Construction and Functional Analysis of the ceRNA Regulatory Network Associated with Muscle Development in Shaanbei White Cashmere Goats
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. RNA Extraction, Library Preparation, and Sequencing
2.3. Identification of LncRNA and Screening of DEls
2.4. Prediction and Functional Analysis of miRNAs Targeted by DEls and Target Genes of Key miRNAs
2.5. Validation of Transcriptome Sequencing Data by qRT-PCR
2.6. Luciferase Assays to Validate miRNA–lncRNA Interactions
2.7. Statistical Analysis
3. Results
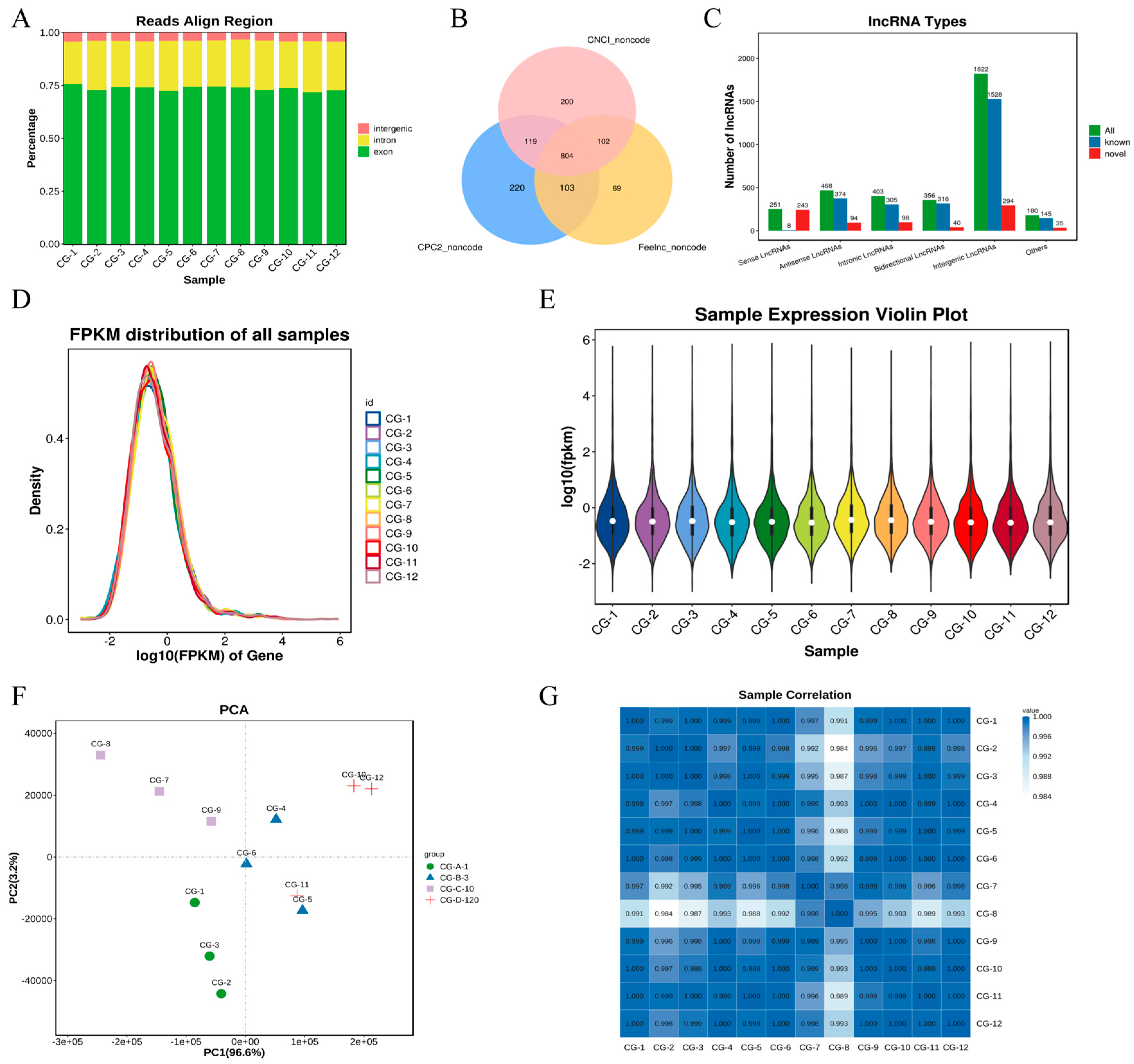
3.1. Sequencing Data: Quality Assessment
3.2. Characteristics of lncRNAs Involved in Muscle Formation
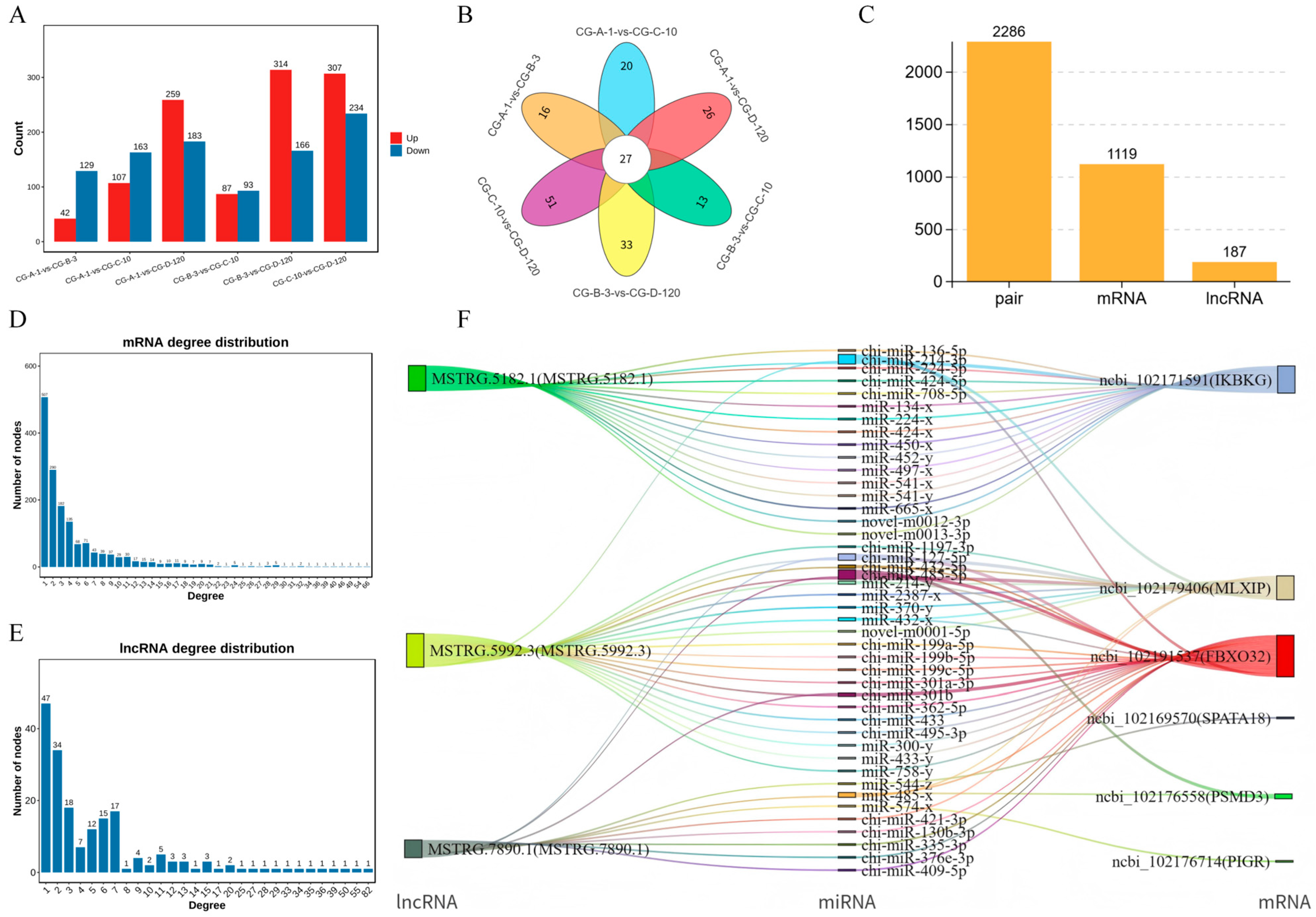
3.3. Differential Expression Analysis of lncRNAs During Myogenesis
3.4. Identification of miRNAs Targeted by lncRNAs in Muscle Growth and Development
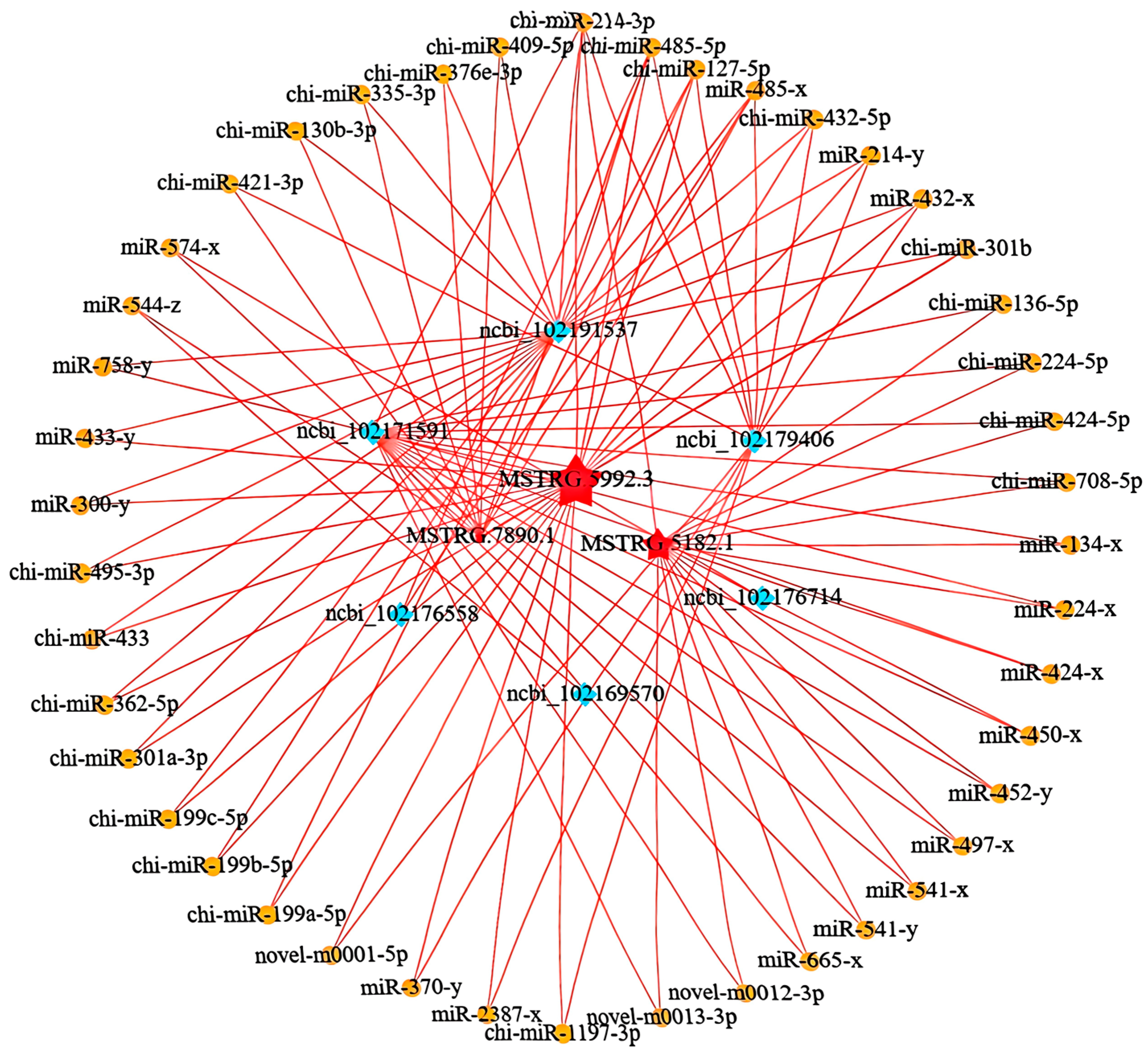
3.5. Analysis of miRNA and ceRNA Regulatory Networks
3.6. Validation of ceRNA Regulatory Network by qRT-PCR
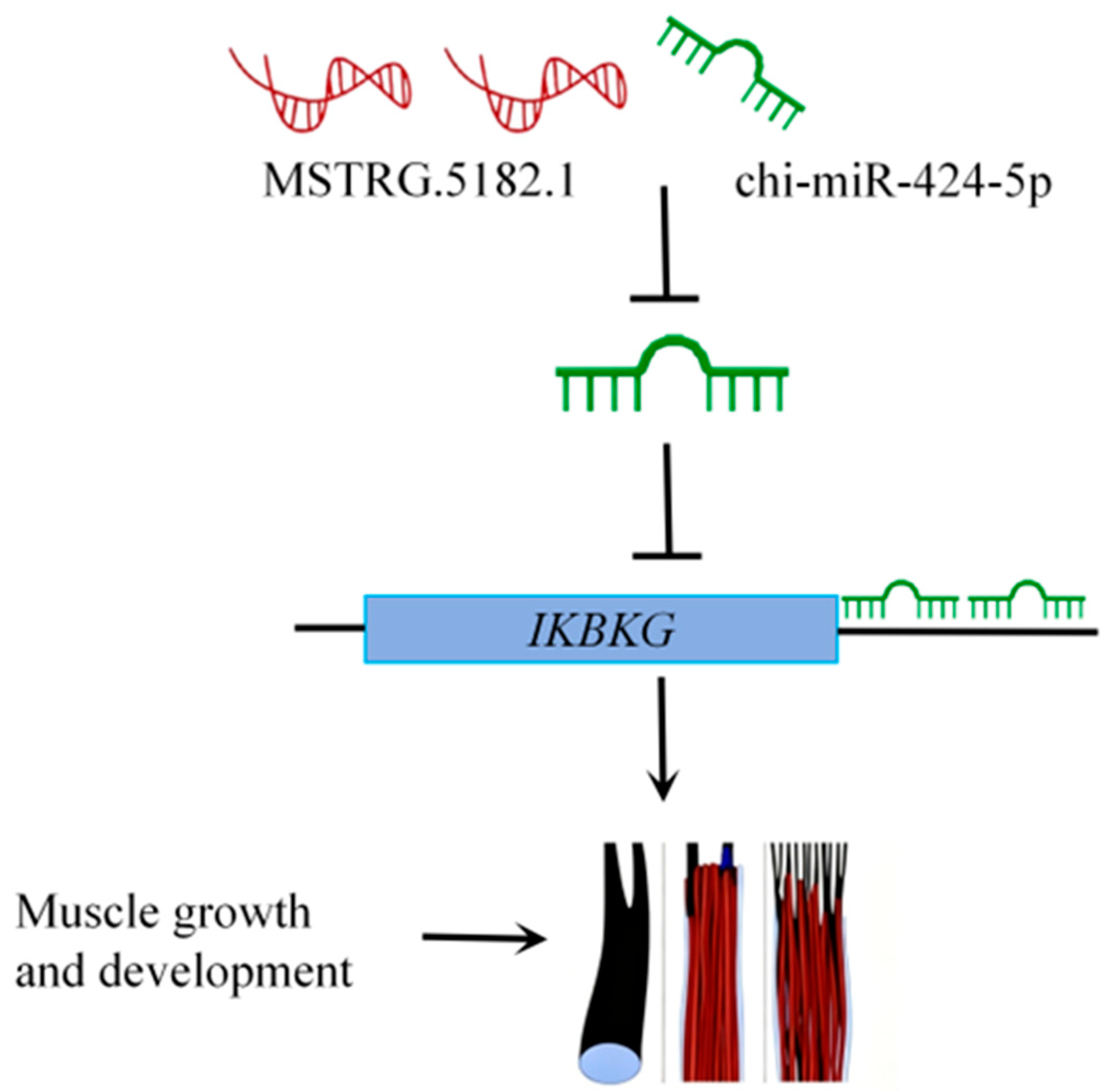
3.7. The Characterization of the MSTRG.5182.1-chi-miR-424-5p/IKBKG Regulatory Axis in Muscle Development
3.8. Functional Analysis of the MSTRG.5182.1-chi-miR-424-5p/IKBKG Regulatory Axis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| lncRNAs | Long non-coding RNAs |
| LDM | Longissimus dorsi muscle |
| SL-RT | stem–loop reverse transcription primer |
| F | forward primer |
| R | reverse primer |
| RIN | RNA Integrity Number |
| miRNAs | microRNAs |
| DEls | Differentially Expressed lncRNAs |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ceRNA | competitive endogenous RNA |
| qRT-PCR | real-time quantitative PCR |
