Cryptic Divergence of Rochia nilotica (Gastropoda: Tegulidae) from Chuuk Lagoon, Federated States of Micronesia, Revealed by Morphological and Mitochondrial Genome Analyses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
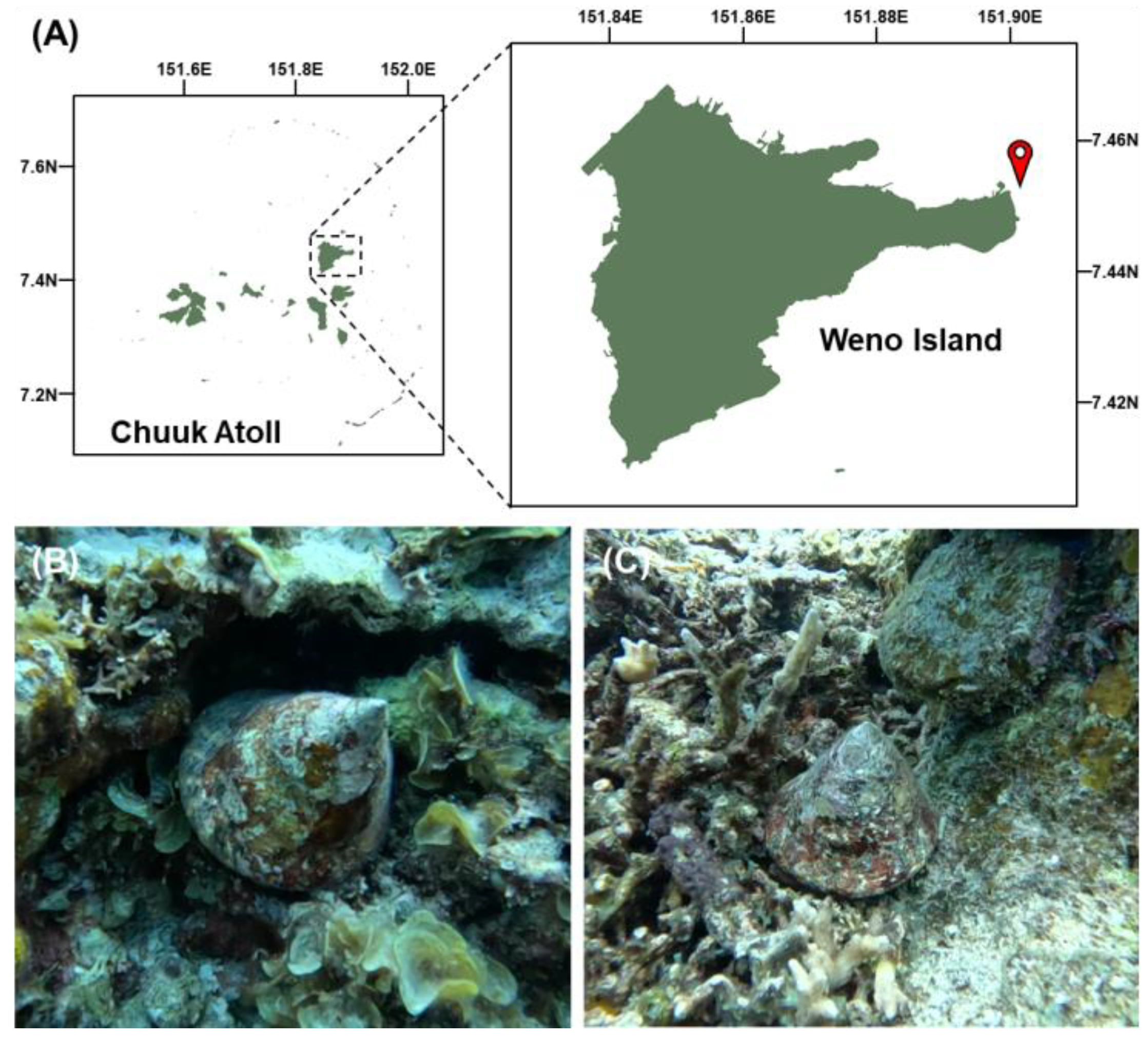
2.1. Study Area and Sampling
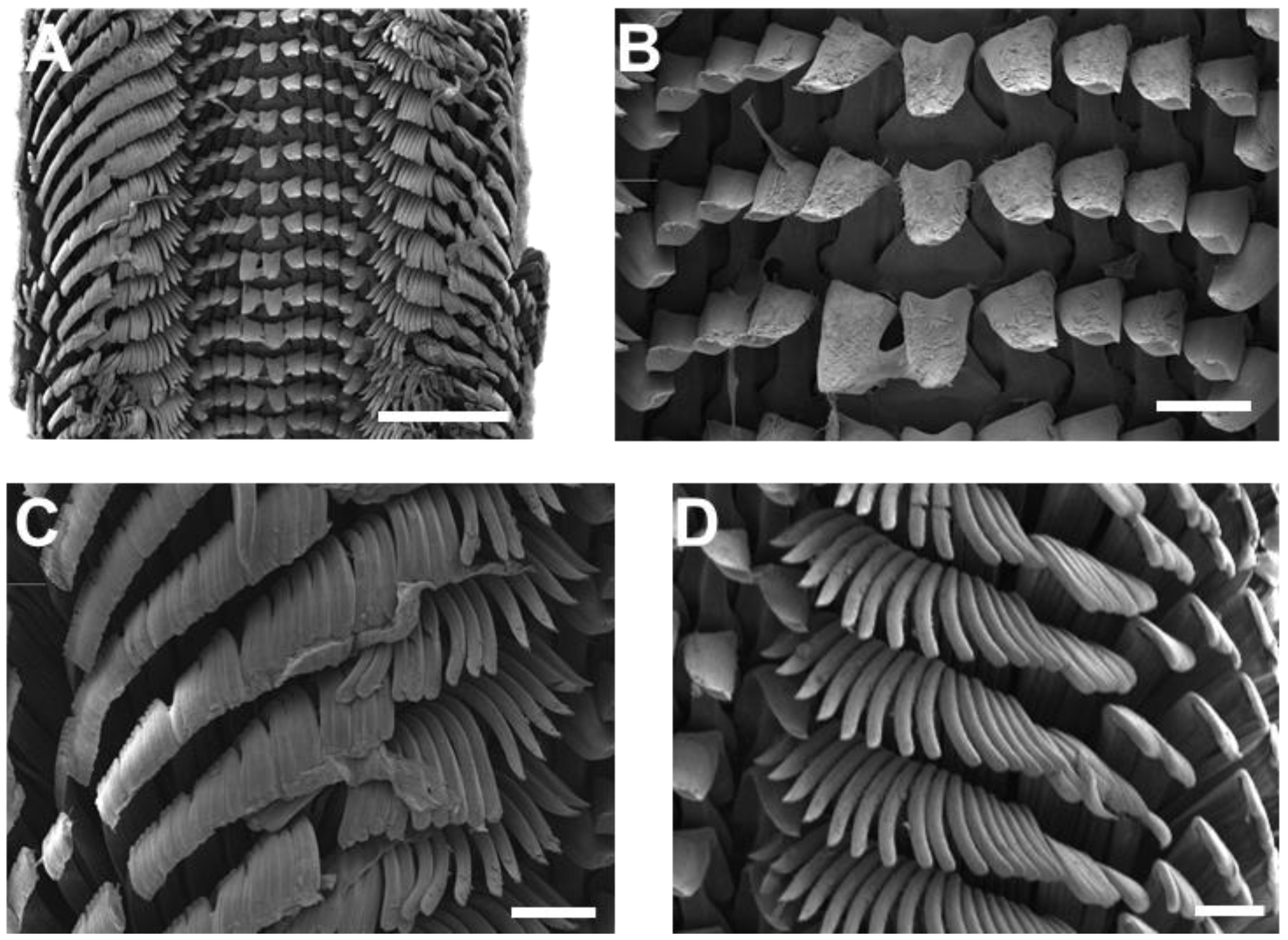
2.2. Morphological Analysis
2.3. Molecular Analysis
2.3.1. DNA Extraction and Sequencing
2.3.2. De Novo Assembly and Annotation
2.4. Phylogenetic Analysis
3. Results and Discussion
3.1. Morphological Characteristics of Rochia nilotica from Chuuk Atoll
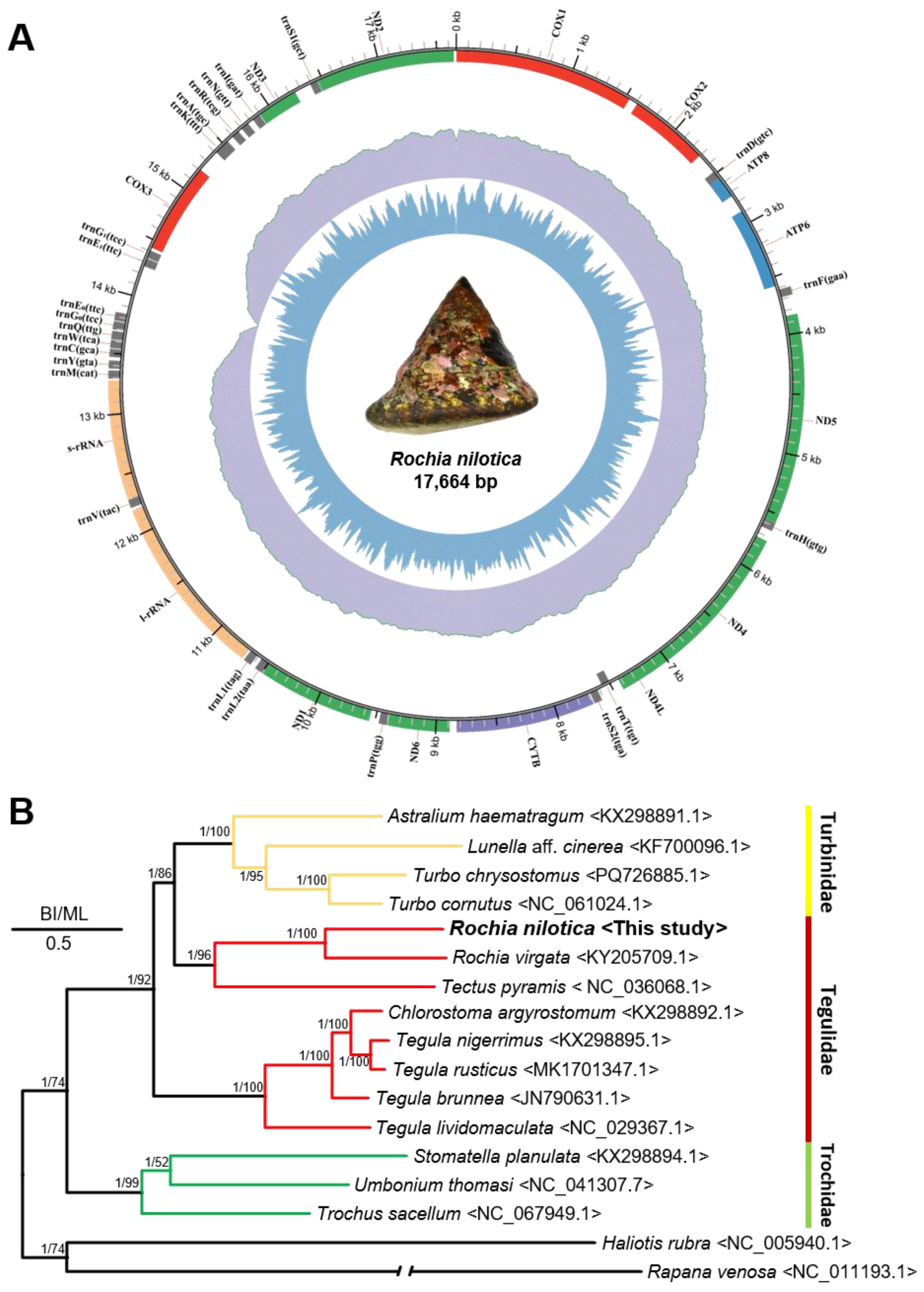
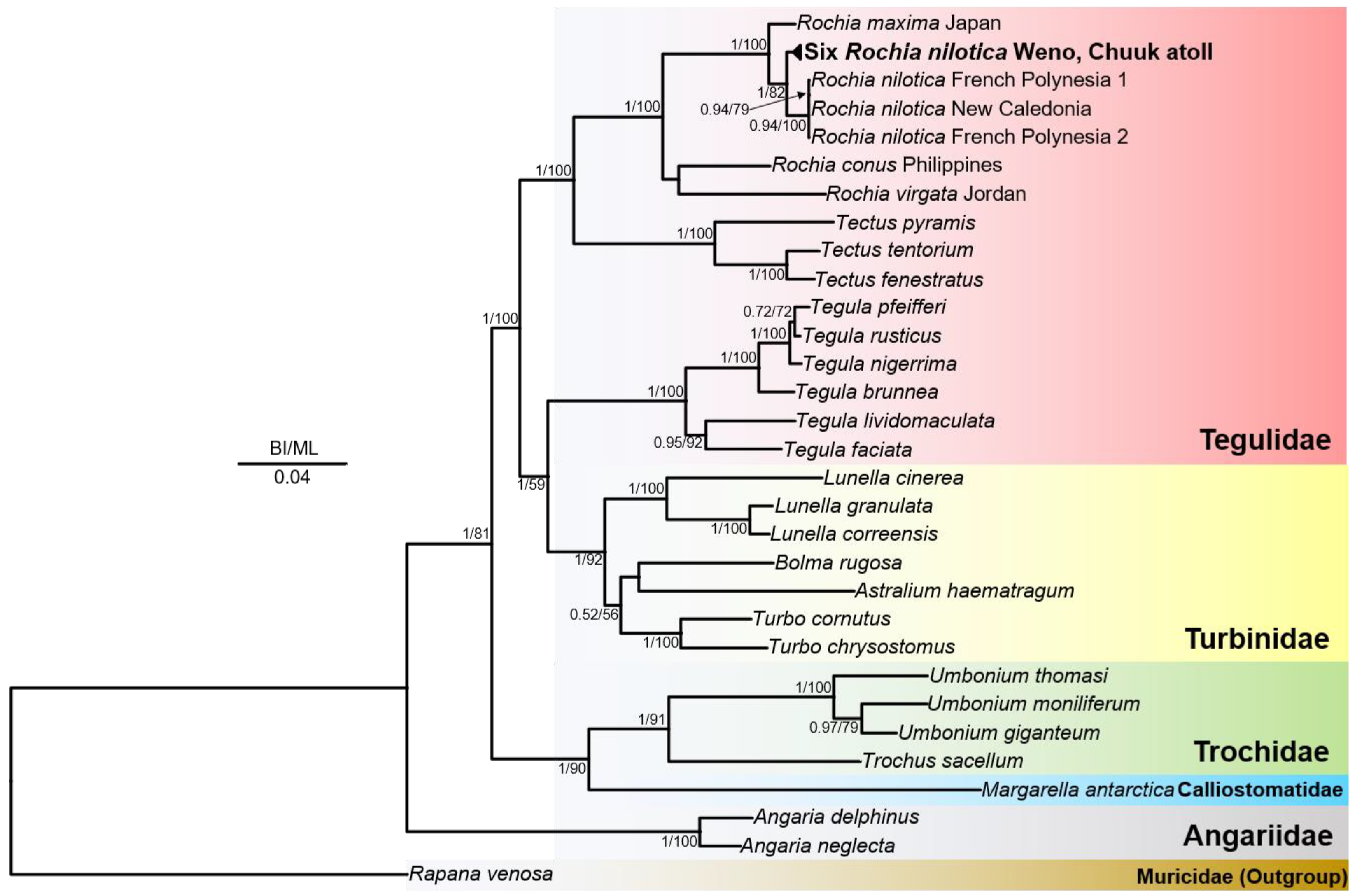
3.2. Mitochondrial Genome Characterization and Phylogeny
3.3. Species Identification Based on Partial COX1 and 16S rRNA Gene Sequences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ulm, S.; McNiven, I.J.; Aird, S.J.; Lambrides, A.B. Sustainable Harvesting of Conomurex luhuanus and Rochia nilotica by Indigenous Australians on the Great Barrier Reef over the Past 2000 Years. J. Archaeol. Sci. Rep. 2019, 28, 102017. [Google Scholar] [CrossRef]
- Berry, O.; Richards, Z.; Moore, G.; Hernawan, U.; Travers, M.; Gruber, B. Oceanic and Coastal Populations of a Harvested Macroinvertebrate Rochia nilotica in North-Western Australia Are Isolated and May Be Locally Adapted. Mar. Freshw. Res. 2019, 71, 782–793. [Google Scholar] [CrossRef]
