Evolutionary Dynamics of Matrix Metalloproteases with Collagenolytic Activity in Teleosts
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis
2.1.1. Database Mining and Sequence Retrieval
2.1.2. Sequence Alignments and Phylogenetic Analysis
2.1.3. Collinearity and Synteny Analysis and Duplicate Gene Origin Classification
2.1.4. Functional Conservation Estimate
2.2. Gene Expression Profiling
2.2.1. Larvae Rearing and Sampling
2.2.2. RNA Extraction and Transcriptome Sequencing
3. Results
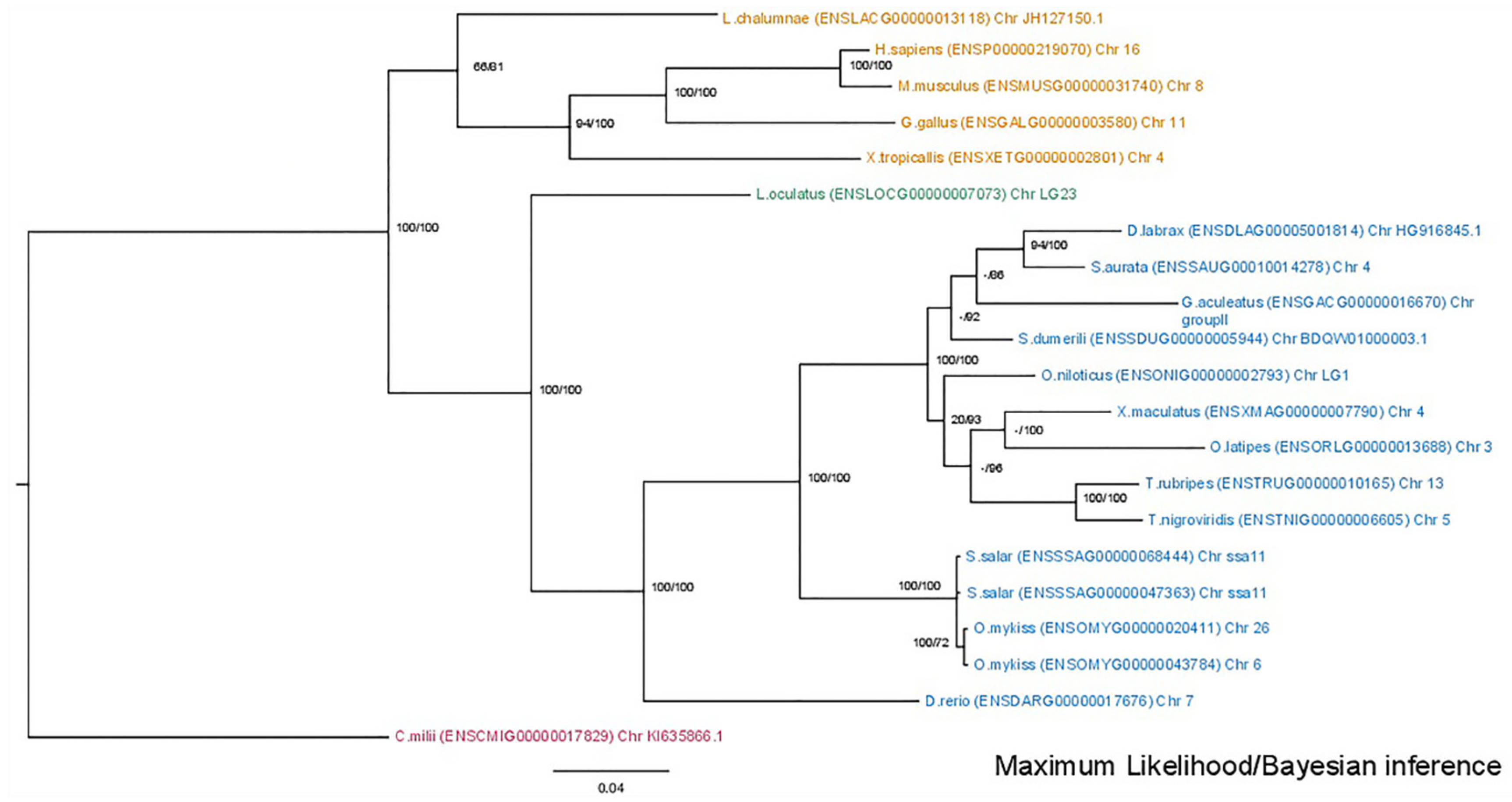
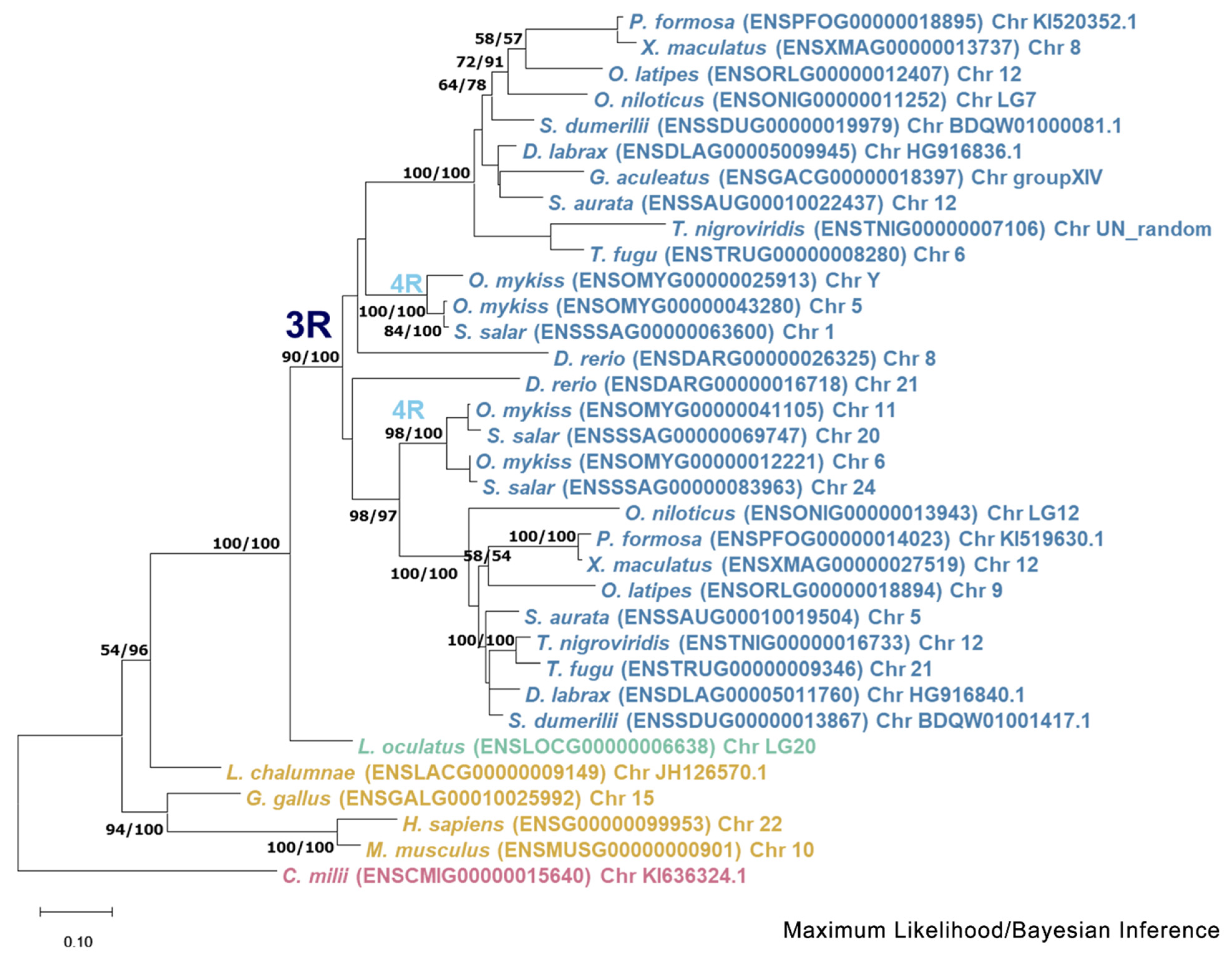
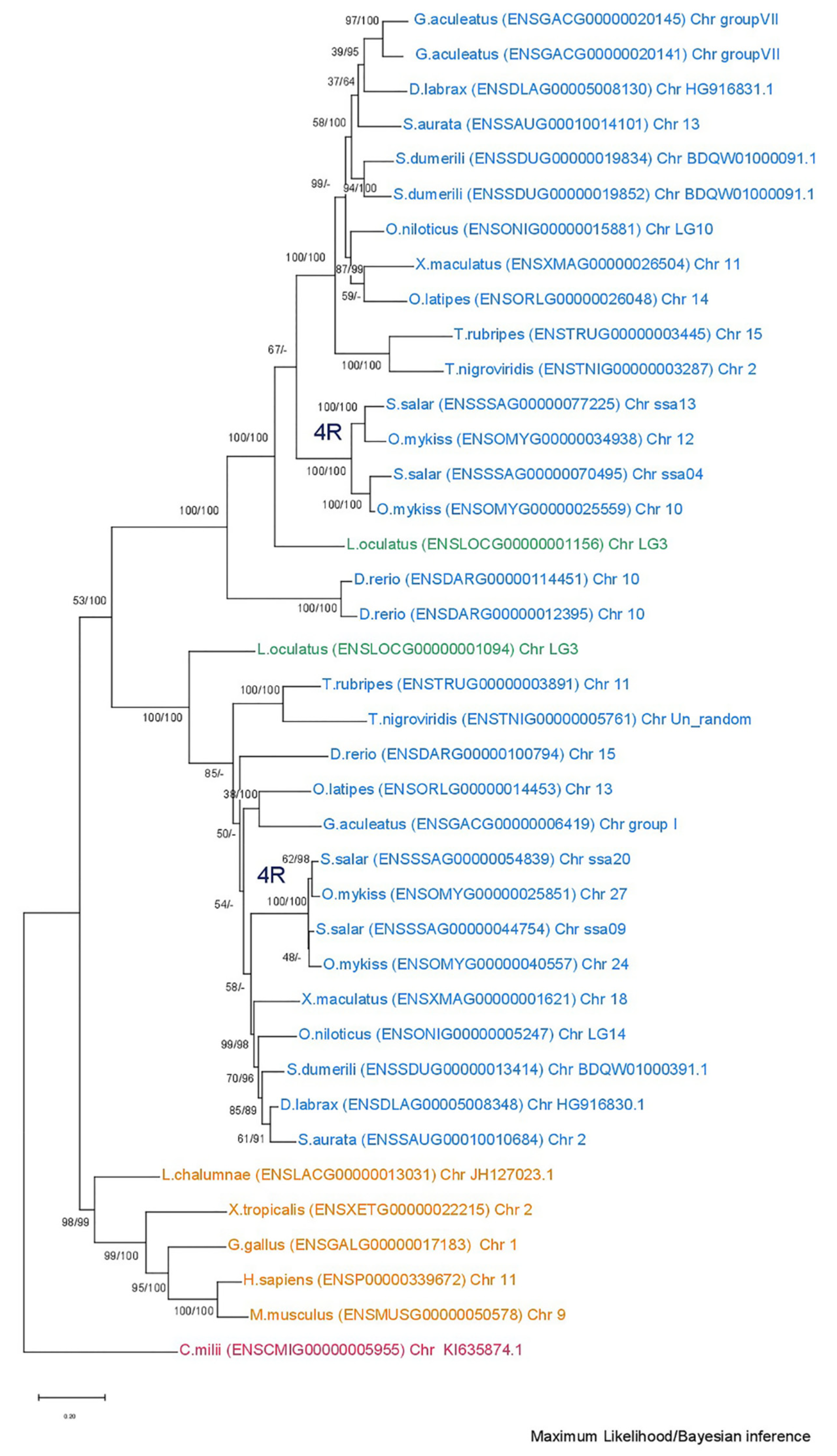
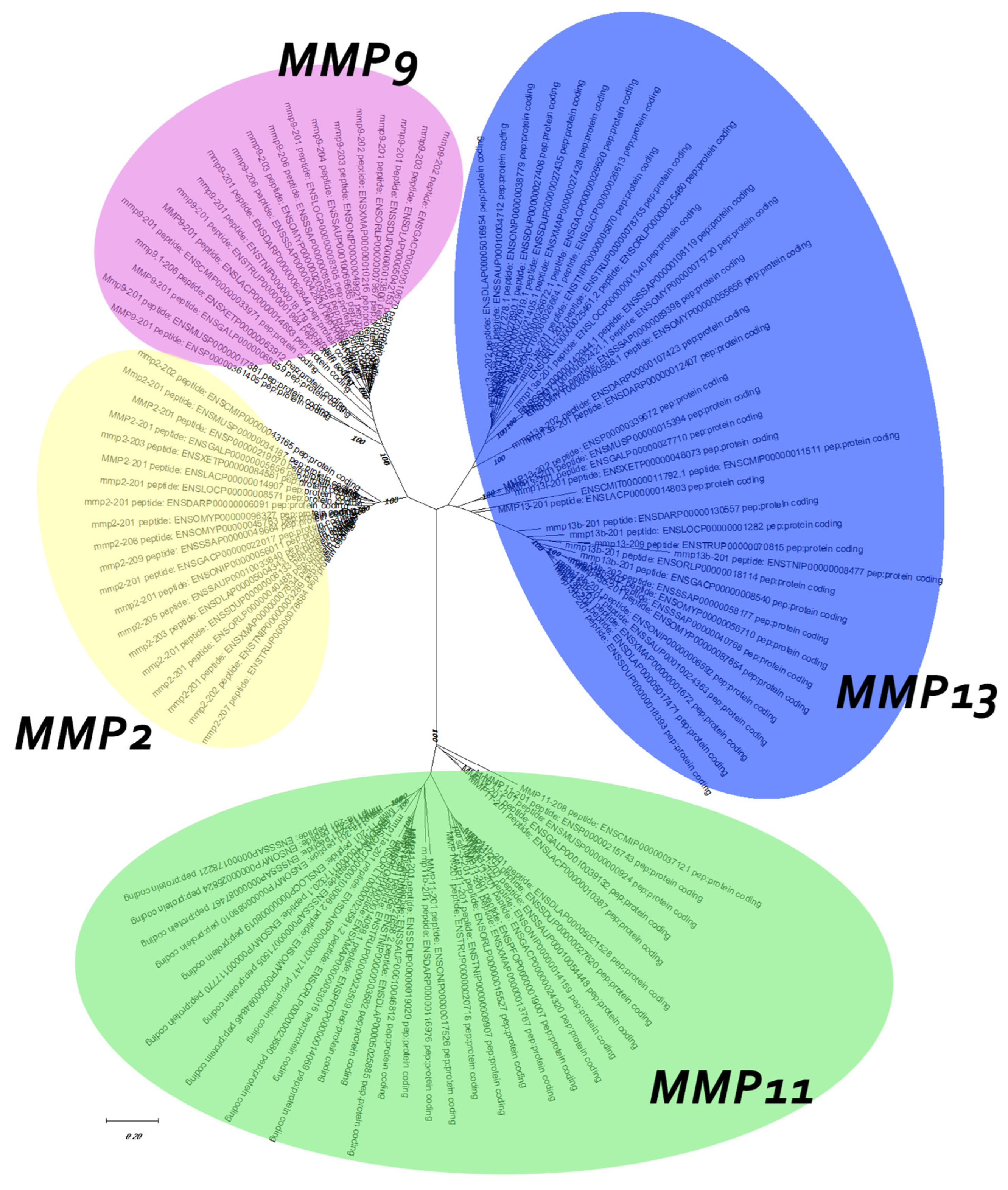