| DMEM | Dulbecco’s Modified Eagle Medium |
References
- Zhang, X.; He, L.; Wang, L.; Wang, Y.; Yan, E.; Wan, B.; Zeng, Q.; Zhang, P.; Zhao, X.; Yin, J. CLIC5 Promotes Myoblast Differentiation and Skeletal Muscle Regeneration via the BGN-Mediated Canonical Wnt/β-Catenin Signaling Pathway. Sci. Adv. 2024, 10, eadq6795. [Google Scholar] [CrossRef]
- Liu, S.; Du, M.; Tu, Y.; You, W.; Chen, W.; Liu, G.; Li, J.; Wang, Y.; Lu, Z.; Wang, T.; et al. Fermented Mixed Feed Alters Growth Performance, Carcass Traits, Meat Quality and Muscle Fatty Acid and Amino Acid Profiles in Finishing Pigs. Anim. Nutr. 2023, 12, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, H.; Wang, Y.; Zhang, Y.; Liu, Y.; Han, R.; Li, H.; Cai, H.; Liu, X.; Kang, X.; et al. The VGLL2 Gene Participates in Muscle Development in Gushi Chickens1. J. Integr. Agric. 2025, 24, 246–260. [Google Scholar] [CrossRef]
- Lindberg, J.; Lundeberg, J. The Plasticity of the Mammalian Transcriptome. Genomics 2010, 95, 1–6. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, Transcriptome and Proteome: The Rise of Omics Data and Their Integration in Biomedical Sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Qimuge, N.; Ma, M.; Wang, Y.; Tang, G.; Zhang, Q.; Sun, Y.; Chen, X.; Yu, T.; Dong, W.; et al. MicroRNA-664-5p Promotes Myoblast Proliferation and Inhibits Myoblast Differentiation by Targeting Serum Response Factor and Wnt1. J. Biol. Chem. 2018, 293, 19177–19190. [Google Scholar] [CrossRef]
