Salinity Effects on Aquatic and Host Intestinal Microbiota Dynamics in Rhinogobio ventralis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
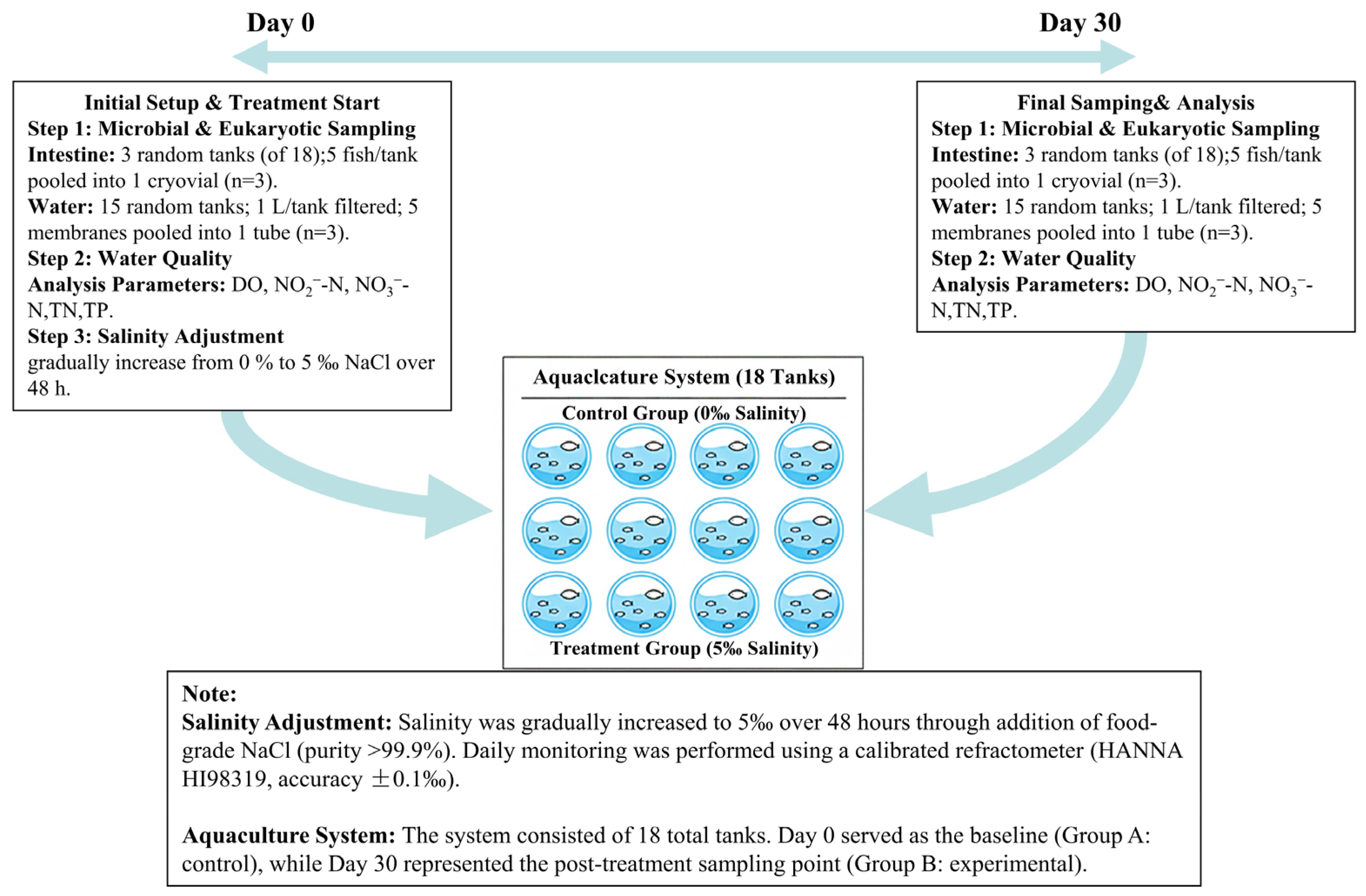
2.2. Experimental Design and Salinity Treatment
2.3. Sampling Protocols
2.3.1. Intestinal Microbial Sampling
2.3.2. Water Microbial and Eukaryotic Community Sampling
2.3.3. Water Quality Analysis
2.4. Bacterial 16S rRNA Target Amplification and PCR Purification
