoring of the Pacific herring (Clupea pallasii) based on real-time PCR assay utilization on environmental water samples. Korean J. Ecol. Environ. 2021, 54, 209–220. [Google Scholar] [CrossRef]
- Gustavson, M.S.; Collins, P.C.; Finarelli, J.A.; Egan, D.; Conchúir, R.Ó.; Wightman, G.D.; King, J.J.; Gauthier, D.T.; Whelan, K.; Carlsson, J.E.; et al. An eDNA assay for Irish Petromyzon marinus and Salmo trutta and field validation in running water. J. Fish. Biol. 2015, 87, 1254–1262. [Google Scholar] [CrossRef]
- Banks, J.C.; Demetras, N.J.; Hogg, I.D.; Knox, M.A.; West, D.W. Monitoring brown trout (Salmo trutta) eradication in a wildlife sanctuary using environmental DNA. N. Z. Nat. Sci. 2016, 41, 1–13. [Google Scholar]
- Carim, K.J.; Wilcox, T.M.; Anderson, M.; Lawrence, D.J.; Young, M.K.; McKelvey, K.S.; Schwartz, M.K. An environmental DNA marker for detecting nonnative brown trout (Salmo trutta). Conserv. Genet. Resour. 2016, 8, 259–261. [Google Scholar] [CrossRef]
- Deutschmann, B.; Müller, A.K.; Hollert, H.; Brinkmann, M. Assessing the fate of brown trout (Salmo trutta) environmental DNA in a natural stream using a sensitive and specific dual-labelled probe. Sci. Total Environ. 2019, 655, 321–327. [Google Scholar] [CrossRef]
- Asahida, T.; Kobayashi, T.; Saitoh, K.; Nakayama, I. Tissue preservation and total DNA extraction form fish stored at ambient temperature using buffers containing high concentration of urea. Fish. Sci. 1996, 62, 727–730. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Crête-Lafrenière, A.; Weir, L.K.; Bernatchez, L. Framing the Salmonidae family phylogenetic portrait: A more complete picture from increased taxon sampling. PLoS ONE 2012, 7, e46662. [Google Scholar] [CrossRef] [PubMed]
- Lodge, D.M.; Turner, C.R.; Jerde, C.L.; Barnes, M.A.; Chadderton, L.; Egan, S.P.; Feder, J.L.; Mahon, A.R.; Pfrender, M.E. Conservation in a cup of water: Estimating biodiversity and population abundance from environmental DNA. Mol. Ecol. 2012, 21, 2555–2558. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Jane, S.F.; Lowe, W.H.; Whiteley, A.R.; Schwartz, M.K. Robust detection of rare species using environmental DNA: The importance of primer specificity. PLoS ONE 2013, 8, e59520. [Google Scholar] [CrossRef]
- National Institute of Ecology. Investigating Ecological Risk of Alien Species in 2020; National Institute of Ecology: Seocheon, Republic of Korea, 2020. (In Korean) [Google Scholar]
- Kim, J.; Hong, D.; Kim, J.; Kim, B.; Kim, H.; Choi, J. Length–weight relationship and condition factor of the invasive fish species brown trout (Salmo trutta) in Soyang River. J. Agric. Life Environ. Sci. 2023, 35, 604–617. (In Korean) [Google Scholar]