- Jin, Z.; Gao, H.; Fu, Y.; Ren, R.; Deng, X.; Chen, Y.; Hou, X.; Wang, Q.; Song, G.; Fan, N.; et al. Whole-Transcriptome Analysis Sheds Light on the Biological Contexts of Intramuscular Fat Deposition in Ningxiang Pigs. Genes 2024, 15, 642. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Dong, J.; Li, J.; Huang, M.; Wang, D.; Tan, X. Novel Insights into the Molecular Mechanisms of Chicken Breast Muscle Development by Integrating Non-Coding RNA and mRNA Profiles. Int. J. Mol. Sci. 2025, 26, 8181. [Google Scholar] [CrossRef]
- Sun, X.; Li, M.; Sun, Y.; Cai, H.; Lan, X.; Huang, Y.; Bai, Y.; Qi, X.; Chen, H. The Developmental Transcriptome Sequencing of Bovine Skeletal Muscle Reveals a Long Noncoding RNA, lncMD, Promotes Muscle Differentiation by Sponging miR-125b. Biochim. Biophys. Acta 2016, 1863, 2835–2845. [Google Scholar] [CrossRef]
- Qiu, J.; Ma, Z.; Hong, Z.; Yin, X.; Chen, Y.; Ahmed, H.Q.; Zan, L.; Li, A. Comparative Analysis of the Whole Transcriptome Landscapes of Muscle and Adipose Tissue in Qinchuan Beef Cattle. BMC Genom. 2025, 26, 32. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, R.; Pan, C.; Chen, H.; Qu, L.; Wu, L.; Guo, Z.; Zhu, H.; Lan, X. Genetic Variations and mRNA Expression of Goat DNAH1 and Their Associations with Litter Size. Cells 2022, 11, 1371. [Google Scholar] [CrossRef]
- Zhan, S.; Xue, Y.; Yang, L.; Li, D.; Dai, H.; Zhong, T.; Wang, L.; Dai, D.; Li, L.; Zhang, H. Transcriptome Analysis Reveals Long Non-Coding Natural Antisense Transcripts Involved in Muscle Development in Fetal Goat (Capra hircus). Genomics 2022, 114, 110284. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; He, Q.; Xia, W.; Rong, G.; Wang, D.; Li, M.; Ji, F.; Sun, W.; Cao, T.; Zhou, H.; et al. Integrated Analysis of lncRNA and Gene Expression in Longissimus Dorsi Muscle at Two Developmental Stages of Hainan Black Goats. PLoS ONE 2022, 17, e0276004. [Google Scholar] [CrossRef]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-kappaB-YY1-miR-29 