Optimizing Nucleic Acid Extraction from Extended Bovine Semen for Endemic and High-Consequence Pathogens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Field and Reference Samples
2.2. Extraction Chemistries and Equipment
2.3. Polymerase Chain Reactions (PCR)
2.4. PCR and Statistical Analysis
3. Results
3.1. Extraction Method Comparison
3.1.1. New Zealand Requirement of 200 µL Semen Input
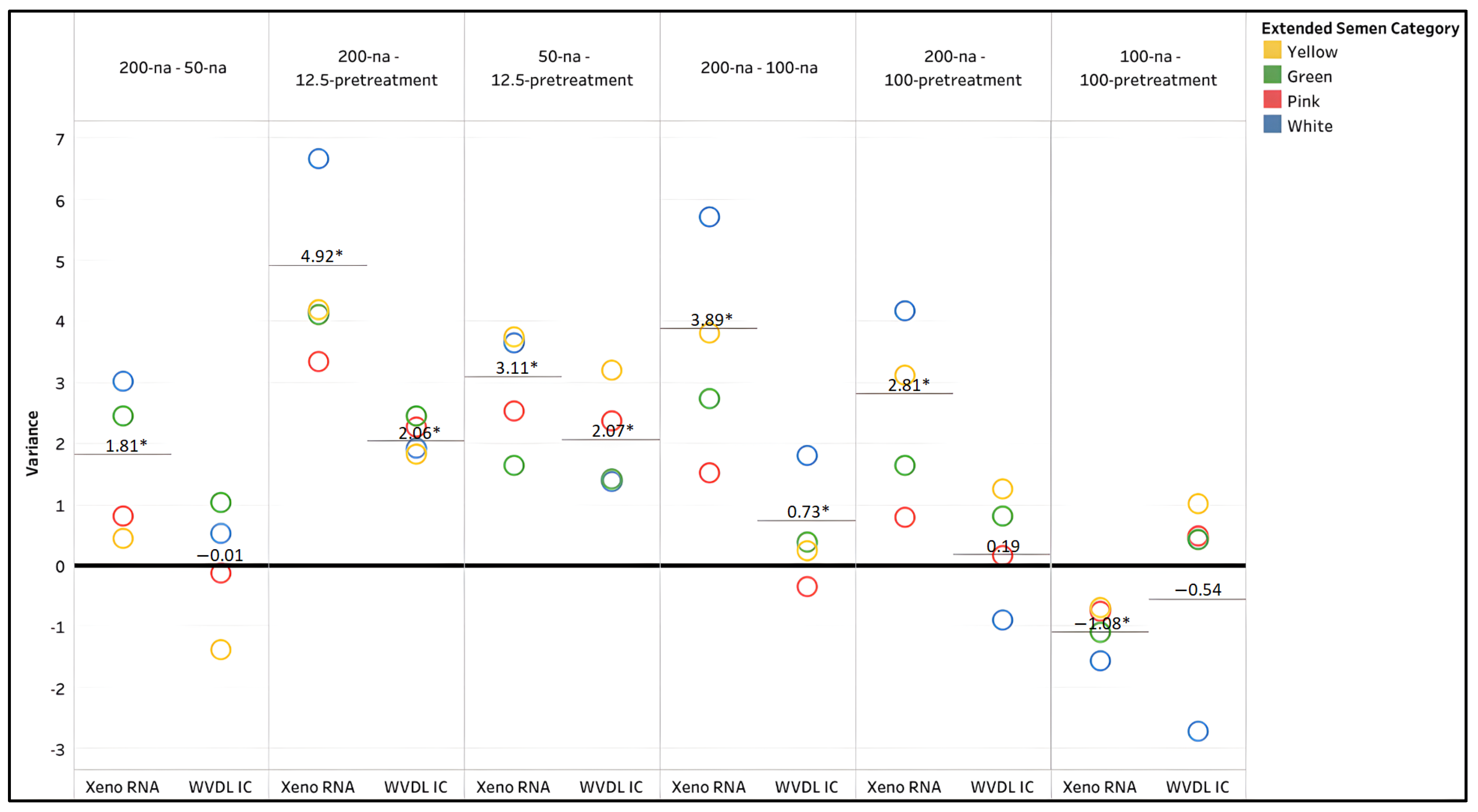
3.1.2. Manufacturer-Suggested Extraction Modifications
3.1.3. In-Depth Analysis of the PCR Internal Controls
3.2. Evaluation of Diagnostic Sensitivity for the Selected Extraction Protocols
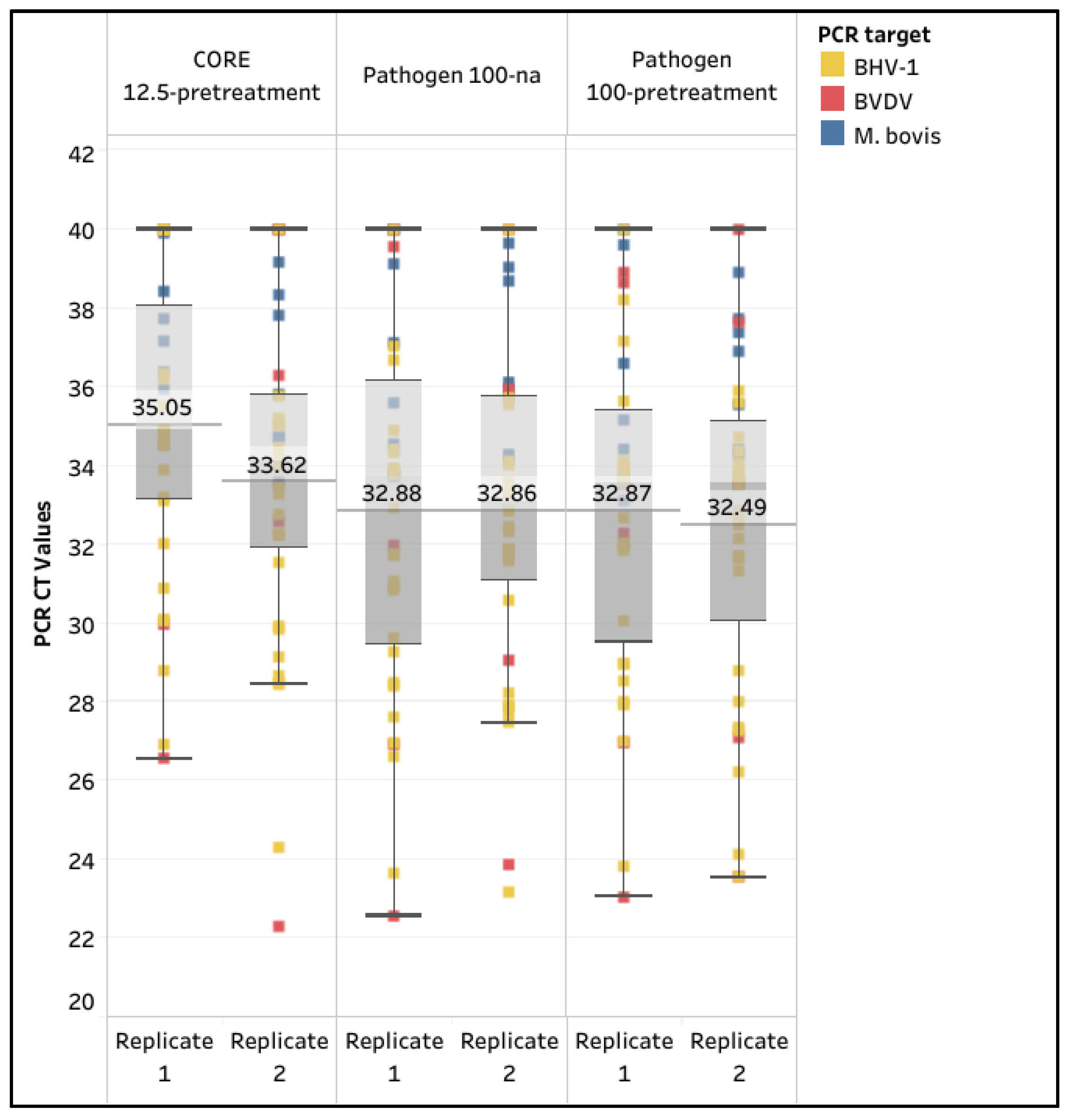
3.3. Evaluation of Analytical Sensitivity with CORE 12.5-Pretreatment, Pathogen 100-na, and Pathogen 100-Pretreatment for M. bovis
3.4. Evaluation of Pathogen 100-na Protocol with the IAV Assays
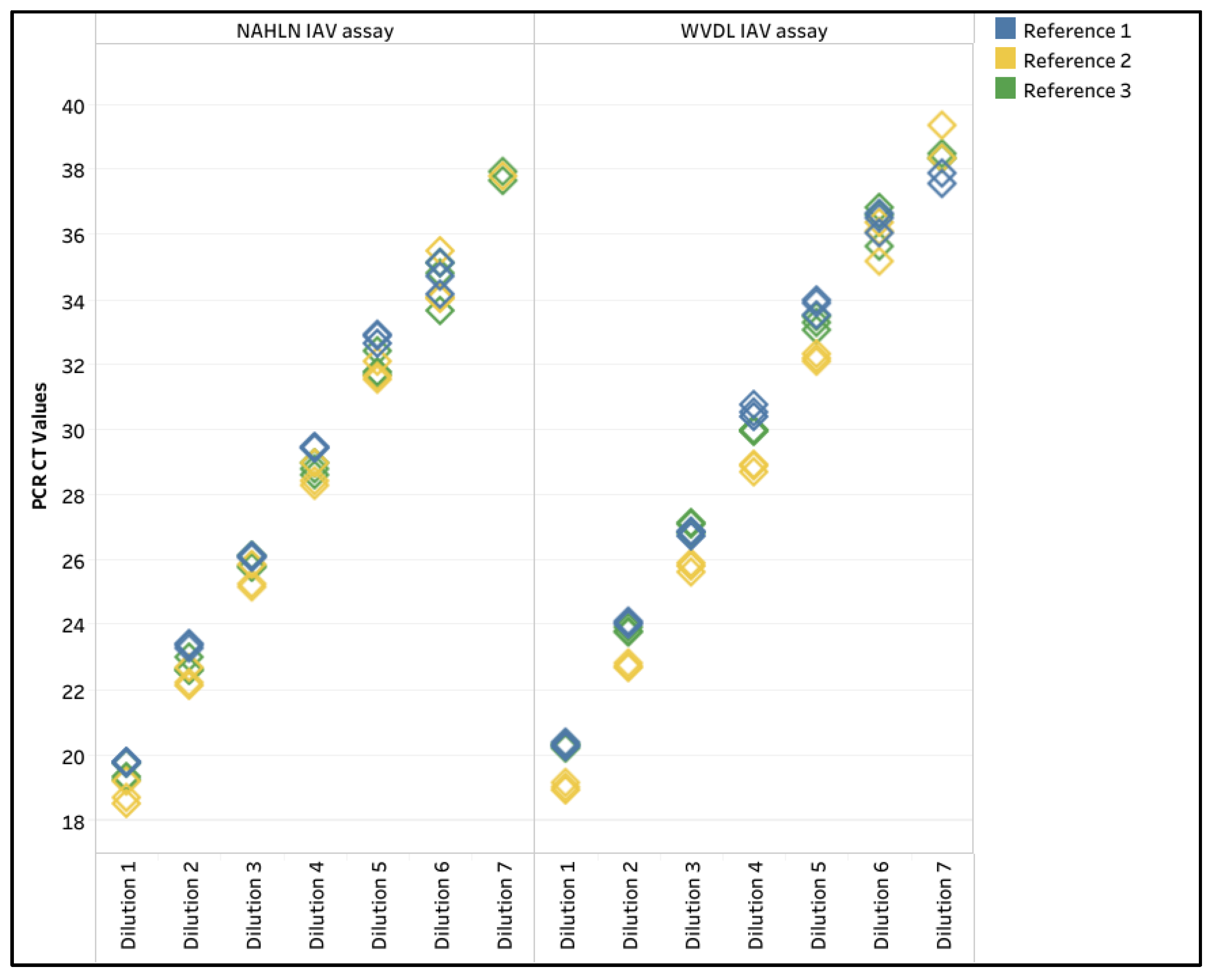
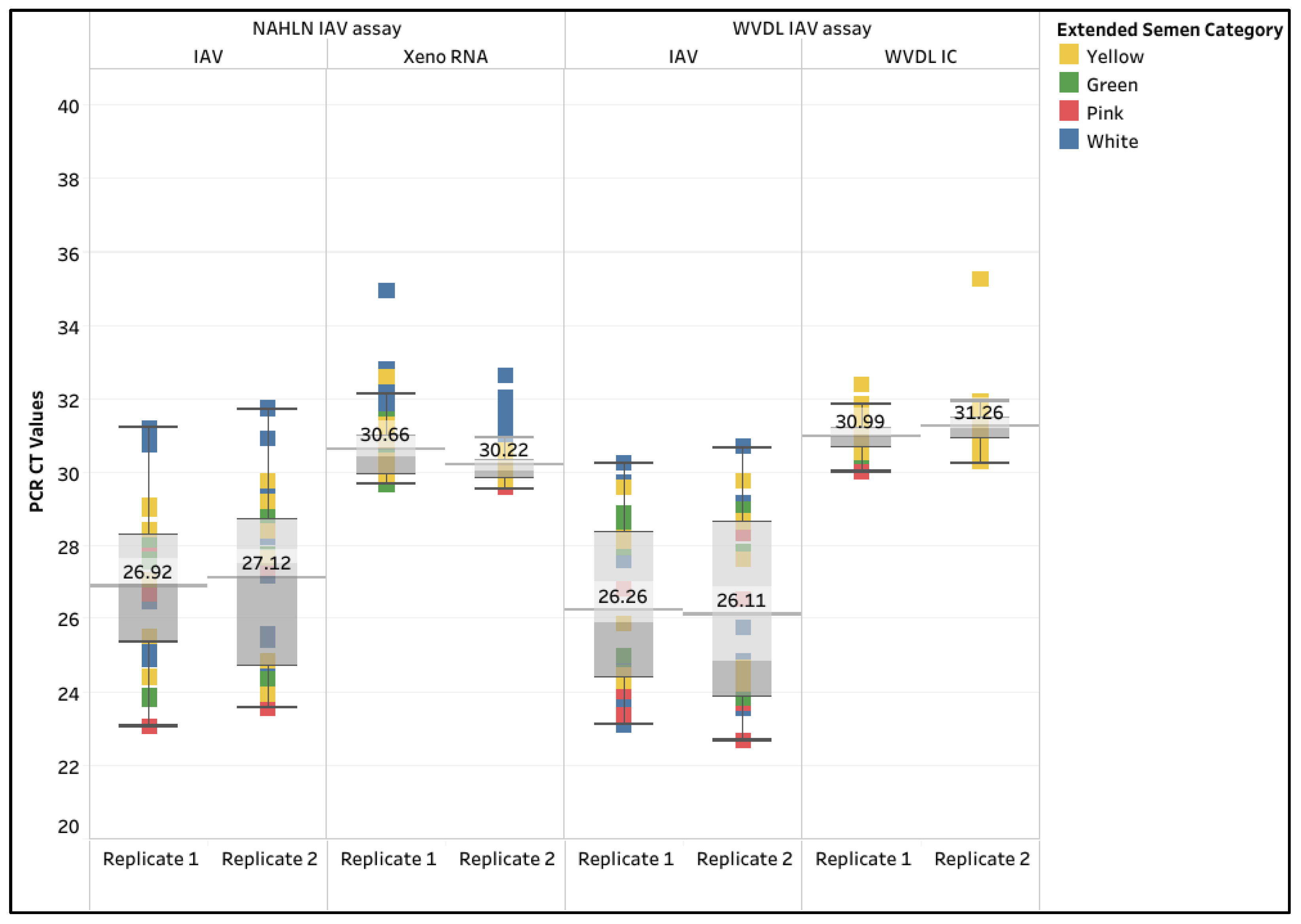
3.4.1. Limit of Detection, R2, and Percent PCR Efficiency
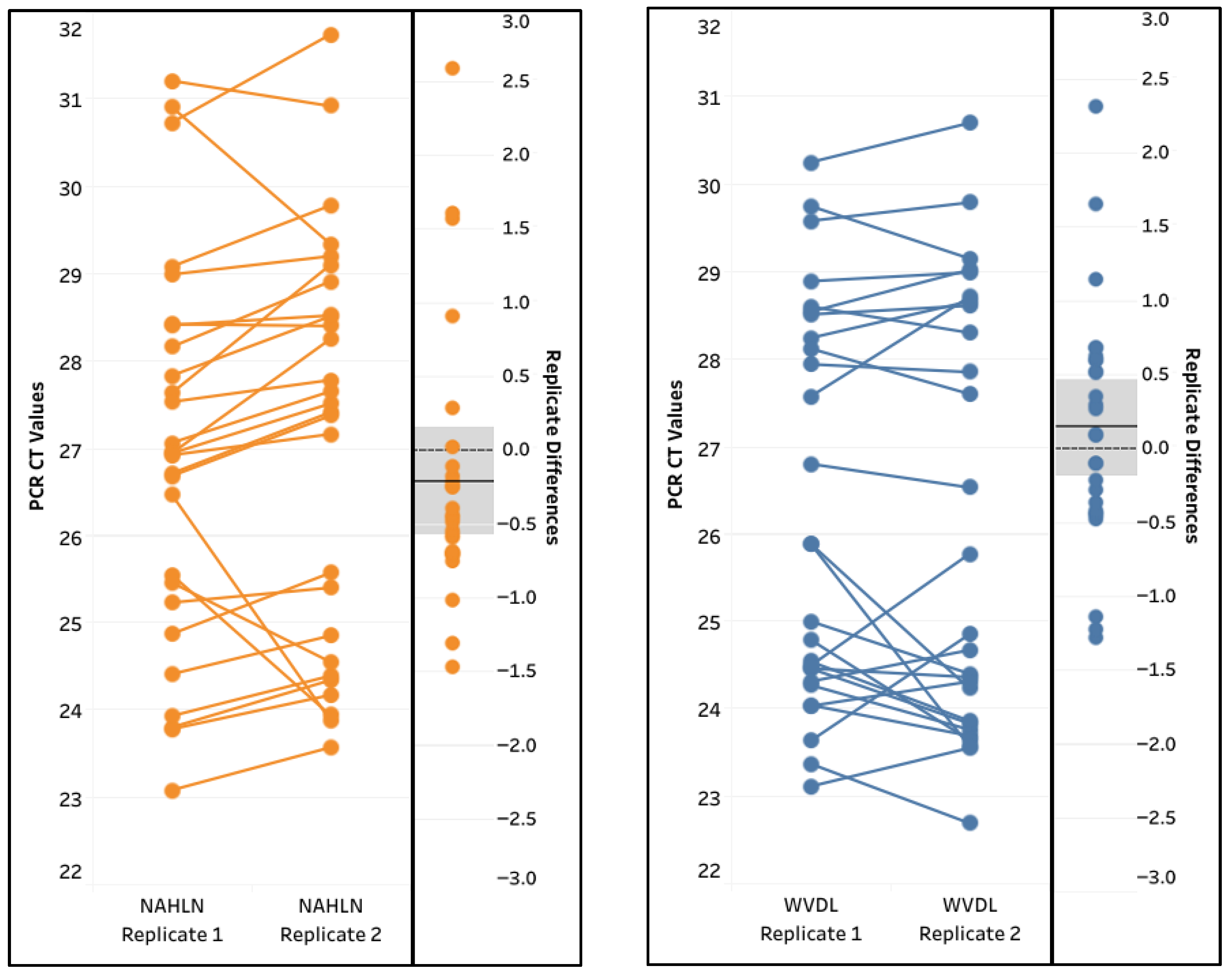
3.4.2. Diagnostic Sensitivity and Specificity, and Inter-Run Repeatability
3.5. Influenza A Virus Surveillance of Semen Samples Used in This Study
4. Discussion
4.1. Extraction Method Optimization
4.2. Evaluation of Diagnostic Sensitivity of the Selected Extraction Protocols
4.3. Evaluation of Analytical Sensitivity with CORE 12.5-Pretreatment, Pathogen 100-na, and Pathogen 100-Pretreatment for M. bovis
4.4. Evaluation of Pathogen 100-na Protocol with the IAV Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of Variance |
| BHV-1 | Bovine herpesvirus-1 |
| BVDV | Bovine viral diarrhea virus |
| CORE | MagMAX CORE Nucleic Acid Purification Kit |
| CT | Cycle threshold |