- Wahyudi, N.D.; Hidayati, D.; Arbi, U.Y.; Ismail, A. Morphometric Study of Lola Rochia nilotica (Linnaeus 1767) Shells from Natural Harvest Found in Indonesian. Biodiversitas J. Biol. Divers. 2023, 24, 4711–4722. [Google Scholar] [CrossRef]
- Gillett, R.; McCoy, M.; Bertram, I.; Kinch, J.; Desurmont, A. Trochus in the Pacific Islands: A Review of the Fisheries, Management and Trade; Fisheries, Aquaculture and Marine Ecosystems Division, Secretariat of the Pacific Community (SPC): Noumea, New Caledonia, 2020. [Google Scholar]
- Balisco, R.A.; Gonzales, B.; Dolorosa, R. Species Composition, Abundance and Conservation Status of Some Economically Important Macrobenthic Invertebrates in Pag-Asa Island, Kalayaan, Palawan, Philippines. Asian Fish. Sci. 2020, 33, 357. [Google Scholar] [CrossRef]
- Lasi, F. Trochus Production in Solomon Islands from 1953 to 2006. SPC Trochus Inf. Bull. 2010, 15, 24–27. [Google Scholar]
- Lincoln, J.M.; Schlanger, S.O. Atoll Stratigraphy as a Record of Sea Level Change: Problems and Prospects. J. Geophys. Res. Solid Earth 1991, 96, 6727–6752. [Google Scholar] [CrossRef]
- George, A.; Luckymis, M.; Palik, S.; Adams, K.; Joseph, E.; Mathias, D.; Malakai, S.; Nakayama, M.R.; Graham, C.; Rikim, K. The State of Coral Reef Ecosystems of the Federated States of Micronesia; National Oceanographic and Atmospheric Administration: Washington, DC, USA, 2008. [Google Scholar]