2.5. Eukaryotic 18S rRNA Target Amplification and PCR Purification
2.6. Data Processing
3. Results
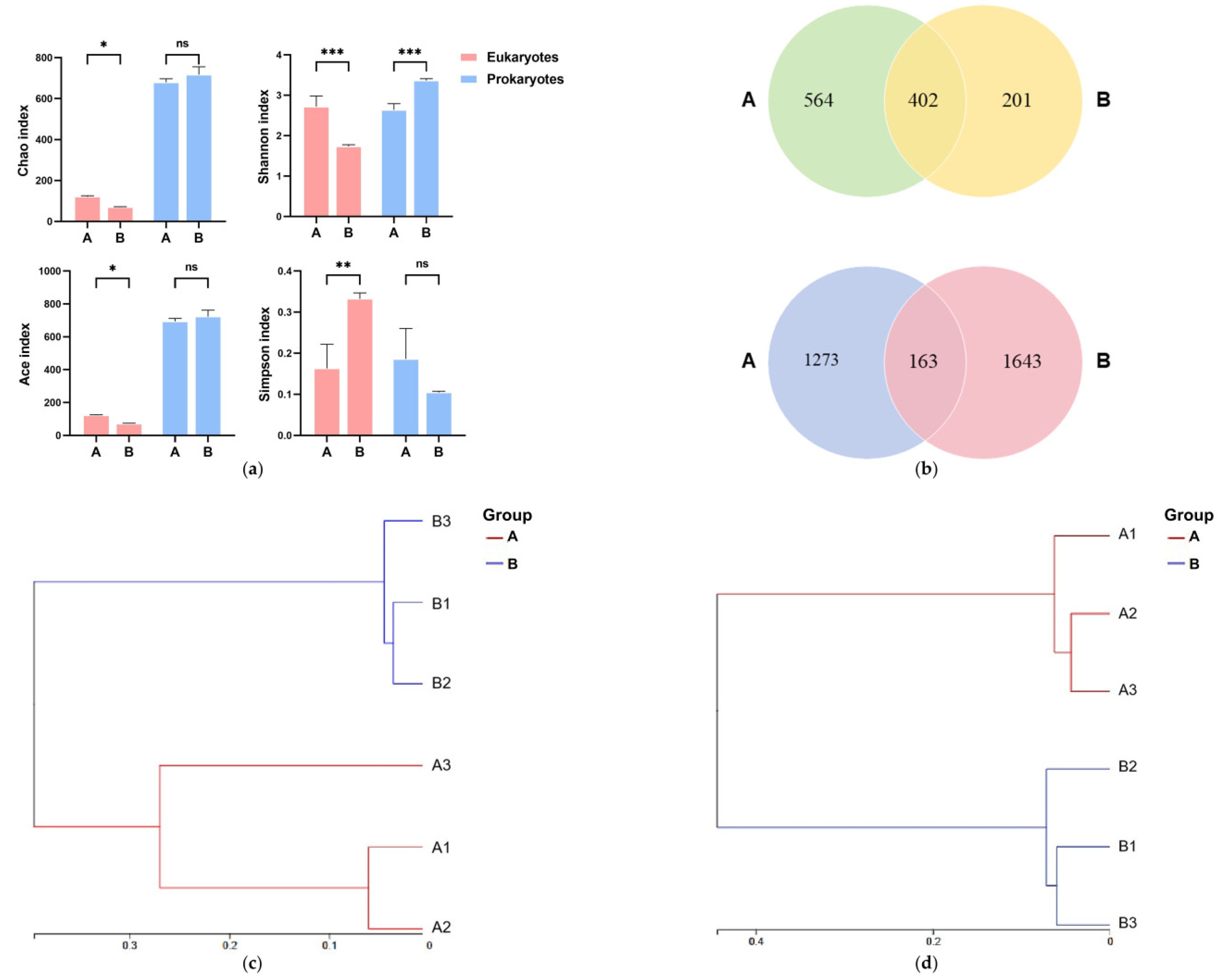
3.1. Water Microbial and Eukaryotic Molecular Signal Dynamics
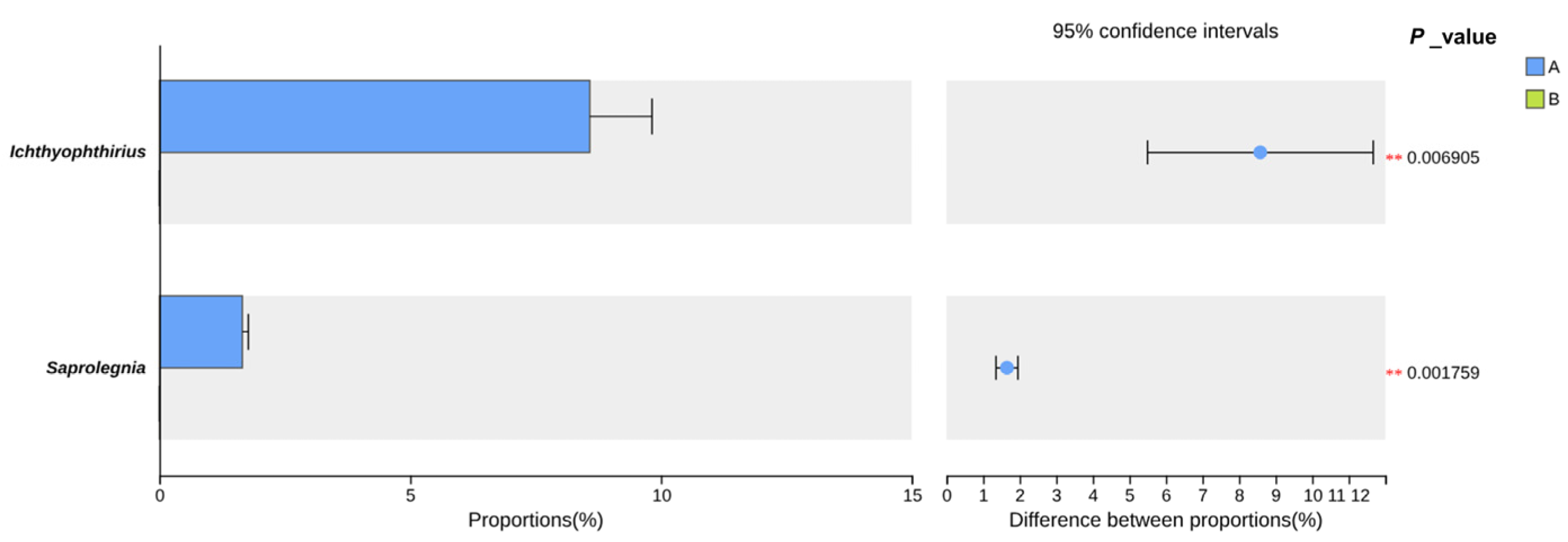
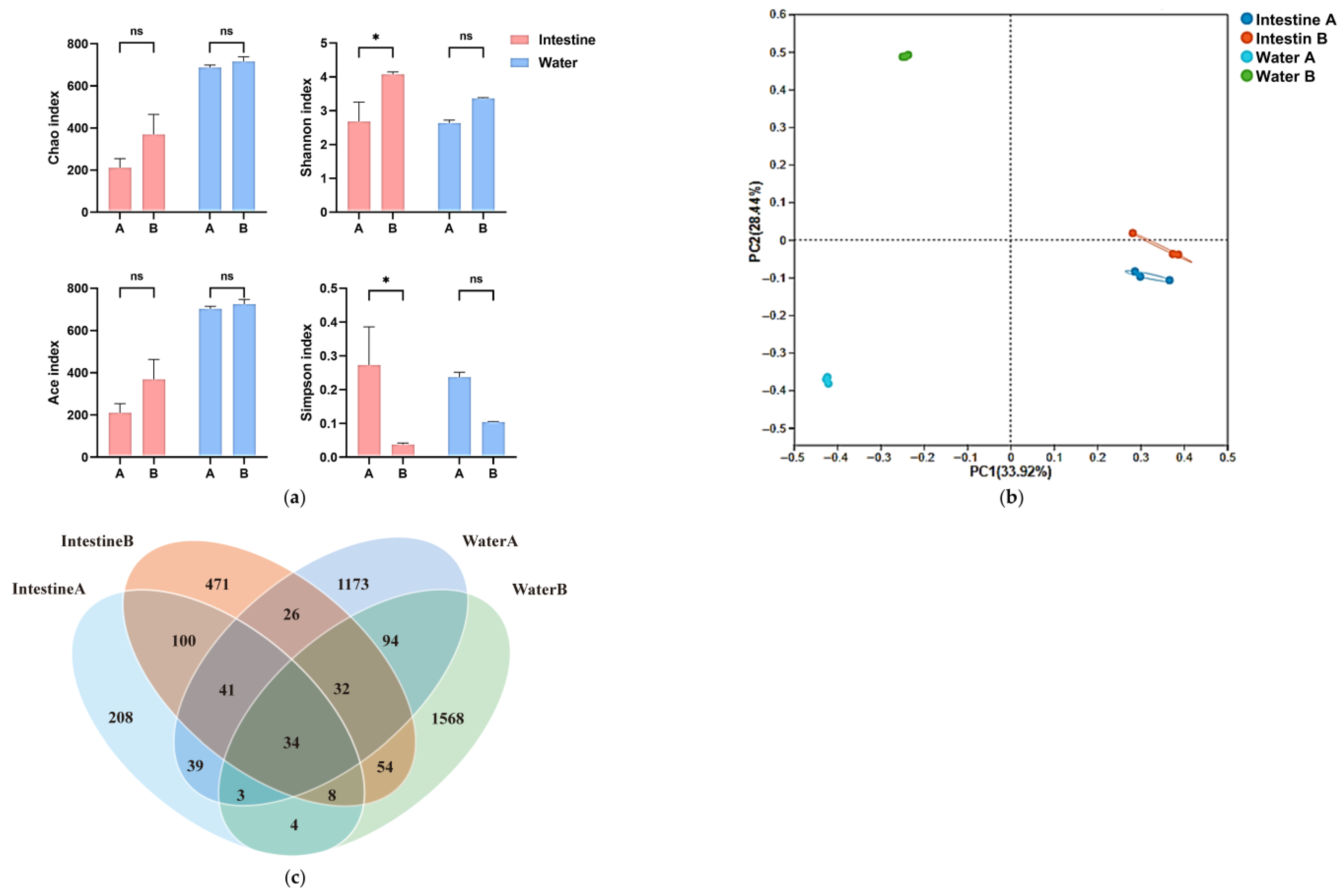
3.1.1. Microbial and Eukaryotic Diversity Response to Salinity
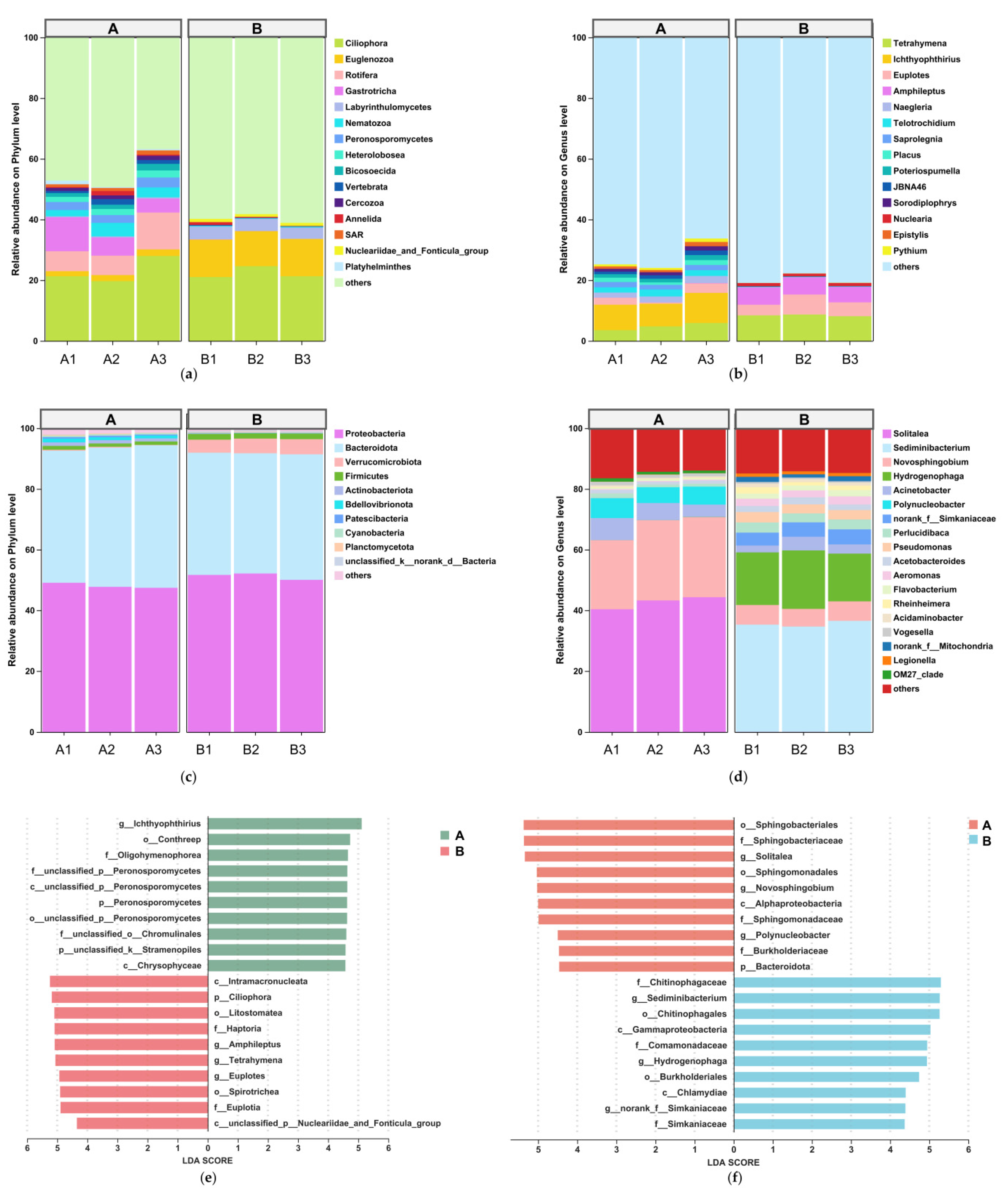
3.1.2. Compositional Shifts in Water Microbial and Eukaryotic Communities
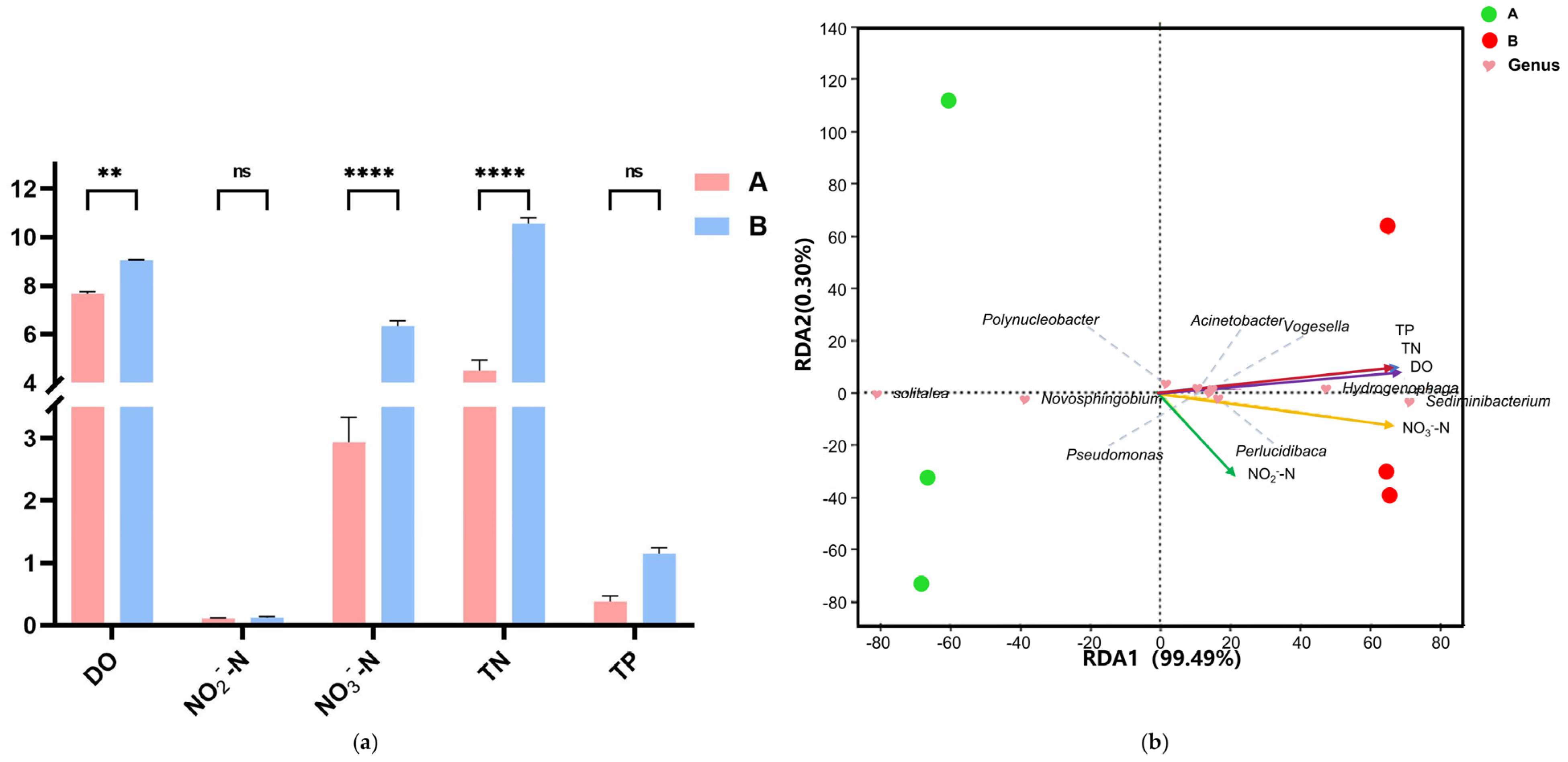