3.1. Phylogenetic Analysis
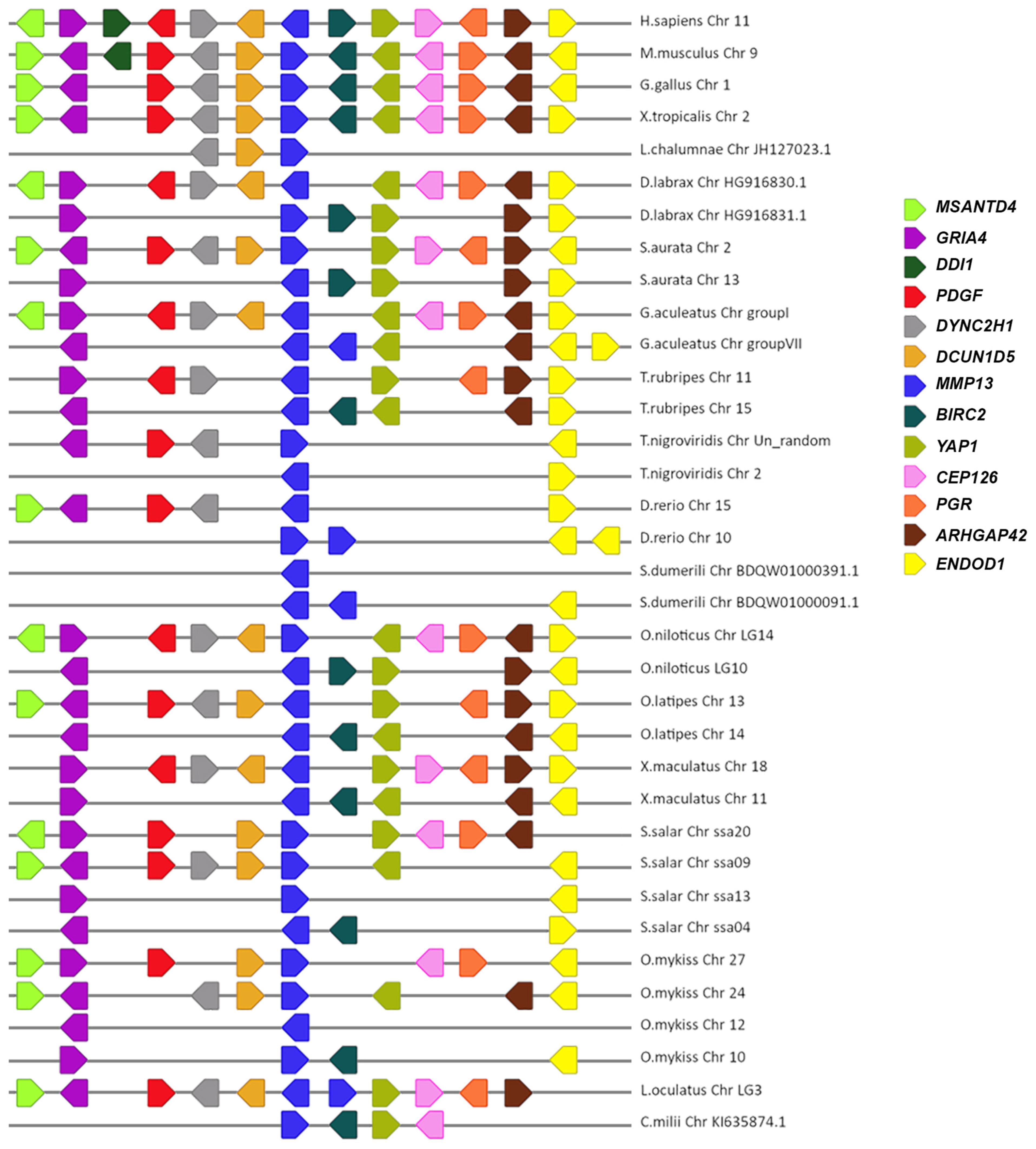
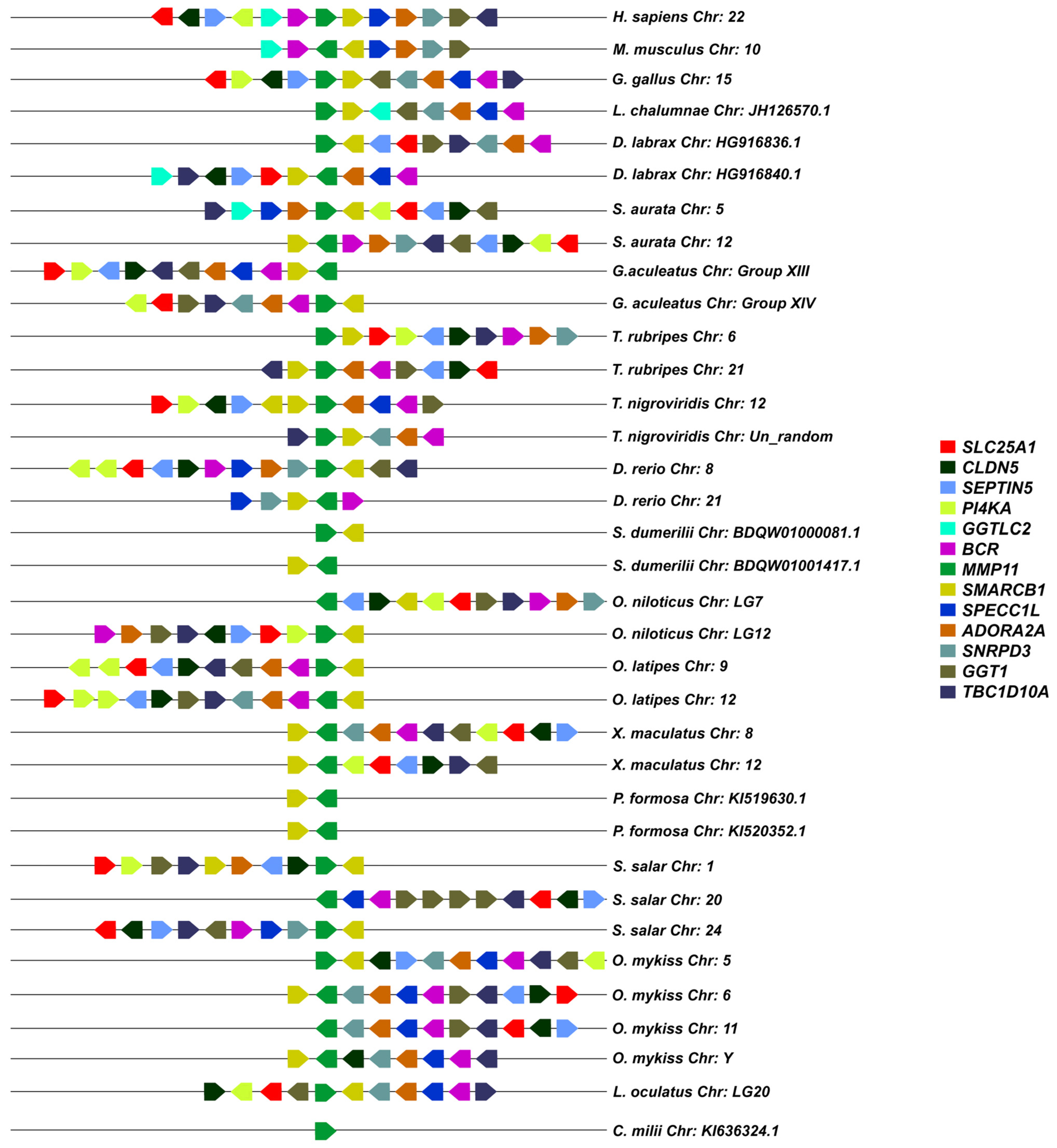