Regulatory Circuitry in Skeletal Myogenesis and Rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-Coding Transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wucher, V.; Legeai, F.; Hédan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: A Tool for Long Non-Coding RNA Annotation and Its Application to the Dog Transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective microRNA Target Sites in Mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.-W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global Analyses of the Effect of Different Cellular Contexts on microRNA Targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive Modeling of microRNA Targets Predicts Functional Non-Conserved and Non-Canonical Sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA Targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, Y.; Ma, D.; Liu, Y.; Xu, Q.; Cheng, D.; Li, G.; Ni, C. ALKBH5 Promotes Lung Fibroblast Activation and Silica-Induced Pulmonary Fibrosis Through miR-320a-3p and FOXM1. Cell. Mol. Biol. Lett. 2022, 27, 26. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, X.; Sun, C.; Yu, L.; Hua, D.; Shi, C.; Wang, Q.; Rao, C.; Luo, W.; Jiang, Z.; et al. The ATPase Pontin Is a Key Cell Cycle Regulator by Amplifying E2F1 Transcription Response in Glioma. Cell Death Dis. 2021, 12, 141. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, X.; Luo, Z.; Zhang, Q.; Liu, J. Identification and Characterization of microRNAs from Chinese Pollination Constant Non-Astringent Persimmon Using High-Throughput Sequencing. BMC Plant Biol. 2015, 15, 11. [Google Scholar] [CrossRef]