3.1.3. Effects of Salinity on Water Physicochemical Parameters
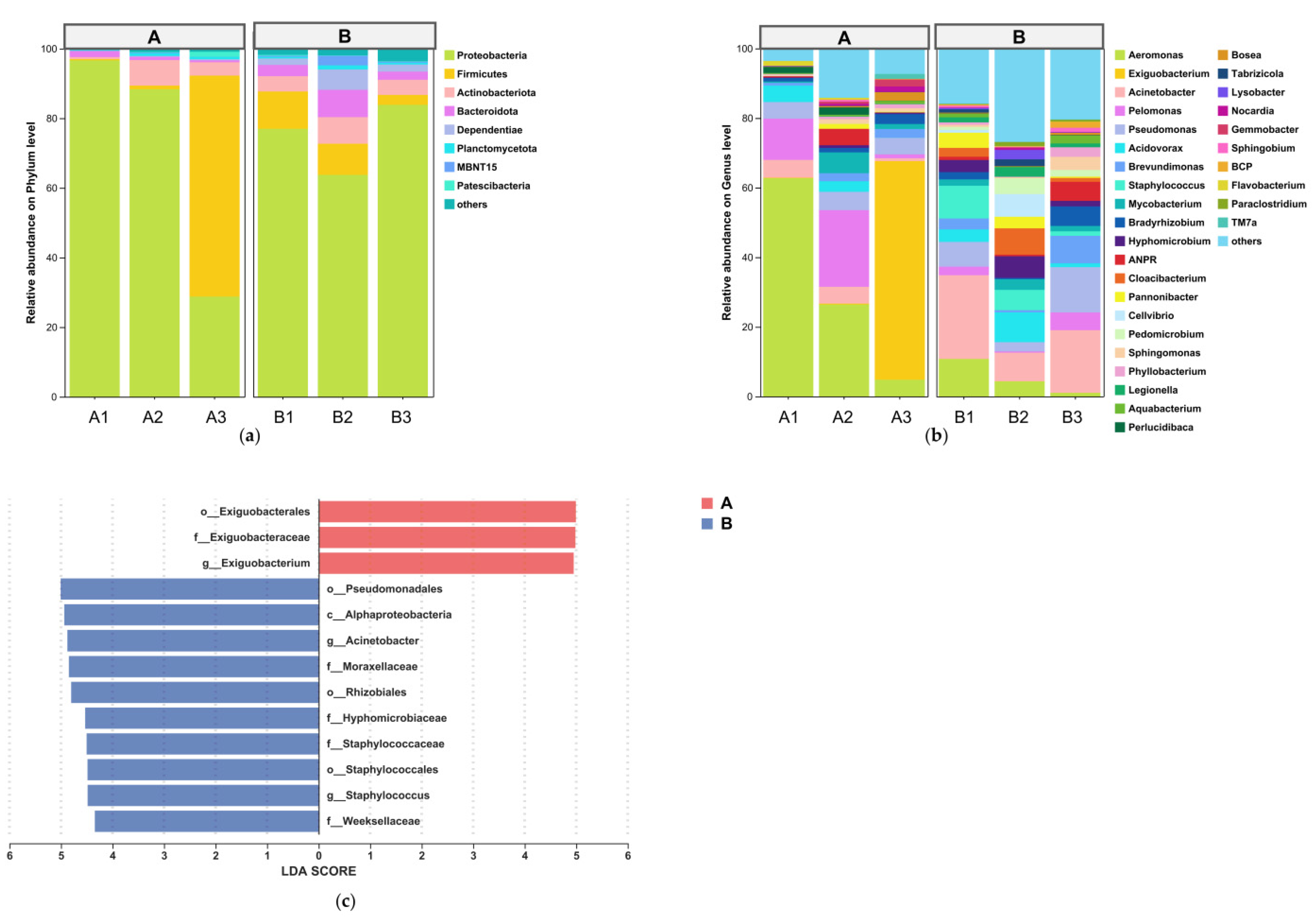
3.2. Compositional Shifts in Intestinal Microbial Communities
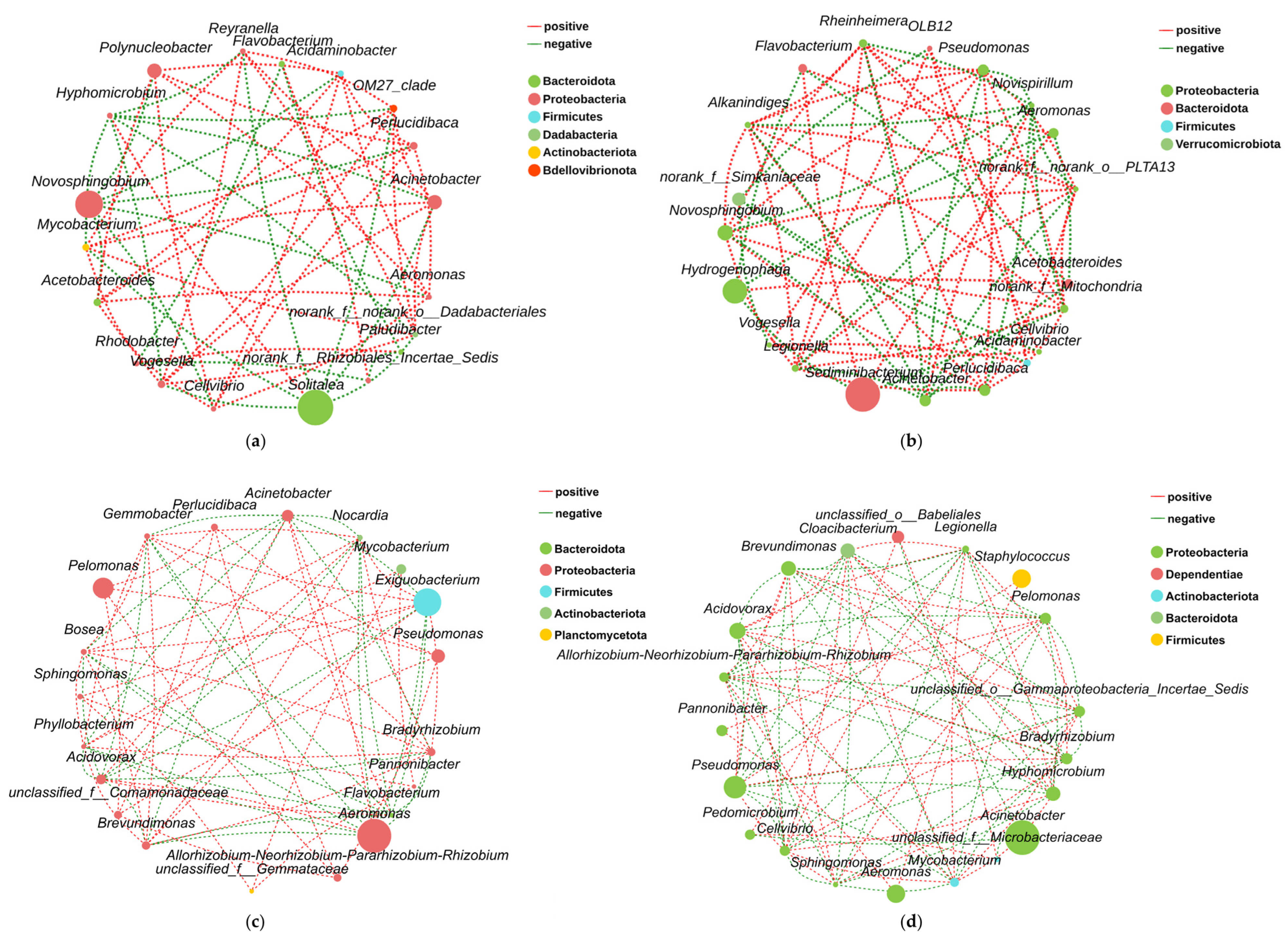
3.3. Water–Intestinal Microbial Interaction Networks
3.3.1. Community Association Patterns
3.3.2. Key Species Interactions in Water and Intestinal Microbiota
4. Discussion
4.1. Structural Impact of 5‰ Salinity on Environmental and Host Microbiomes
4.1.1. Aquatic Ecological Response
4.1.2. Water Quality Changes and Impairment of Biogeochemical Function
4.2. Salinity Impacts on Intestinal Microbial Consortia
4.2.1. Salinity-Driven Modulation of Intestinal Microbial Diversity
4.2.2. Enrichment of Potentially Opportunistic Taxa in the Intestinal Microbiota
4.2.3. Impact of Salinity on Potential Beneficial Taxa
4.2.4. Salinity-Driven Aquatic-Intestinal Microbial Cross-Domain Interactions
5. Conclusions
- (1)
- In the aquatic environment, increased prokaryotic diversity and network complexity coincided with impaired self-purification capacity, evidenced by elevated dissolved oxygen, nitrate nitrogen, and total nitrogen.
- (2)
- In the host, the intestinal microbiome underwent significant destabilization; this was characterized by a simplification of its interaction network, a surge in potentially opportunistic taxa, and a decline in potential beneficial probiotics, all collectively indicating a state of dysbiosis.