- Kim, T.; Lee, D.-W.; Kim, H.-J.; Jung, Y.-H.; Choi, Y.-U.; Oh, J.-H.; Kim, T.-H.; Kang, D.-H.; Park, H.-S. Estimation of the Benthic Habitat Zonation by Photo-Quadrat Image Analysis along the Fringing Reef of Weno Island, Chuuk, Micronesia. J. Mar. Sci. Eng. 2022, 10, 1643. [Google Scholar] [CrossRef]
- Jin, Y.-S.; Park, Y.-J.; Kim, H.-J.; Na, O.-S.; Song, Y.-B.; Lee, C.-H.; Choi, M.-S.; Rho, S.; Lee, Y.-D. Reproductive Cycle of Top Shell, Trochus niloticus in Chuuk Island, Micronesia. Korean J. Malacol. 2004, 20, 65–73. [Google Scholar]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kim, J.G.; Kwon, O.-N.; Park, J.J.C.; Lee, K.-W.; Choi, Y.-U. On the Species Identification of Korean Geoduck Clam (Panopea sp. 1) Based on the Morphological and Molecular Evidence. J. Mar. Sci. Eng. 2023, 11, 2115. [Google Scholar] [CrossRef]
- Kim, M.; Choi, H.; Kim, H.; Kang, J.; Jeong, H.G.; Eyun, S.; Kang, J.-H. Characterization of the Mitochondrial Genome, Ecological Distribution, and Morphological Features of the Marine Gastropod Mollusc Lophocochlias parvissimus (Gastropoda, Tornidae). J. Mar. Sci. Eng. 2023, 11, 2307. [Google Scholar] [CrossRef]
- Shin, J.-S.; Song, C.; Choi, H.; Yang, S.H.; Kwon, K.K.; Eyun, S.; Choi, K.-S. The Complete Mitochondrial Genome of the Chemosymbiotic Lucinid Bivalve Pillucina pisidium (Dunker, 1860) Occurring in Seagrass Zostera marina Bed in a Lagoon in Jeju Island, Korea. J. Mar. Sci. Eng. 2024, 12, 847. [Google Scholar] [CrossRef]