- Trick, A.Y.; Chen, F.-E.; Schares, J.A.; Freml, B.E.; Lor, P.; Yun, Y.; Wang, T.-H. High Resolution Estimates of Relative Gene Abundance with Quantitative Ratiometric Regression PCR (qRR-PCR). Analyst 2021, 146, 6463–6469. [Google Scholar] [CrossRef]
- Goes, C.P.; Vieceli, F.M.; De La Cruz, S.M.; Simões-Costa, M.; Yan, C.Y.I. Scratch2, a Snail Superfamily Member, Is Regulated by miR-125b. Front. Cell Dev. Biol. 2020, 8, 769. [Google Scholar] [CrossRef]
- Xu, X.-W.; Zhou, X.-H.; Wang, R.-R.; Peng, W.-L.; An, Y.; Chen, L.-L. Functional Analysis of Long Intergenic Non-Coding RNAs in Phosphate-Starved Rice Using Competing Endogenous RNA Network. Sci. Rep. 2016, 6, 20715. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Zhu, Y.; Zhan, S.; Chao, Z.; Zhong, T.; Guo, J.; Wang, Y.; Li, L.; Zhang, H. Genome-Wide Identification and Characterization of Long Noncoding RNAs of Brown to White Adipose Tissue Transformation in Goats. Cells 2019, 8, 904. [Google Scholar] [CrossRef]
- Kern, C.; Wang, Y.; Chitwood, J.; Korf, I.; Delany, M.; Cheng, H.; Medrano, J.F.; Van Eenennaam, A.L.; Ernst, C.; Ross, P.; et al. Genome-Wide Identification of Tissue-Specific Long Non-Coding RNA in Three Farm Animal Species. BMC Genom. 2018, 19, 684. [Google Scholar] [CrossRef]
- Han, H.; Wang, X.; Li, W.; Liu, J.; Fan, Y.; Zhang, H.; Yang, J.; Gao, Y.; Liu, Y. Identification and Characterization of lncRNAs Expression Profile Related to Goat Skeletal Muscle at Different Development Stages. Animals 2022, 12, 2683. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Sun, J.; Malyar, R.M.; Shi, F. Analysis of lncRNAs and Their Regulatory Network in Skeletal Muscle Development of the Yangtze River Delta White Goat. Animals 2024, 14, 3125. [Google Scholar] [CrossRef] [PubMed]