- Yi, Y.K.; Lee, H.S.; Baek, H.J.; Kim, Y.D. Temperature variation of release water of Soyang Reservoir. In Convention 2006 Civil Expo & Conference; Korean Society of Civil Engineers: Seoul, Republic of Korea, 2006; pp. 165–166. (In Korean) [Google Scholar]
- Kondolf, G.M.; Wolman, M.G. The sizes of salmonid spawning gravels. Water Resour. Res. 1993, 29, 2275–2285. [Google Scholar] [CrossRef]
- Young, M.K. Conservation Assessment for Inland Cutthroat Trout; Rocky Mountain Forest and Range Experiment Station: Fort Collins, CO, USA, 1995; Volume 256. [Google Scholar]
- Armstrong, J.D.; Kemp, P.S.; Kennedy, G.J.A.; Ladle, M.; Milner, N.J. Habitat requirements of Atlantic salmon and brown trout in rivers and streams. Fish. Res. 2003, 62, 143–170. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Strickler, K.M.; Pilliod, D.S. Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol. Conserv. 2015, 183, 1–3. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Yates, M.C.; Fraser, D.J.; Derry, A.M. Meta-analysis supports further refinement of eDNA for monitoring aquatic species-specific abundance in nature. Environ. DNA 2019, 1, 5–13. [Google Scholar] [CrossRef]


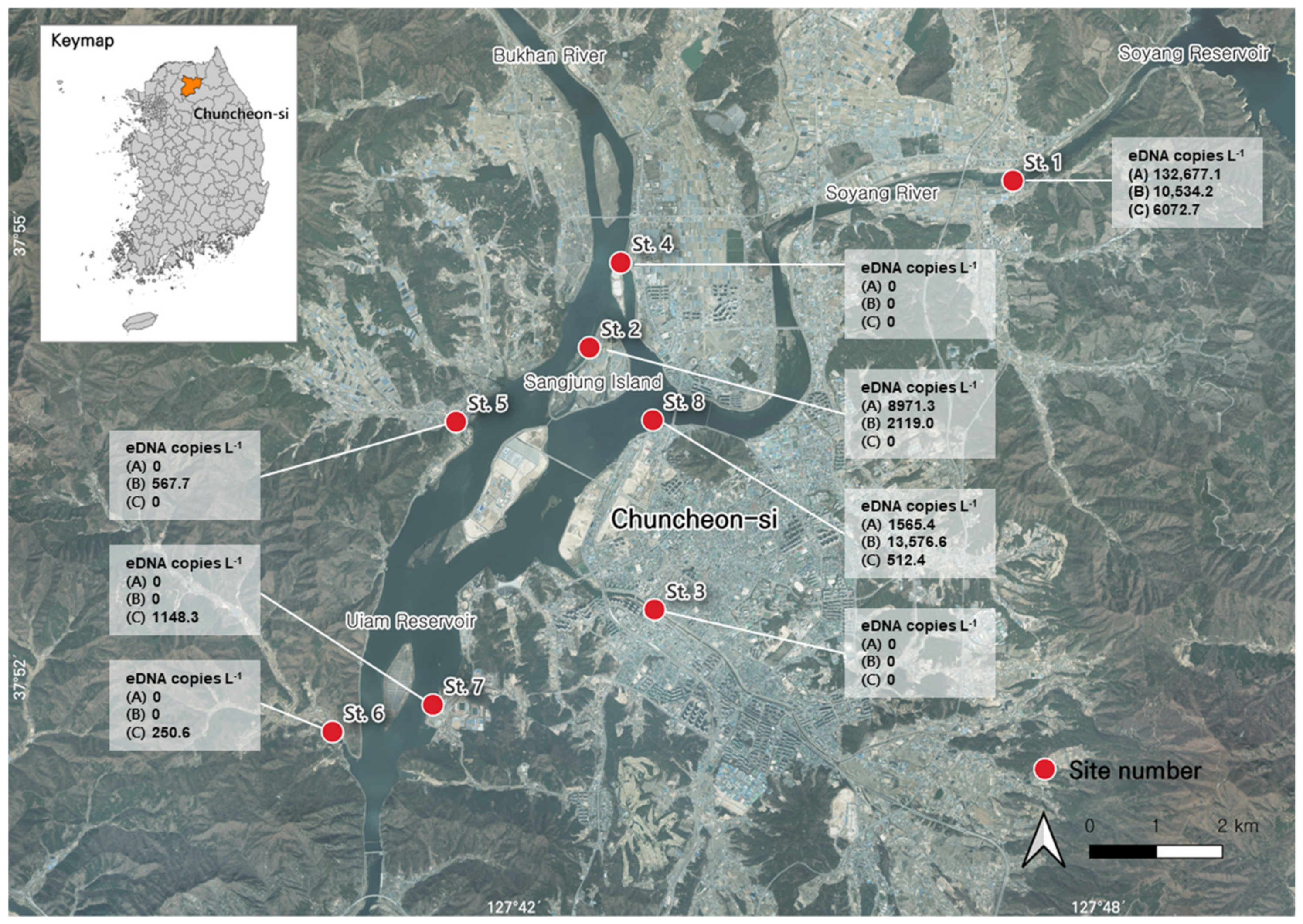
| Station | GPS Coordinate | Location |
|---|---|---|
| St. 1 | 37°55′37.17″ N 127°47′07.03″ E | Upstream of the Soyang River |
| St. 2 | 37°54′15.93″ N 127°42′46.12″ E | The Sangjung Island |
| St. 3 | 37°52′08.34″ N 127°43′26.22″ E | The Gongji Stream |
| St. 4 | 37°54′57.30″ N 127°43′05.38″ E | Downstream of the Bukhan River |
| St. 5 | 37°53′39.78″ N 127°41′23.97″ E | Small stream flowing into the Uiam Reservoir |
| St. 6 | 37°51′08.81″ N 127°40′07.78″ E | Small stream flowing into the Uiam Reservoir |
| St. 7 | 37°51′21.98″ N 127°41′09.69″ E | Downstream of the Uiam Reservoir |
| St. 8 | 37°53′40.60″ N 127°43′25.11″ E | Downstream of the Soyang River |
| Oligonucleotide Name 1 | Sequence (5′ → 3′) | G + C (%) | Nearest Neighbor Tm (°C) | References |
|---|---|---|---|---|
| Forward primer | ||||
| Str-cyb-0294f | CGCCCGAGGACTCTACTATGGT | 59.09 | 67.87 | Carim et al. [26] |
| Str-cyb-0297f | CCGAGGACTCTACTATGGT | 52.63 | 60.07 | This study |
| Reverse primer | ||||
| Str-cyb-0382r | GGAAGAACGTAGCCCACGAA | 55.00 | 65.00 | Carim et al. [26] |
| Str-cyb-0384r | GGAAGAACGTAGCCCACG | 61.11 | 62.94 | This study |
| Hydrolysis probe | ||||
| Str-cyb-0345p | CGGAGTCGTACTGCTAC | 58.82 | 58.83 | Carim et al. [26] |
| Str-cyb-0341p | ATATCGGAGTCGTACTGCTA | 45.00 | 60.01 | This study |
| Water Sample | Volume (mL) | Quantification Cycle (Cq) Value | Total Volume Equivalent (Copies L−1) | Average | Standard Deviation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Replicate | 1 | 2 | 3 | 1 | 2 | 3 | |||
| January 2023 | |||||||||
| St. 1 | 2000 | 30.757 | 30.809 | 30.665 | 131,416 | 127,015 | 139,600 | 132,677.1 | 6386.3 |