- Jiang, D.; Liu, Z.; Zeng, X.; Tang, X.; Yang, S.; Zheng, X. The Complete Mitochondrial Genome of Rochia nilotica (Gastropoda: Tegulidae). Mitochondrial DNA Part B 2019, 4, 1121–1122. [Google Scholar] [CrossRef]
- Smith, B.D. Growth Rate, Distribution and Abundance of the Introduced Topshell Trochus niloticus Linnaeus on Guam, Mariana Islands. Bull. Mar. Sci. 1987, 41, 466–474. [Google Scholar]
- Krueger, F. Trim Galore!: A Wrapper around Cutadapt and FastQC to Consistently Apply Adapter and Quality Trimming to FastQ Files, with Extra Functionality for RRBS Data; Babraham Institute: Cambridge, UK, 2015. [Google Scholar]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC Results into a Web-Based, Interactive, and Extensible FASTQ Quality Control Tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A Toolkit for Animal Mitochondrial Genome Assembly, Annotation and Visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A Fast and Scalable Metagenome Assembler Driven by Advanced Methodologies and Community Practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Okutani, T. Marine Mollusks in Japan; Tokai University Press: Hiratsuka, Japan, 2000. [Google Scholar]
- Dharma, B.; Pringgenies, D. Differences in Shell Morphology of the Species Rochia nilotica (Linnaeus, 1767) and Rochia maxima (F. C. L. Koch, 1844) (Gastropoda: Tegulidae). J. Moluska Indones. 2023, 7, 36–42. [Google Scholar] [CrossRef]
- Pilsbry, H.A. New Japanese Marine Mollusca: Gastropoda. Proc. Acad. Nat. Sci. USA 1904, 56, 3–37. [Google Scholar]