- Connolly, M.; Paul, R.; Farre-Garros, R.; Natanek, S.A.; Bloch, S.; Lee, J.; Lorenzo, J.P.; Patel, H.; Cooper, C.; Sayer, A.A.; et al. miR-424-5p Reduces Ribosomal RNA and Protein Synthesis in Muscle Wasting. J. Cachexia Sarcopenia Muscle 2018, 9, 400–416. [Google Scholar] [CrossRef]
- Ji, L.L. Nuclear Factor κB Signaling Revisited: Its Role in Skeletal Muscle and Exercise. Free Radic. Biol. Med. 2025, 232, 158–170. [Google Scholar] [CrossRef]
- Bakkar, N.; Wang, J.; Ladner, K.J.; Wang, H.; Dahlman, J.M.; Carathers, M.; Acharyya, S.; Rudnicki, M.A.; Hollenbach, A.D.; Guttridge, D.C. IKK/NF-kappaB Regulates Skeletal Myogenesis via a Signaling Switch to Inhibit Differentiation and Promote Mitochondrial Biogenesis. J. Cell Biol. 2008, 180, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shen, K.; Lou, H.; Zhou, L.; An, Y.; Zhao, X.; Ding, Y. Clinical Relevance of Loss-of-Function Mutations of NEMO/IKBKG. Genes Dis. 2025, 12, 101531. [Google Scholar] [CrossRef]
- Straughn, A.R.; Hindi, S.M.; Xiong, G.; Kumar, A. Canonical NF-κB Signaling Regulates Satellite Stem Cell Homeostasis and Function During Regenerative Myogenesis. J. Mol. Cell Biol. 2019, 11, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Rugowska, A.; Starosta, A.; Konieczny, P. Epigenetic Modifications in Muscle Regeneration and Progression of Duchenne Muscular Dystrophy. Clin. Epigenet. 2021, 13, 13. [Google Scholar] [CrossRef] [PubMed]




| Name | NCBI Login Number | Primer Sequence (5′ → 3′) | Fragment Size, bp |
|---|---|---|---|
| β-Actin | F: GGACTTCGAGCAGGAGATGG | 104 | |
| R: CCAGGAAGGAAGGCTGGAAG | |||
| IKBKG | XM_013976823.2 | F: CTGAAGACATGCCAGCAGATG | 126 |
| R: CAAAGCCTGGCGCTCCTTAG | |||