3.2. Synteny Analysis
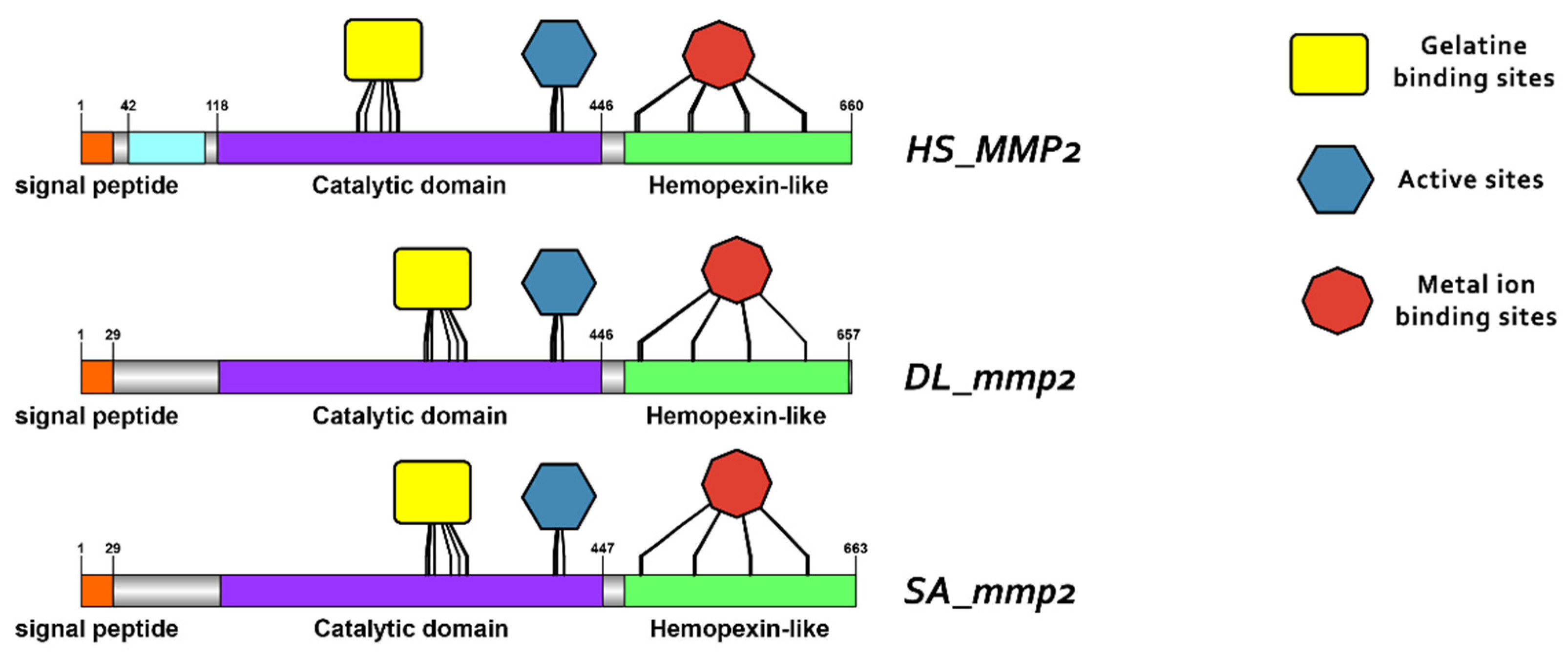
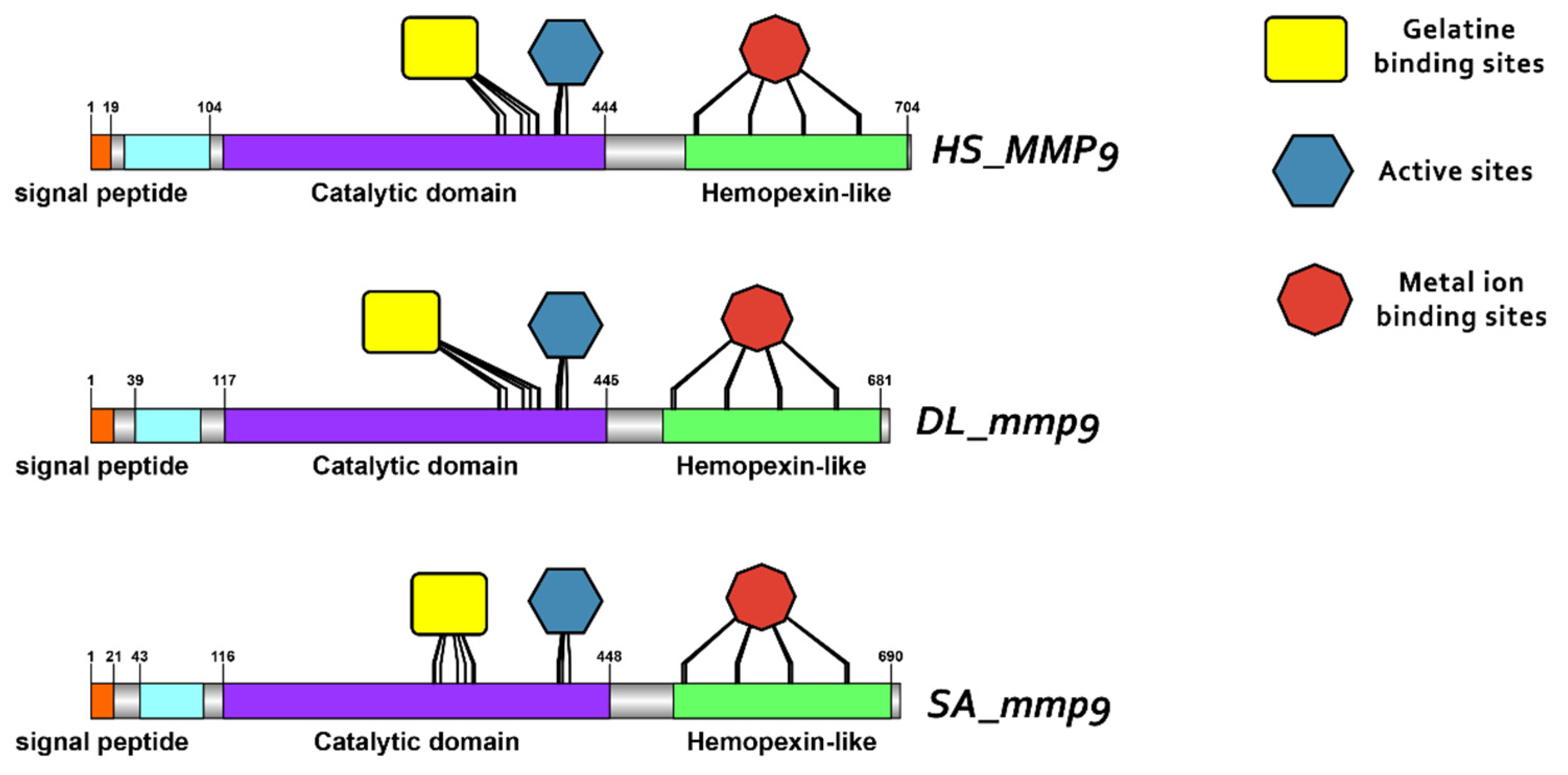
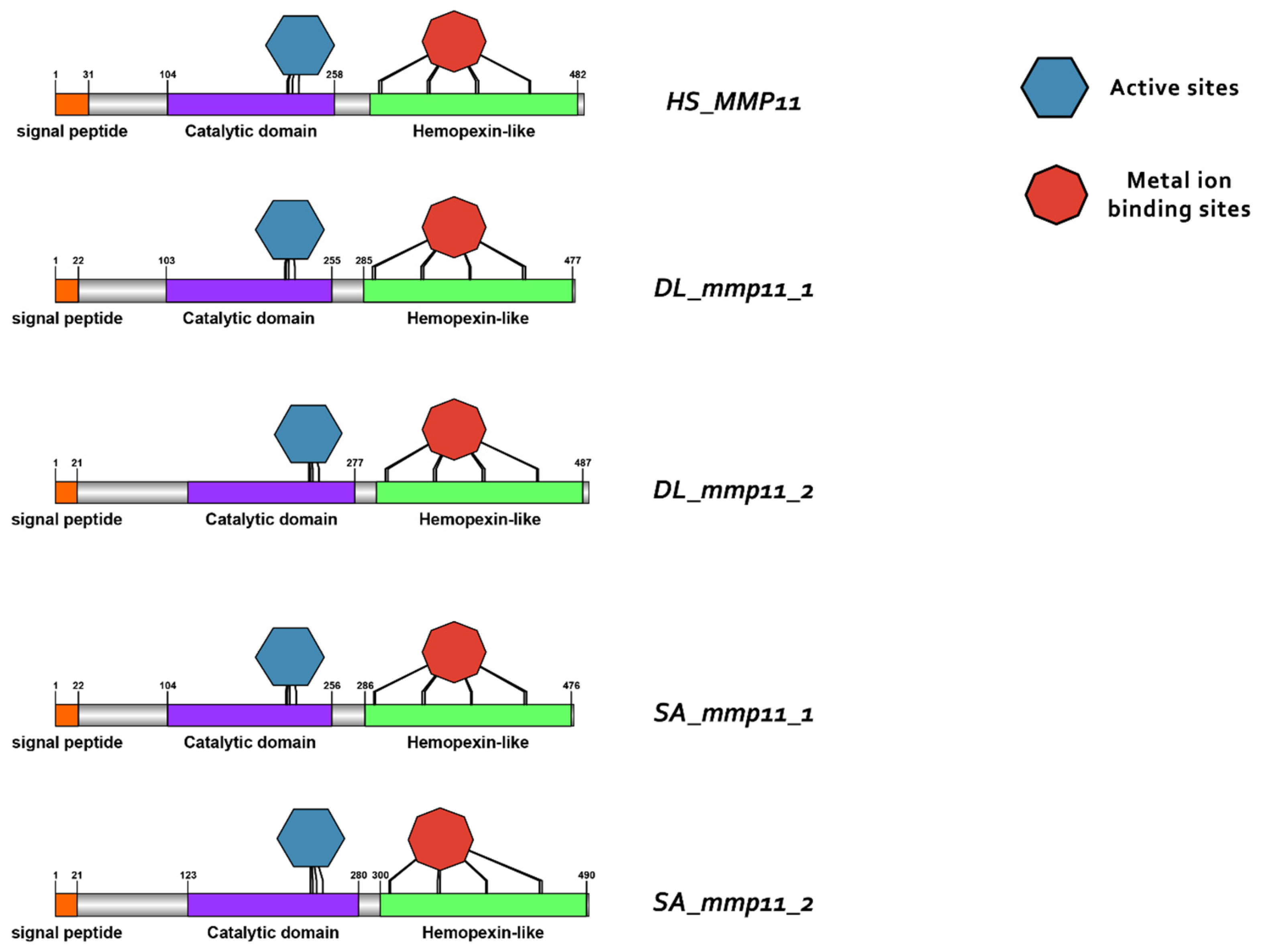