| FMDV | Foot and mouth disease virus |
| HPAI | Highly Pathogenic Avian Influenza |
| IAV | Influenza A virus |
| IC | Internal control |
| LOD | Limit of detection |
| LPAI | Low Pathogenic Avian Influenza |
| MagMAX CORE | MagMAX CORE Nucleic Acid Purification Kit |
| M. bovis | Mycoplasma bovis |
| NAHLN | National Animal Health Laboratory Network |
| NAHLN assay | NAHLN IAV Matrix PCR assay |
| NVSL | National Veterinary Service Laboratories |
| Pathogen | IndiMag Pathogen Kit |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reactions |
| R2 | Coefficient of determination of the standard curve |
| Xeno RNA | VetMAX Xeno Internal Positive Control RNA |
| WVDL | Wisconsin Veterinary Diagnostic Laboratory |
| WVDL assay | WVDL in-house influenza A Matrix PCR assay |
| WVDL IC | WVDL internal control |
Appendix A
| IAV | Subtype | Strain ID |
|---|---|---|
| IAV reference 1 | H9N2 | Influenza A Virus A/Turkey/CA/6889/1980 |
| IAV reference 2 | H5N9 | Influenza A Virus A/Turkey/Wisconsin/1968 |
| IAV reference 3 | H7N3 | Influenza A Virus A/Turkey/Oregon/1977 |
| LPAI 1 | H3N8 | Influenza A Virus A/Equine/Miami/1/63 |
| LPAI 2 | H1N7 | Influenza A Virus A/NJ/8/76/EQ-1 |
| LPAI 3 | HON3 | Influenza A Virus A/NWS-NOV2 |
| LPAI 4 | H10N7 | Influenza A Virus A/CK/GERM/49 |
| LPAI 5 | H2N3 | Influenza A Virus A/Mallard/A16/77 |
| LPAI 6 | H4N8 | Influenza A Virus A/MYNAH/Mass/71 |
| LPAI 7 | H7N3 | Influenza A Virus A/TY/ORE |
| LPAI 8 | H4N8 | Influenza A Virus A/DK/England/62 |
| LPAI 9 | H3N8 | Influenza A Virus A/DK/Ukraine/1/63 |
| Extraction Platform | Protocol | Description of Methodology |
|---|---|---|
| MagMAX CORE (CORE) | 200-na | 200 µL sample |
| 50-na | 50 µL sample and 150 µL PBS | |