| MLXIP | XM_018061302.1 | F: TTTAAAGCAGAACCGGCAGAT | 165 |
| R: TGCAGCTTGGTGATGTACTCC | |||
| FBXO32 | XM_005688865.3 | F: AAGCTGATCCATGCGAGTGT | 100 |
| R: CCTTTCCTCAAACGCTGTGC | |||
| MSTRG.5182.1 | F: GTTTACAGGGAGCAGAGCGT | 157 | |
| R: TAACCGTTCTGCAGGGACAC | |||
| MSTRG.5992.3 | F: GCTCCTGGAATTGGGGTCTC | 197 | |
| R: CAGATTGGGGGAAGCAGAGG | |||
| MSTRG.7890.1 | F: TCCCCATAGACTGAGGAGCC | 223 | |
| R: CACAGGTGGGAGAGTAGGGA |
| Name | Primer Type | Primer Sequence (5′ → 3′) | Fragment Size, bp |
|---|---|---|---|
| U6 | F | GGAACGATACAGAGAAGATTAGC | 68 |
| R | TGGAACGCTTCACGAATTTGCG | ||
| chi-miR-424-5p | SL-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTTTGA | - |
| F | GCGCCAGCAGCAATTCATG | 65 | |
| R | GTCGTATCCAGTGCAGGGT | ||
| chi-miR-22-3p | SL-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGAAC | - |
| F | GCGCAAGCTGCCAGTTG | 67 | |
| R | GTCGTATCCAGTGCAGGGT | ||
| chi-miR-145-3p | SL-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTTCTT | - |
| F | GCGCGCATTCCTGGAAATACT | 71 | |
| R | GTCGTATCCAGTGCAGGGT |
| Sample | Raw Reads | Clean Reads | Error Rate | Q20 /% | Q30 /% | GC Content | Total Mapped | Mapped Ratio |
|---|---|---|---|---|---|---|---|---|
| CG-1 | 90,671,620 | 90,270,588 | 0.06 | 98.00 | 94.27 | 56.96 | 89,631,538 | 83.02 |
| CG-2 | 95,248,262 | 94,803,262 | 0.09 | 97.77 | 93.77 | 57.31 | 94,367,874 | 78.39 |
| CG-3 | 97,739,300 | 97,350,874 | 0.06 | 97.90 | 94.04 | 56.91 | 96,925,510 | 82.34 |
| CG-4 | 109,992,834 | 109,499,310 | 0.07 | 97.78 | 93.81 | 57.91 | 108,985,938 | 78.03 |
| CG-5 | 88,231,856 | 87,945,132 | 0.05 | 97.94 | 94.04 | 57.69 | 87,572,092 | 76.57 |
| CG-6 | 96,761,602 | 96,367,518 | 0.06 | 97.90 | 94.04 | 57.96 | 95,923,486 | 77.52 |