- Poppe, G.T. Descriptions of Spectacular New Species from the Philippines. Visaya 2004, 1, 4–19. [Google Scholar]
- Williams, S.T. Advances in Molecular Systematics of the Vetigastropod Superfamily Trochoidea. Zool. Scr. 2012, 41, 571–595. [Google Scholar] [CrossRef]
- Guo, E.; Yang, Y.; Kong, L.; Yu, H.; Liu, S.; Liu, Z.; Li, Q. Mitogenomic Phylogeny of Trochoidea (Gastropoda: Vetigastropoda): New Insights from Increased Complete Genomes. Zool. Scr. 2021, 50, 43–57. [Google Scholar] [CrossRef]
- Cunha, T.J.; Reimer, J.D.; Giribet, G. Investigating Sources of Conflict in Deep Phylogenomics of Vetigastropod Snails. Syst. Biol. 2022, 71, 1009–1022. [Google Scholar] [CrossRef]
- James, J.E.; Lanfear, R.; Eyre-Walker, A. Molecular Evolutionary Consequences of Island Colonization. Genome Biol. Evol. 2016, 8, 1876–1888. [Google Scholar] [CrossRef] [PubMed]
- Mayer, L.; Jakobsson, M.; Allen, G.; Dorschel, B.; Falconer, R.; Ferrini, V.; Lamarche, G.; Snaith, H.; Weatherall, P. The Nippon Foundation—GEBCO Seabed 2030 Project: The Quest to See the World’s Oceans Completely Mapped by 2030. Geosciences 2018, 8, 63. [Google Scholar] [CrossRef]
- Foale, S.; Day, R. Stock Assessment of Trochus (Trochus niloticus) (Gastropoda: Trochidae) Fisheries at West Nggela, Solomon Islands. Fish. Res. 1997, 33, 1–16. [Google Scholar] [CrossRef]
- Heslinga, G.A.; Hillmann, A. Hatchery Culture of the Commercial Top Snail Trochus niloticus in Palau, Caroline Islands. Aquaculture 1981, 22, 35–43. [Google Scholar] [CrossRef]