- (3)
- A fundamental divergence in microbial adaptation strategies was observed: the environmental community enhanced its resilience through increased cooperation, whereas the host-associated intestinal community shifted toward a stressed, antagonistic state.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| R. ventralis | Rhinogobio ventralis |
| I. multifiliis | Ichthyophthirius multifiliis |
| RAS | Recirculating aquaculture system |
| OTUs | Operational taxonomic units |
| ASVs | Amplicon sequence variants |
| RDA | Redundancy analysis |
| PCoA | Principal co-ordinates analysis |
| DO | Dissolved oxygen |
| NO3−-N | Nitrate nitrogen |
| TN | Total nitrogen |
Appendix A

References
- Bao, X.; Xie, W.; Huang, D.; Zhu, B. Analysis of meat percentage and nutritional components of the long fin gill snail in the jinsha river. J. Hydroecol. 2011, 32, 142–148. [Google Scholar]
- Jiang, Z.G.; Jiang, J.P.; Wang, Y.Z. China’s Red List of Vertebrates: Volume 1—Vertebrate Overview; Report; Ministry of Environmental Protection & Chinese Academy of Sciences: Beijing, China, 2016. [Google Scholar]
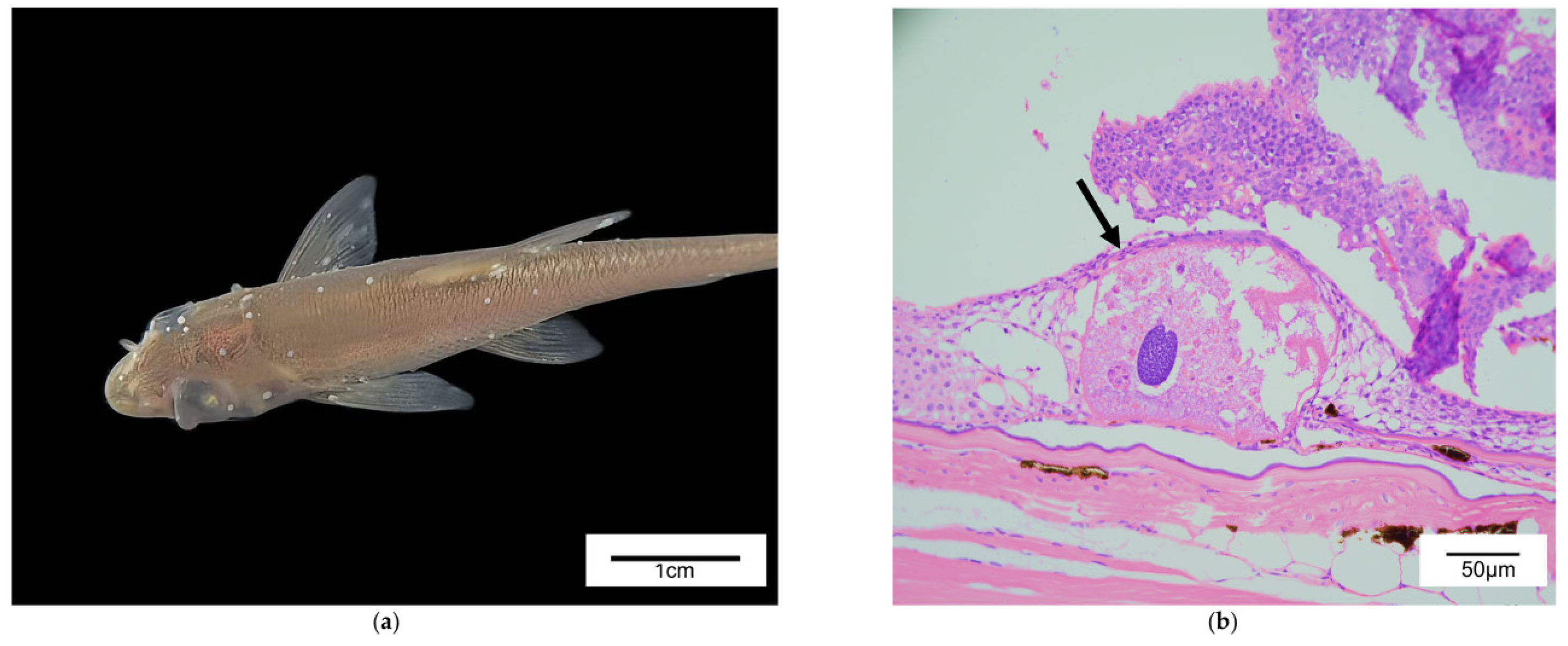
- Zhao, Q.; Li, S.; Wang, S.; Zhao, Z.; Li, X.; Sun, H.; Wu, X.; Wang, Z.; Li, F. Response of rhinogobio ventralis skin to ichthyophthirius multifiliis infection: Pathological, transcriptomic and metabolomic analyses. Fish. Shellfish. Immunol. 2025, 167, 110679. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, X.; He, Y.; Yang, D. Study on broodstock culture, induced spawning and artificial incubation of Rhinogobio ventralis (sauvage et dabry) in recirculating aquaculture system. Freshw. Fish. 2018, 48, 101–106. [Google Scholar]
- Huang, K.; Wang, R.; Hu, G.; Zhou, W.; Li, W.; Zou, H.; Wang, G.; Li, M. Immune response of Rhinogobio ventralis to Ichthyophthirius multifiliis infection: Insights from histopathological and real-time gene expression analyses. Fish. Shellfish. Immunol. 2024, 153, 109801. [Google Scholar] [CrossRef]
- Ward, D.L. Salinity of the little colorado river in grand canyon confers anti-parasitic properties on a native fish. West. N. Am. Nat. 2012, 72, 334–338. [Google Scholar] [CrossRef]
- Aihua, L.; Buchmann, K. Temperature-and salinity-dependent development of a nordic strain of Ichthyophthirius multifiliis from rainbow trout. J. Appl. Ichthyol. 2001, 17, 273–276. [Google Scholar] [CrossRef]
- Sformo, T.L.; De la Bastide, P.Y.; LeBlanc, J.; Givens, G.H.; Adams, B.; Seigle, J.C.; Kunaknana, S.C.; Moulton, L.L.; Hintz, W.E. Temperature response and salt tolerance of the opportunistic pathogen saprolegnia parasitica: Implications for the broad whitefish subsistence fishery. Arct. Antarct. Alp. Res. 2021, 53, 271–285. [Google Scholar] [CrossRef]
- Bal, A.; Pati, S.G.; Panda, F.; Mohanty, L.; Paital, B. Low salinity induced challenges in the hardy fish Heteropneustes fossilis; Future prospective of aquaculture in near coastal zones. Aquaculture 2021, 543, 737007. [Google Scholar] [CrossRef]
- Jia, Y.; Du, J.; Xi, R.; Zhang, Q.; Li, L.; Li, D.; Takagi, Y.; Zhang, X. Effects of different culture salinities on the growth and muscle quality of grass carp (Ctenopharyngodon idellus). J. Anim. Sci. 2024, 102, skae281. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, Y.; Cui, X.; Yue, P.; Li, K.; Liu, X.; Tripathi, B.M.; Chu, H. Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems 2019, 4, 11. [Google Scholar] [CrossRef]
- Mkulo, E.M.; Iddrisu, L.; Yohana, M.A.; Zheng, A.; Zhong, J.; Jin, M.; Danso, F.; Wang, L.; Zhang, H.; Tang, B.; et al. Exploring salinity adaptation in teleost fish, focusing on omics perspectives on osmoregulation and gut microbiota. Front. Mar. Sci. 2025, 12, 1559871. [Google Scholar] [CrossRef]
- Li, X.; Wang, A.; Wan, W.; Luo, X.; Zheng, L.; He, G.; Huang, D.; Chen, W.; Huang, Q. High salinity inhibits soil bacterial community mediating nitrogen cycling. Appl. Environ. Microbiol. 2021, 87, e01366-21. [Google Scholar] [CrossRef] [PubMed]
- Kanika, N.H.; Liaqat, N.; Chen, H.; Ke, J.; Lu, G.; Wang, J.; Wang, C. Fish gut microbiome and its application in aquaculture and biological conservation. Front. Microbiol. 2025, 15, 1521048. [Google Scholar] [CrossRef]
- Floyd, R.F. Reed, Ichthyophthirius multifiliis (White Spot) Infections in Fish; Fisheries and Aquatic Sciences Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida: Gainesville, FL, USA, 2009; CIR 920. [Google Scholar]