| CG-7 | 77,733,584 | 77,478,014 | 0.05 | 97.98 | 94.16 | 56.49 | 77,079,874 | 76.81 |
| CG-8 | 81,516,648 | 81,160,334 | 0.06 | 97.73 | 93.67 | 56.32 | 80,736,506 | 76.29 |
| CG-9 | 80,439,174 | 80,129,922 | 0.04 | 97.89 | 94.04 | 57.31 | 79,693,318 | 78.01 |
| CG-10 | 90,861,758 | 90,559,984 | 0.06 | 98.19 | 94.62 | 58.86 | 90,093,198 | 84.88 |
| CG-11 | 80,538,374 | 80,276,974 | 0.06 | 98.14 | 94.48 | 58.22 | 79,957,986 | 86.23 |
| CG-12 | 83,032,174 | 82,815,918 | 0.05 | 98.30 | 94.83 | 58.50 | 82,474,292 | 82.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Wang, F.; Zhou, L.; Ren, Z.; Shang, S.; Qu, L.; Zhu, H.; Zhang, L. Construction and Functional Analysis of the ceRNA Regulatory Network Associated with Muscle Development in Shaanbei White Cashmere Goats. Animals 2025, 15, 3568. https://doi.org/10.3390/ani15243568
Liu L, Wang F, Zhou L, Ren Z, Shang S, Qu L, Zhu H, Zhang L. Construction and Functional Analysis of the ceRNA Regulatory Network Associated with Muscle Development in Shaanbei White Cashmere Goats. Animals. 2025; 15(24):3568. https://doi.org/10.3390/ani15243568
Chicago/Turabian StyleLiu, Lina, Fenghong Wang, Long Zhou, Zhaofei Ren, Shutao Shang, Lei Qu, Haijing Zhu, and Lei Zhang. 2025. "Construction and Functional Analysis of the ceRNA Regulatory Network Associated with Muscle Development in Shaanbei White Cashmere Goats" Animals 15, no. 24: 3568. https://doi.org/10.3390/ani15243568
APA StyleLiu, L., Wang, F., Zhou, L., Ren, Z., Shang, S., Qu, L., Zhu, H., & Zhang, L. (2025). Construction and Functional Analysis of the ceRNA Regulatory Network Associated with Muscle Development in Shaanbei White Cashmere Goats. Animals, 15(24), 3568. https://doi.org/10.3390/ani15243568