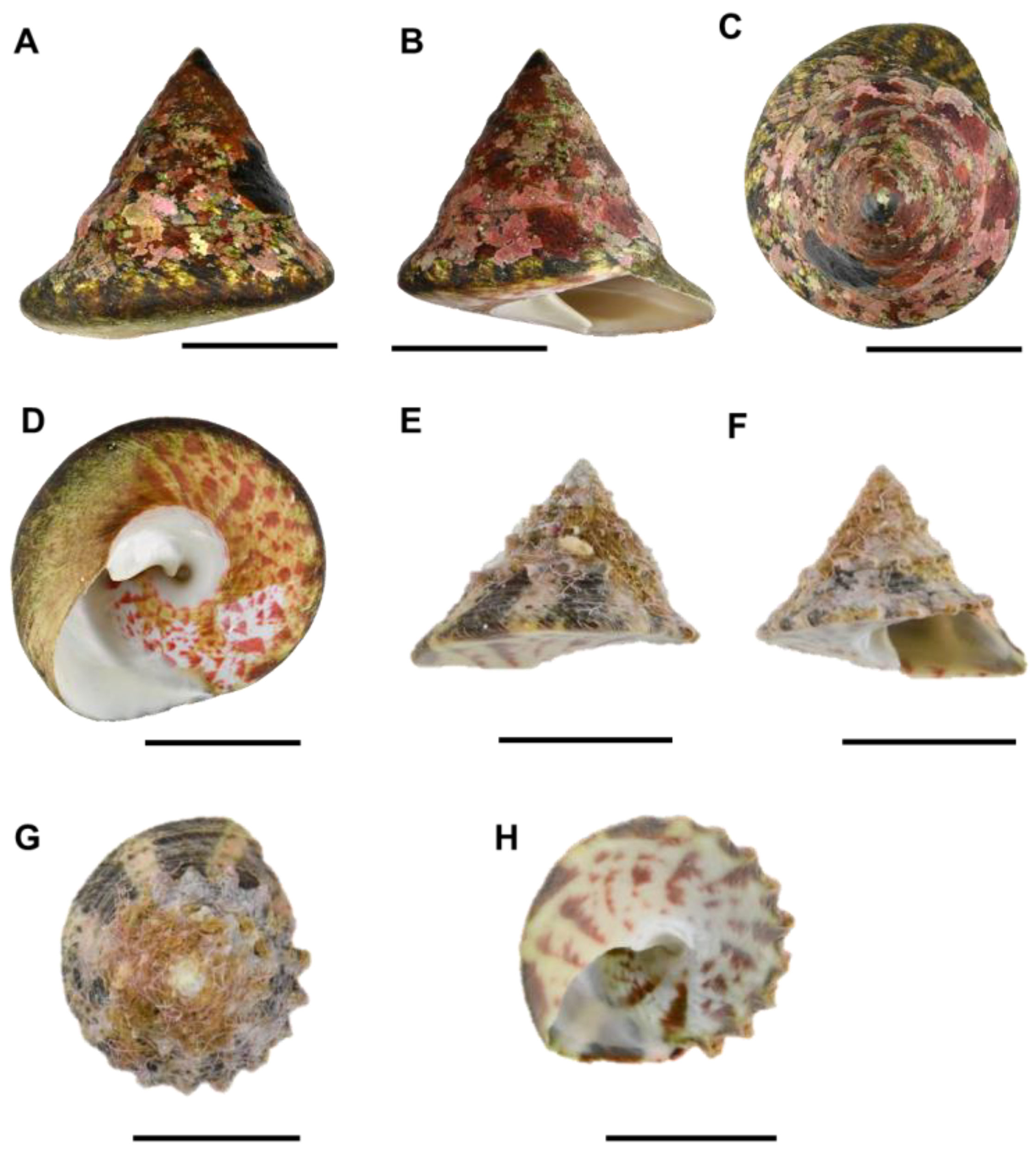
| Voucher Number | SH (mm) | SW (mm) | H/W |
|---|---|---|---|
| HNIBRIV 18781 | 101.39 | 104.83 | 0.97 |
| HNIBRIV 18782 | 101.34 | 104.37 | 0.97 |
| HNIBRIV 18783 | 91.73 | 100.85 | 0.91 |
| HNIBRIV 18784 | 86.94 | 96.34 | 0.90 |
| HNIBRIV 18785 | 92.58 | 96.27 | 0.96 |
| HNIBRIV 18786 | 102.70 | 98.57 | 1.04 |
| HNIBRIV 18787 | 90.70 | 91.99 | 0.99 |
| HNIBRIV 18788 | 86.01 | 92 | 0.93 |
| HNIBRIV 18789 | 88.49 | 94.53 | 0.94 |
| HNIBRIV 18790 | 89.07 | 95.94 | 0.93 |
| Average | 93.10 | 97.57 | 0.95 |
| Gene Name | Location | Size | Start Codon | Stop Codon | Intergenic Region |
|---|---|---|---|---|---|
| Cytochrome c oxidase subunit 1 | 1–1536 | 1536 | ATG | TAA | 74 |
| Cytochrome c oxidase subunit 2 | 1611–2297 | 687 | ATG | TAA | 179 |
| tRNA-Asp | 2477–2545 | 69 | - | - | 0 |
| ATP synthase F0 subunit 8 | 2546–2734 | 189 | ATG | TAA | 149 |
| ATP synthase F0 subunit 6 | 2884–3579 | 696 | ATG | TAG | 36 |
| tRNA-Phe | 3616–3683 | 68 | - | - | 159 |
| NADH dehydrogenase subunit 5 | 3843–5576 | 1734 | ATG | TAA | 0 |
| tRNA-His | 5577–5643 | 67 | - | - | 90 |
| NADH dehydrogenase subunit 4 | 5734–7131 | 1398 | ATG | TAG | −7 |
| NADH dehydrogenase subunit 4L | 7125–7424 | 300 | ATG | TAA | 73 |
| tRNA-Thr | 7498–7568 | 71 | - | - | 48 |
| tRNA-Ser | 7617–7683 | 67 | - | - | 8 |