| 50-pretreatment | 50 µL sample input volume with 10 µL of proteinase K, 5 µL of 1M DTT, and 35 µL of 2% SDS and heat at 60 °C for 5 min. 100 µL of PBS after heating for extraction | |
| 12.5-pretreatment | 12.5 µL sample input volume and 35 µL of PBS, with 10 µL of proteinase K, 5 µL of 1M DTT, and 35 µL 2% SDS heat at 60 °C for 5 min. 100 µL of PBS after heating for extraction | |
| IndiMag Pathogen (Pathogen) | 200-na | 200 µL sample |
| 100-na | 100 µL sample and 100 µL PBS | |
| 100-pretreatment | 100 µL semen with 20 µL of proteinase K, 90 µL of Buffer ATL and heating and mixing at 56 °C for 10 min |
| Reference Strain | NAHLN IAV Assay | WVDL IAV Assay | |||||
|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | Replicate 1 | Replicate 2 | Replicate 3 | ||
| Limit of Detection | 1 | 6 | 6 | 6 | 6 | 6 | 6 |
| 2 | 7 | 6 | 6 | 6 | 7 | 7 | |
| 3 | 7 | 6 | 7 | 6 | 7 | 7 | |
| R2 value | 1 | 0.997 | 0.990 | 0.994 | 0.997 | 0.995 | 0.999 |
| 2 | 0.999 | 0.998 | 0.998 | 0.999 | 0.999 | 0.996 | |
| 3 | 0.997 | 0.995 | 0.999 | 0.997 | 0.995 | 0.996 | |
| PCR Efficiency (%) | 1 | 110.6 | 118.0 | 113.3 | 102.1 | 104.8 | 103.8 |
| 2 | 98.5 | 109.0 | 101.3 | 106.2 | 97.8 | 101.5 | |
| 3 | 100.6 | 120.7 | 110.9 | 116.4 | 112.2 | 109.9 | |
References
- Givens, M.D. Review: Risks of disease transmission through semen in cattle. Animal 2018, 12, s165–s171. [Google Scholar] [CrossRef]
- Haapala, V.; Pohjanvirta, T.; Vähänikkilä, N.; Halkilahti, J.; Simonen, H.; Pelkonen, S.; Soveri, T.; Simojoki, H.; Autio, T. Semen as a source of Mycoplasma bovis mastitis in dairy herds. Vet. Microbiol. 2018, 216, 60–66. [Google Scholar] [CrossRef]