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association: Washington, DC, USA, 2017; ISBN 978-0-87553-287-0. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, Q.; Guo, T.; Wang, P. Effect of sodium chloride on the expression of genes involved in the salt tolerance of bacillus sp. Strain “ SX4 “ isolated from salinized greenhouse soil. Open Chem. 2021, 19, 9–22. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitas, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Mo, Y.; Peng, F.; Gao, X.; Xiao, P.; Logares, R.; Jeppesen, E.; Ren, K.; Xue, Y.; Yang, J. Low shifts in salinity determined assembly processes and network stability of microeukaryotic plankton communities in a subtropical urban reservoir. Microbiome 2021, 9, 128. [Google Scholar] [CrossRef]
- Li, C.; Jin, L.; Zhang, C.; Li, S.; Zhou, T.; Hua, Z.; Wang, L.; Ji, S.; Wang, Y.; Gan, Y.; et al. Destabilized microbial networks with distinct performances of abundant and rare biospheres in maintaining networks under increasing salinity stress. IMETA 2023, 2, e79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Symonds, J.E.; Walker, S.P.; Steiner, K.; Carter, C.G.; Bowman, J.P.; Nowak, B.F. Salinity and fish age affect the gut microbiota of farmed chinook salmon (Oncorhynchus tshawytscha). Aquaculture 2020, 528, 735539. [Google Scholar] [CrossRef]
- Wang, J. Preliminary Study on Artificial Propagation Technique and the Saprolegniasis of Spawned of Schizothorax waltoni. Master’s Thesis, Southwest University, Chongqing, China, 2020. [Google Scholar]
- Ali, E.H. Morphological and biochemical alterations of oomycete fish pathogen saprolegnia parasitica as affected by salinity, ascorbic acid and their synergistic action. Mycopathologia 2005, 159, 231–243. [Google Scholar] [CrossRef]
- Patureau, D.; Zumstein, E.; Delgenes, J.P.; Moletta, R. Aerobic denitrifiers isolated from diverse natural and managed ecosystems. Microb. Ecol. 2000, 39, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; Busse, J.; Goor, M.; Pot, B.; Falsen, E.; Jantzen, E.; Hoste, B.; Gillis, M.; Kersters, K.; Auling, G.; et al. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes hydrogenophaga flava comb. Nov. (Formerly pseudomonas flava), hydrogenophaga palleronii (formerly pseudomonas palleronii), hydrogenophaga pseudoflava (formerly pseudomonas pseudoflava and “pseudomonas carboxy do flava”), and hydrogenophaga taeniospiralis (formerly pseudomonas taeniospiralis). Int. J. Syst. Evol. Microbiol. 1989, 39, 319–333. [Google Scholar]
- Cohen, N.R.; Krinos, A.I.; Kell, R.M.; Chmiel, R.J.; Moran, D.M.; Mcilvin, M.R.; Lopez, P.Z.; Barth, A.J.; Stone, J.P.; Alanis, B.A.; et al. Microeukaryote metabolism across the western north atlantic ocean revealed through autonomous underwater profiling. Nat. Commun. 2024, 15, 7325. [Google Scholar] [CrossRef]
- Hernandez, D.J.; David, A.S.; Menges, E.S.; Searcy, C.A.; Afkhami, M.E. Environmental stress destabilizes microbial networks. ISME J. 2021, 15, 1722–1734. [Google Scholar] [CrossRef]
- Mougin, J.; Joyce, A. Fish disease prevention via microbial dysbiosis-associated biomarkers in aquaculture. Rev. Aquac. 2023, 15, 579–594. [Google Scholar] [CrossRef]
- Xavier, R.; Severino, R.; Silva, S.M. Signatures of dysbiosis in fish microbiomes in the context of aquaculture. Rev. Aquac. 2024, 16, 706–731. [Google Scholar] [CrossRef]
- Sledge, D.G.; Danieu, P.K.; Bolin, C.A.; Bolin, S.R.; Lim, A.; Anderson, B.C.; Kiupel, M. Outbreak of neonatal diarrhea in farmed mink kits (Mustella vison) associated with enterotoxigenic Staphylococcus delphini. Vet. Pathol. 2010, 47, 751–757. [Google Scholar] [CrossRef]
- Medina-Felix, D.; Vargas-Albores, F.; Garibay-Valdez, E.; Martinez-Cordova, L.R.; Martinez-Porchas, M. Oreochromis niloticus gastrointestinal microbiota affected by the infection with Staphylococcus haemolyticus and Providencia vermicola, two emerging pathogens in fish aquaculture. Aquaculture 2024, 582, 740529. [Google Scholar] [CrossRef]
- Huete-Perez, J.A.; Quezada, F. Genomic approaches in marine biodiversity and aquaculture. Biol. Res. 2013, 46, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Youmans, B.P.; Ajami, N.J.; Jiang, Z.; Campbell, F.; Wadsworth, W.D.; Petrosino, J.F.; DuPont, H.L.; Highlander, S.K. Characterization of the human gut microbiome during travelers’ diarrhea. Gut Microbes 2015, 6, 110–119. [Google Scholar] [CrossRef]
- Liu, H.; Fu, S.; Zhang, S.; Ding, M.; Wang, A. Lead induces structural damage, microbiota dysbiosis and cell apoptosis in the intestine of juvenile bighead carp (Hypophthalmichthys nobilis). Aquaculture 2020, 528, 735573. [Google Scholar] [CrossRef]
- Chen, C.; Fang, H.; Zhang, X. Identification of acinetobacter junii morphovar from diseased stone flounder (Kareius bicoloratus). Microbiol. China 2005, 34–39. [Google Scholar]