| St. 2 | 2000 | 34.607 | 34.628 | 35.457 | 10,517 | 10,375 | 6022 | 8971.3 | 2555.0 |
| St. 3 | 2000 | ND 1 | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 4 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 5 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 6 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 7 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 8 | 2000 | 35.836 | ND | ND | 4696 | 0 | 0 | 1565.4 | 2711.3 |
| February 2023 | |||||||||
| St. 1 | 2000 | 34.116 | 34.961 | 34.886 | 14,510 | 8335 | 8758 | 10,534.2 | 3449.3 |
| St. 2 | 2000 | 36.464 | 36.399 | ND | 3111 | 3246 | 0 | 2119.0 | 1836.4 |
| St. 3 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 4 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 5 | 2000 | 37.382 | ND | ND | 1703 | 0 | 0 | 567.7 | 983.2 |
| St. 6 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 7 | 1000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 8 | 2000 | 35.039 | 34.237 | 33.673 | 7920 | 13,408 | 19,402 | 13,576.6 | 5742.7 |
| March 2023 | |||||||||
| St. 1 | 2000 | 35.469 | 34.946 | 36.150 | 5976 | 8420 | 3822 | 6072.7 | 2300.9 |
| St. 2 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 3 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 4 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 5 | 2000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
| St. 6 | 2000 | ND | ND | 38.629 | 0 | 0 | 752 | 250.6 | 334.1 |
| St. 7 | 1000 | 37.365 | ND | ND | 3445 | 0 | 0 | 1148.3 | 1989.0 |
| St. 8 | 2000 | ND | ND | 37.539 | 0 | 0 | 1537 | 512.4 | 887.5 |
| Negative control | 1000 | ND | ND | ND | 0 | 0 | 0 | 0.0 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-H.; Lee, S.-I.; Lee, S.-H.; Jo, S.-E.; Kim, K.-Y. The Molecular Monitoring of an Invasive Freshwater Fish, Brown Trout (Salmo trutta), Using Real-Time PCR Assay and Environmental Water Samples. Animals 2025, 15, 659. https://doi.org/10.3390/ani15050659
Kim S-H, Lee S-I, Lee S-H, Jo S-E, Kim K-Y. The Molecular Monitoring of an Invasive Freshwater Fish, Brown Trout (Salmo trutta), Using Real-Time PCR Assay and Environmental Water Samples. Animals. 2025; 15(5):659. https://doi.org/10.3390/ani15050659
Chicago/Turabian StyleKim, Su-Hwan, Soo-In Lee, Sang-Hun Lee, So-Eun Jo, and Keun-Yong Kim. 2025. "The Molecular Monitoring of an Invasive Freshwater Fish, Brown Trout (Salmo trutta), Using Real-Time PCR Assay and Environmental Water Samples" Animals 15, no. 5: 659. https://doi.org/10.3390/ani15050659
APA StyleKim, S.-H., Lee, S.-I., Lee, S.-H., Jo, S.-E., & Kim, K.-Y. (2025). The Molecular Monitoring of an Invasive Freshwater Fish, Brown Trout (Salmo trutta), Using Real-Time PCR Assay and Environmental Water Samples. Animals, 15(5), 659. https://doi.org/10.3390/ani15050659