- Jordan, A.; Sadler, R.J.; Sawford, K.; van Andel, M.; Ward, M.; Cowled, B. Mycoplasma bovis outbreak in New Zealand cattle: An assessment of transmission trends using surveillance data. Transbound. Emerg. Dis. 2021, 68, 3381–3395. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Subramaniam, S.; De, A.; Das, B.; Dash, B.; Sanyal, A.; Misra, A.; Pattnaik, B. Detection of foot-and-mouth disease virus in semen of infected cattle bulls. Indian J. Anim. Sci. 2012, 82, 1472–1476. [Google Scholar] [CrossRef]
- Alexandersen, S.; Zhang, Z.; Donaldson, A.I.; Garland, A.J.M. The Pathogenesis and Diagnosis of Foot-and-Mouth Disease. J. Comp. Pathol. 2003, 129, 1–36. [Google Scholar] [CrossRef]
- Burrough, E.R.; Magstadt, D.R.; Petersen, B.; Timmermans, S.J.; Gauger, P.C.; Zhang, J.; Siepker, C.; Mainenti, M.; Li, G.; Thompson, A.C.; et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024. Emerg. Infect. Dis. 2024, 30, 1335–1343. [Google Scholar] [CrossRef]
- Caserta, L.C.; Frye, E.A.; Butt, S.L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L.M.; Thompson, A.C.; Koscielny, M.P.; Cronk, B.; Johnson, A.; et al. Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle. Nature 2024, 634, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.; Poulsen, K.; Caserta, L.C.; Guan, L.; Opgenorth, E.; Beal, M.P.; Eisfeld, A.J.; Kawaoka, Y.; Diel, D.G. Unexpected Detection of Highly Pathogenic Avian Influenza (HPAI) H5N1 virus in bovine semen from a bull used for natural breeding on an affected dairy farm. bioRxiv 2025. [Google Scholar] [CrossRef]
- Seekford, Z.; Alexander Morris, E.R.; Smith, D.; Chaki, S.; Kahl-McDonagh, M.; Zuelke, K.; Swinford, A.K.; Da Silveira, B.P.; Schwarz, E.; Gomez, G.; et al. Inoculation of dairy bulls with highly pathogenic avian influenza virus H5N1 2025. In Proceedings of the American Association of Veterinary Laboratory Diagnosticians Annual Meeting, Aurora, CO, USA, 2 November 2025. [Google Scholar]