| Cytochrome b | 7692–8831 | 1140 | ATG | TAA | 55 |
| NADH dehydrogenase subunit 6 | 8887–9393 | 507 | ATG | TAA | 4 |
| tRNA-Pro | 9398–9467 | 70 | - | - | 79 |
| NADH dehydrogenase subunit 1 | 9547–10,494 | 948 | ATA | TAA | 4 |
| tRNA-Leu | 10,499–10,566 | 68 | - | - | 45 |
| tRNA-Leu | 10,612–10,679 | 68 | - | - | 15 |
| 16S ribosomal RNA | 10,695–12,202 | 1508 | - | - | 22 |
| tRNA-Val | 12,225–12,294 | 70 | - | - | 3 |
| 12S ribosomal RNA | 12,298–13,280 | 983 | - | - | 12 |
| tRNA-Met | 13,293–13,360 | 68 | - | - | 15 |
| tRNA-Tyr | 13,376–13,441 | 66 | - | - | 28 |
| tRNA-Cys | 13,470–13,535 | 66 | - | - | 6 |
| tRNA-Trp | 13,542–13,608 | 67 | - | - | 5 |
| tRNA-Gln | 13,614–13,682 | 69 | - | - | 11 |
| tRNA-Gly | 13,694–13,761 | 68 | - | - | 6 |
| tRNA-Glu | 13,768–13,837 | 70 | - | - | 426 |
| tRNA-Glu | 14,264–14,333 | 70 | - | - | 6 |
| tRNA-Gly | 14,340–14,407 | 68 | - | - | 24 |
| cytochrome c oxidase subunit 3 | 14,432–15,211 | 780 | ATG | TAG | 195 |
| tRNA-Lys | 15,407–15,478 | 72 | - | - | −5 |
| tRNA-Ala | 15,474–15,541 | 68 | - | - | 55 |
| tRNA-Arg | 15,597–15,665 | 69 | - | - | 34 |
| tRNA-Asn | 15,700–15,770 | 71 | - | - | 50 |
| tRNA-Ile | 15,821–15,889 | 69 | - | - | 4 |
| NADH dehydrogenase subunit 3 | 15,894–16,247 | 354 | ATG | TAG | 142 |
| tRNA-Ser | 16,390–16,457 | 68 | - | - | 3 |