- Chen, C.; Zhang, X.; Fang, H.; Al, E. Isolation and identification of Acinetobacter junii from ornamental carp (Cyprinus carpio). J. Hydroecol. 2009, 30, 86–90. [Google Scholar]
- Gasparotto, P.H.G.; de Freitas, H.T.; Dantas Filho, J.V.; Cavali, J.; Pontuschka, R.; da Silva Francisco, R.; Daudt, C. Detection of Pseudomonas sp. In pirarucu (Arapaima gigas): A case report in the western amazon. Acta Vet. Bras. 2020, 14, 209–214. [Google Scholar] [CrossRef]
- Zhang, M.; Feng, Y.; Zhong, Z.; Du, Q.; Yu, W.; Wu, J.; Huang, X.; Huang, Z.; Xie, G.; Shu, H. Host gut-derived probiotic, Exiguobacterium acetylicum g1-33, improves growth, immunity, and resistance to Vibrio harveyi in hybrid grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂). Microorganisms 2024, 12, 1688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yang, H.; Cai, G.; Nie, Q.; Sun, Y. The interactions between the host immunity and intestinal microorganisms in fish. Appl. Microbiol. Biotechnol. 2024, 108, 30. [Google Scholar] [CrossRef]
- Rappaport, H.B.; Oliverio, A.M. Lessons from extremophiles: Functional adaptations and genomic innovations across the eukaryotic tree of life. Genome Biol. Evol. 2024, 16, evae160. [Google Scholar] [CrossRef]
- Webster, T.M.U.; Consuegra, S.; de Leaniz, C.G. Early life stress causes persistent impacts on the microbiome of Atlantic salmon. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100888. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liang, Y.; Lian, K.; Zhang, C.; Han, M.; Wang, M.; Shao, H.; McMinn, A.; Wang, H. Correlation with viruses enhances network complexity and stability of co-occurrence prokaryotes across the oceans. mSystems 2025, 10, e0053925. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Nazir, S.; Ijaz, F.; Zahid, M.U.; Mushtaq, M.; Khan, M.; Rahman, A.; Rahman, M.A.U. Dietary supplementation of bacillus subtilis as probiotic influenced the growth performance, hematological parameters, immune function, antioxidant status, and digestive enzyme activity of nile tilapia fingerlings (Oreochromis niloticus). Animals 2025, 15, 1256. [Google Scholar] [CrossRef] [PubMed]
- Neves, N.O.D.S.; De Dea Lindner, J.; Stockhausen, L.; Delziovo, F.R.; Bender, M.; Serzedello, L.; Cipriani, L.A.; Ha, N.; Skoronski, E.; Gisbert, E.; et al. Fermentation of plant-based feeds with lactobacillus acidophilus improves the survival and intestinal health of juvenile nile tilapia (Oreochromis niloticus) reared in a biofloc system. Animals 2024, 14, 332. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Sun, Y.; Wang, A.; Zhou, Z. Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 2018, 9, 2429. [Google Scholar] [CrossRef]
| Sample ID | Prokaryotes | Eukaryotes | ||||
|---|---|---|---|---|---|---|
| Sequences (×103) | ASV Number | Coverage | Sequences (×103) | OTU Number | Coverage | |
| A1 | 53.64 | 690 | 0.998446 | 99.64 | 741 | 0.999909 |
| A2 | 52.72 | 654 | 0.998446 | 97.24 | 711 | 0.999909 |
| A3 | 59.14 | 656 | 0.997579 | 88.14 | 673 | 0.999943 |
| B1 | 60.76 | 700 | 0.998356 | 95.63 | 459 | 0.999897 |
| B2 | 52.28 | 685 | 0.999863 | 99.09 | 454 | 0.99992 |
| B3 | 55.24 | 759 | 0.999641 | 104.07 | 469 | 0.99992 |
| Parameter | Intestine Control | Intestine Salinity | Water Control | Water Salinity |
|---|---|---|---|---|
| Nodes | 19 | 19 | 19 | 20 |
| Edges | 61 | 54 | 56 | 66 |
| Positive Edges | 40 | 26 | 34 | 44 |
| Negative Edges | 21 | 28 | 22 | 22 |
| Positive/Negative Ratio | 1.9 | 0.93 | 1.55 | 2 |
| Average Degree | 6.42 | 5.68 | 5.89 | 6.6 |
| Avg. Clustering Coefficient | 0.62 | 0.79 | 0.58 | 0.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Zhao, Q.; Jin, T.; Li, X.; Sun, H.; Wu, X.; Ge, H.; Li, F. Salinity Effects on Aquatic and Host Intestinal Microbiota Dynamics in Rhinogobio ventralis. Animals 2025, 15, 3407. https://doi.org/10.3390/ani15233407
Liu K, Zhao Q, Jin T, Li X, Sun H, Wu X, Ge H, Li F. Salinity Effects on Aquatic and Host Intestinal Microbiota Dynamics in Rhinogobio ventralis. Animals. 2025; 15(23):3407. https://doi.org/10.3390/ani15233407
Chicago/Turabian StyleLiu, Kaixuan, Qiang Zhao, Tianzhi Jin, Xuemei Li, Hanchang Sun, Xingbing Wu, Hailong Ge, and Fang Li. 2025. "Salinity Effects on Aquatic and Host Intestinal Microbiota Dynamics in Rhinogobio ventralis" Animals 15, no. 23: 3407. https://doi.org/10.3390/ani15233407
APA StyleLiu, K., Zhao, Q., Jin, T., Li, X., Sun, H., Wu, X., Ge, H., & Li, F. (2025). Salinity Effects on Aquatic and Host Intestinal Microbiota Dynamics in Rhinogobio ventralis. Animals, 15(23), 3407. https://doi.org/10.3390/ani15233407