- European Food Safety Authority (EFSA); Alvarez, J.; Bortolami, A.; Ducatez, M.; Guinat, C.; Stegeman, J.A.; Broglia, A.; Jensen, H.; Kryemadhi, K.; Gervelmeyer, A. Risk posed by the HPAI virus H5N1, Eurasian lineage goose/Guangdong clade 2.3.4.4b. genotype B3.13, currently circulating in the US. EFSA J. 2025, 23, e9508. [Google Scholar] [CrossRef]
- Samanta, L.; Parida, R.; Dias, T.R.; Agarwal, A. The enigmatic seminal plasma: A proteomics insight from ejaculation to fertilization. Reprod. Biol. Endocrinol. 2018, 16, 41. [Google Scholar] [CrossRef]
- Gautier, C.; Aurich, C. “Fine feathers make fine birds”—The mammalian sperm plasma membrane lipid composition and effects on assisted reproduction. Anim. Reprod. Sci. 2022, 246, 106884. [Google Scholar] [CrossRef]
- Raheja, N.; Choudhary, S.; Grewal, S.; Sharma, N.; Kumar, N. A review on semen extenders and additives used in cattle and buffalo bull semen preservation. J. Entomol. Zool. Stud. 2018, 6, 239–245. [Google Scholar]
- Bodu, M.; Hitit, M.; Greenwood, O.C.; Murray, R.D.; Memili, E. Extender development for optimal cryopreservation of buck sperm to increase reproductive efficiency of goats. Front. Vet. Sci. 2025, 12, 1554771. [Google Scholar] [CrossRef]
- Chandaka, D.; Regula, V.; Krovvidi, S.; Dhulipalla, N.; Kamisetty, A.K. Comparative assessment of five distinct DNA extraction protocols from frozen buffalo semen. Explor. Anim. Med. Res. 2024, 14, 249–254. [Google Scholar] [CrossRef]
- Foley, C.; O’Farrelly, C.; Meade, K.G. Technical note: Comparative analyses of the quality and yield of genomic DNA from invasive and noninvasive, automated and manual extraction methods. J. Dairy Sci. 2011, 94, 3159–3165. [Google Scholar] [CrossRef]