| NADH dehydrogenase subunit 2 | 16,461–17,645 | 1185 | ATG | TAG | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.-S.; Park, Y.-J.; Lee, C.; Park, H.-S.; Kim, D.; Song, C.-u.; Kwon, K.; Hur, S.-W.; Min, B.-H.; Kim, J.; et al. Cryptic Divergence of Rochia nilotica (Gastropoda: Tegulidae) from Chuuk Lagoon, Federated States of Micronesia, Revealed by Morphological and Mitochondrial Genome Analyses. Animals 2025, 15, 3471. https://doi.org/10.3390/ani15233471
Shin J-S, Park Y-J, Lee C, Park H-S, Kim D, Song C-u, Kwon K, Hur S-W, Min B-H, Kim J, et al. Cryptic Divergence of Rochia nilotica (Gastropoda: Tegulidae) from Chuuk Lagoon, Federated States of Micronesia, Revealed by Morphological and Mitochondrial Genome Analyses. Animals. 2025; 15(23):3471. https://doi.org/10.3390/ani15233471
Chicago/Turabian StyleShin, Jong-Seop, Yeong-Ji Park, Changju Lee, Heung-Sik Park, Dongsung Kim, Chi-une Song, Kyungman Kwon, Sang-Woo Hur, Byung-Hwa Min, June Kim, and et al. 2025. "Cryptic Divergence of Rochia nilotica (Gastropoda: Tegulidae) from Chuuk Lagoon, Federated States of Micronesia, Revealed by Morphological and Mitochondrial Genome Analyses" Animals 15, no. 23: 3471. https://doi.org/10.3390/ani15233471
APA StyleShin, J.-S., Park, Y.-J., Lee, C., Park, H.-S., Kim, D., Song, C.-u., Kwon, K., Hur, S.-W., Min, B.-H., Kim, J., & Yang, H.-S. (2025). Cryptic Divergence of Rochia nilotica (Gastropoda: Tegulidae) from Chuuk Lagoon, Federated States of Micronesia, Revealed by Morphological and Mitochondrial Genome Analyses. Animals, 15(23), 3471. https://doi.org/10.3390/ani15233471