- Taylor, E.; Deeney, A.; Birch, C.; Mayne, G.; Ridley, A. Comparison of DNA extraction procedures for detection of Mycoplasma bovis directly from extended bovine semen straw samples using a commercial M. bovis PCR. BMC Vet. Res. 2024, 20, 491. [Google Scholar] [CrossRef] [PubMed]
- Standard Operating Procedure for Nucleic Acid Extraction Using the INDICAL BIOSCIENCE IndiMag Pathogen Kit on a Magnetic Particle Processor (NVSL-SOP-0645). National Veterinary Services Laboratories NAHLN Document List. Available online: https://www.aphis.usda.gov/media/document/15399/file (accessed on 26 September 2025).
- Standard Operating Procedure for Nucleic Acid Extraction Using the MagMAX Core Nucleic Acid Purification Kit on a Magnetic Particle Processor. (NVSL-SOP-0643). National Veterinary Services Laboratories NAHLN Document List. Available online: https://www.aphis.usda.gov/media/document/15399/file (accessed on 26 September 2025).
- Standard Operating Procedure for Real-time RT-PCR Detection of Influenza A and Avian Paramyxovirus Type-1. (NVSL-SOP-0068). National Veterinary Services Laboratories NAHLN Document List. Available online: https://www.aphis.usda.gov/media/document/15399/file (accessed on 26 September 2025).
- Vandenburg-Carroll, A.; Marthaler, D.G.; Lim, A. Enhancing Diagnostic Resilience: Evaluation of Extraction Platforms and IndiMag Pathogen Kits for Rapid Animal Disease Detection. Microbiol. Res. 2025, 16, 80. [Google Scholar] [CrossRef]
- Ministry for Primary Industries NZ. Import Health Standard: Bovine Germplasm. Available online: www.mpi.govt.nz/dmsdocument/46537-Bovine-Germplasm-2025-Import-Health-Standard/ (accessed on 18 October 2025).
- Yan, L.; Toohey-Kurth, K.L.; Crossley, B.M.; Bai, J.; Glaser, A.L.; Tallmadge, R.L.; Goodman, L.B. Inhibition monitoring in veterinary molecular testing. J. Vet. Diagn. Investig. 2020, 32, 758–766. [Google Scholar] [CrossRef]
- Jaramillo, D.; Foxwell, J.; Burrows, L.; Snell, A. Mycoplasma bovis testing for the screening of semen imported into New Zealand. N. Z. Vet. J. 2023, 71, 200–208. [Google Scholar] [CrossRef]
- Lee, S.M.; Balakrishnan, H.K.; Doeven, E.H.; Yuan, D.; Guijt, R.M. Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review. Biosensors 2023, 13, 980. [Google Scholar] [CrossRef]
- Schellhammer, S.K.; Hudson, B.C.; Cox, J.O.; Dawson Green, T. Alternative direct-to-amplification sperm cell lysis techniques for sexual assault sample processing. J. Forensic Sci. 2022, 67, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Hennekens, C.M.; Cooper, E.S.; Cotton, R.W.; Grgicak, C.M. The effects of differential extraction conditions on the premature lysis of spermatozoa. J. Forensic Sci. 2013, 58, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sekiguchi, K.; Mizuno, N.; Kasai, K.; Sakai, I.; Sato, H.; Seta, S. The modified method of two-step differential extraction of sperm and vaginal epithelial cell DNA from vaginal fluid mixed with semen. Forensic Sci. Int. 1995, 72, 25–33. [Google Scholar] [CrossRef] [PubMed]
- ThermoFisher SCIENTIFIC. MagMAXTM CORE Nucleic Acid Purification Kit USER GUIDE, Revision D00 2024; ThermoFisher SCIENTIFIC: Waltham, MA, USA, 2024. Available online: https://documents.thermofisher.com/TFS-Assets/LSG/manuals/MAN0015944_MagMAXCORE_NA_Kit_UG.pdf (accessed on 22 October 2025).
- Alexander Morris, E.R.; Da Silveira, B.P.; Pohler, K.; Dimitrov, K.M. Verfication of use of raw and extended semen for detection of avian influenza by real-time RT-PCR 2025. In Proceedings of the American Association of Veterinary Laboratory Diagnosticians Annual Meeting, Aurora, CO, USA, 31 October 2025. [Google Scholar]
- Miller, M.R.; Braun, E.; Ip, H.S.; Tyson, G.H. Domestic and wild animal samples and diagnostic testing for SARS-CoV-2. Vet. Q. 2023, 43, 1–11. [Google Scholar] [CrossRef]
- Hsiao, C.-C.; Lin, C.-C.; Chen, Y.-M.; Cheng, M.-C.; Davison, S.; Ma, J.; Dai, H.-L. Quantitative Real-Time PCR Detection of Inactivated H5 Avian Influenza Virus in Raw Milk Samples by Miniaturized Instruments Designed for On-Site Testing. bioRxiv 2025. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Foot and Mouth Disease; World Organisation for Animal Health: Paris, France, 2009. Available online: https://www.woah.org/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/2.01.05_FMD.pdf (accessed on 26 September 2025).
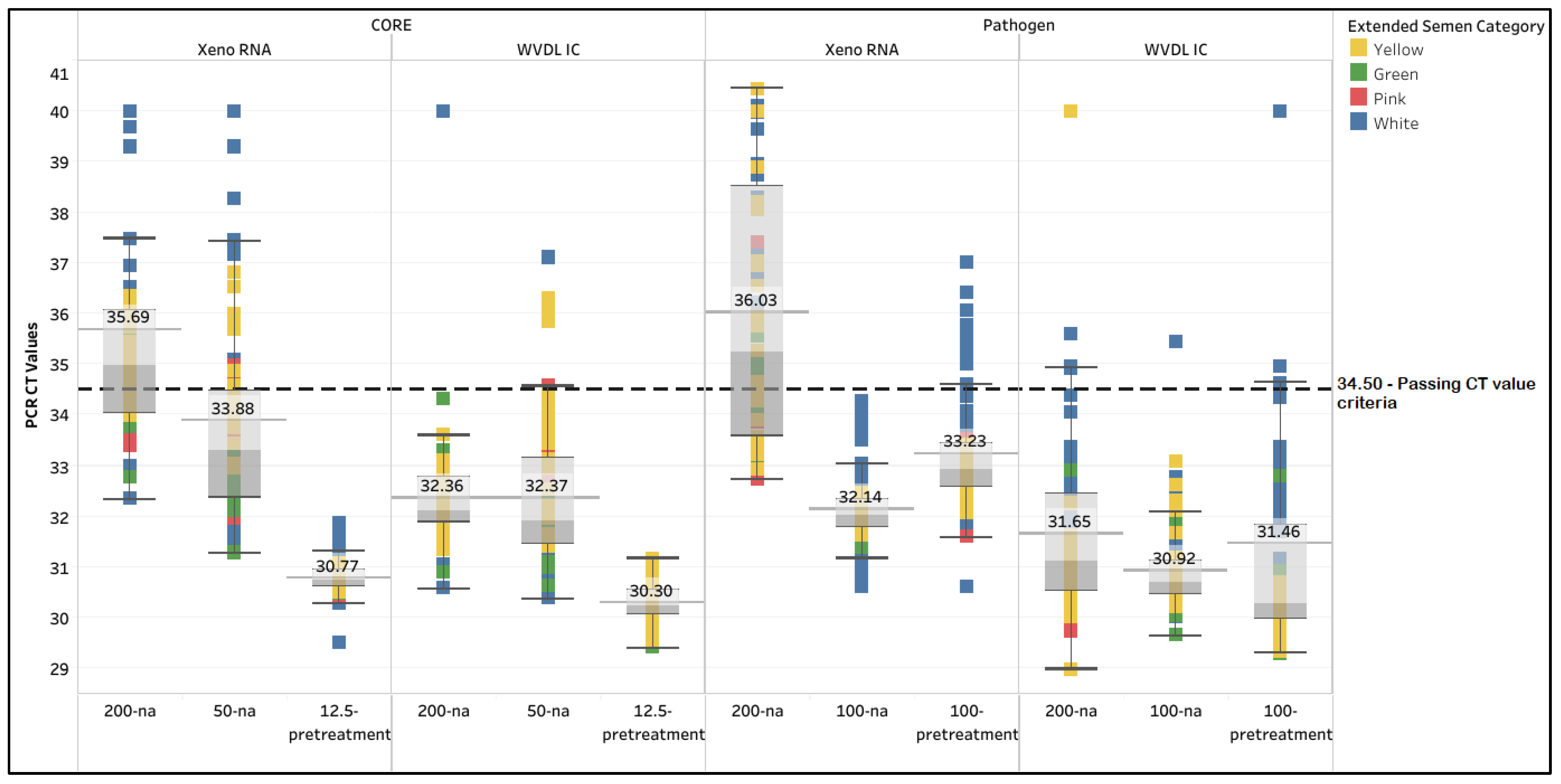

| Extraction Kit | CORE | Pathogen | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Internal Control | Xeno RNA | WVDL IC | Xeno RNA | WVDL IC | ||||||||
| Semen input & modification | 200-na | 50-na | 12.5-pre-treatment | 200-na | 50-na | 12.5-pretreatment | 200-na | 100-na | 100-pretreatment | 200-na | 100-na | 100-pretreatment |
| Yellow (n =24) | 25.0 | 62.5 | 100.0 | 100.0 | 83.3 | 100.0 | 41.7 | 100.0 | 100.0 | 95.8 | 100.0 | 100.0 |
| Green (n = 16) | 12.5 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 50.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| Pink (n = 16) | 81.3 | 87.5 | 100.0 | 100.0 | 93.8 | 100.0 | 87.5 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| White (n = 32) | 21.9 | 62.5 | 100.0 | 93.8 | 96.9 | 100.0 | 3.1 | 100.0 | 59.4 | 83.3 | 96.9 | 68.8 |
| Overall % Passing rate (n = 88) | 31.8 | 73.9 | 100.0 | 97.7 | 93.2 | 100.0 | 37.5 | 100.0 | 85.2 | 94.3 | 98.9 | 88.6 |
| CORE 12.5-Pretreatment | Pathogen 100-na | Pathogen 100-Pretreatment | ||||
|---|---|---|---|---|---|---|
| Replicate | 1 | 2 | 1 | 2 | 1 | 2 |
| M. bovis (n = 8) | 75.0% | 87.5% | 75.0% | 100.0% | 87.5% | 100.0% |
| BVDV (n = 5) | 60.0% | 60.0% | 80.0% | 80.0% | 100.0% | 80.0% |
| BHV-1 (n = 23) | 87.0% | 95.7% | 95.7% | 95.7% | 95.7% | 100.0% |
| Sensitivity (n = 36) | 80.6% | 88.9% | 88.9% | 94.4% | 94.4% | 97.2% |
| CORE 12.5-Pretreatment | Pathogen 100-na | Pathogen 100-Pretreatment | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Replicate | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| Limit of Detection | 5 | 5 | 6 | 6 | 7 | 7 | 5 | 5 | 5 |
| R2 value | 0.996 | 0.956 | 0.986 | 1.000 | 1.000 | 0.999 | 0.994 | 0.988 | 0.981 |
| PCR Efficiency (%) | 81.6 | 84.7 | 133.5 | 90.4 | 93.2 | 94.1 | 92.8 | 76.8 | 71.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimmerman, A.; Vandenburg-Carroll, A.; Marthaler, D.G.; Lim, A. Optimizing Nucleic Acid Extraction from Extended Bovine Semen for Endemic and High-Consequence Pathogens. Animals 2025, 15, 3411. https://doi.org/10.3390/ani15233411
Zimmerman A, Vandenburg-Carroll A, Marthaler DG, Lim A. Optimizing Nucleic Acid Extraction from Extended Bovine Semen for Endemic and High-Consequence Pathogens. Animals. 2025; 15(23):3411. https://doi.org/10.3390/ani15233411
Chicago/Turabian StyleZimmerman, Amanda, Anne Vandenburg-Carroll, Douglas G. Marthaler, and Ailam Lim. 2025. "Optimizing Nucleic Acid Extraction from Extended Bovine Semen for Endemic and High-Consequence Pathogens" Animals 15, no. 23: 3411. https://doi.org/10.3390/ani15233411
APA StyleZimmerman, A., Vandenburg-Carroll, A., Marthaler, D. G., & Lim, A. (2025). Optimizing Nucleic Acid Extraction from Extended Bovine Semen for Endemic and High-Consequence Pathogens. Animals, 15(23), 3411. https://doi.org/10.3390/ani15233411