3.3. Comparative Protein Structure Analysis
3.4. Gene Expression Analysis
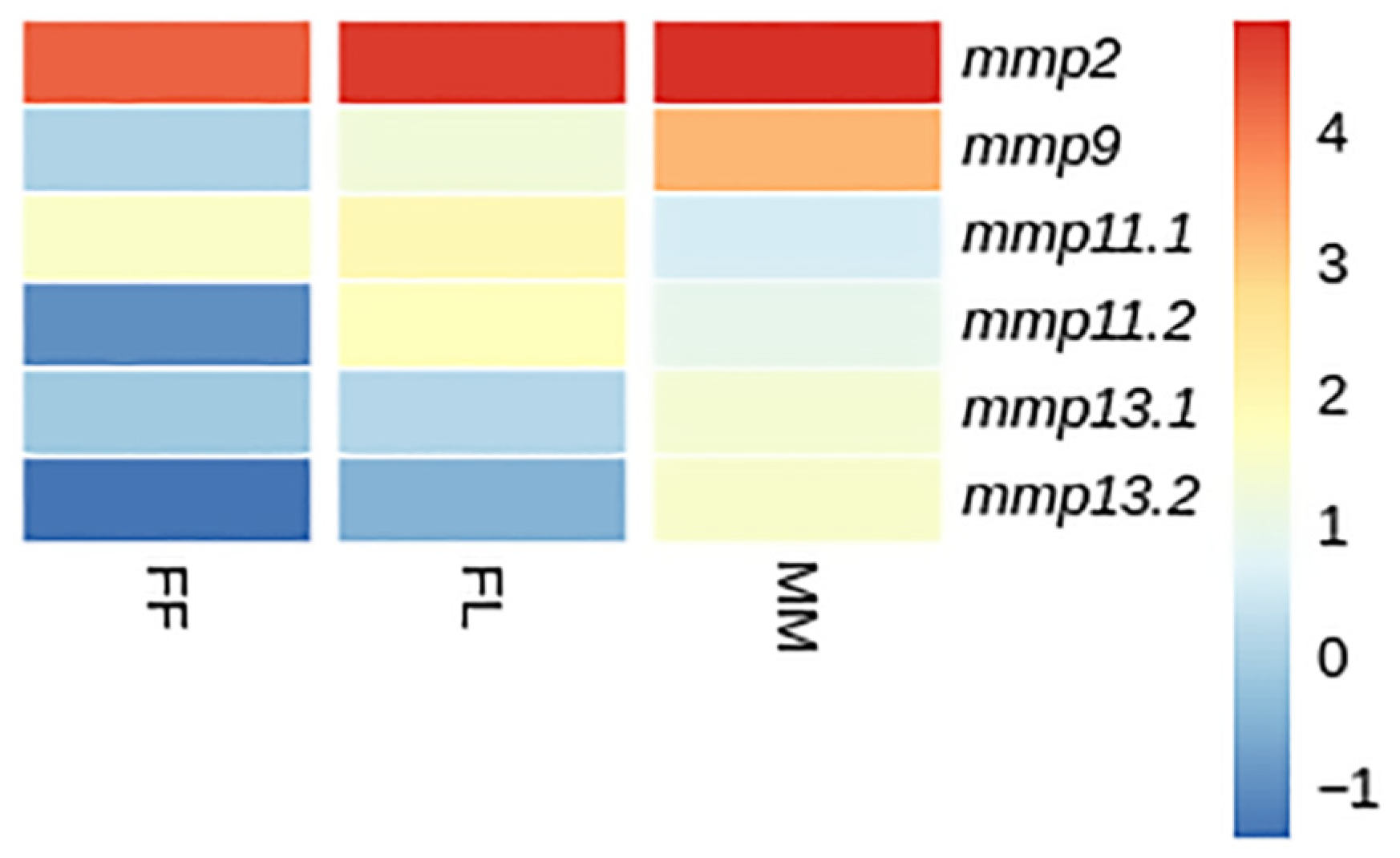
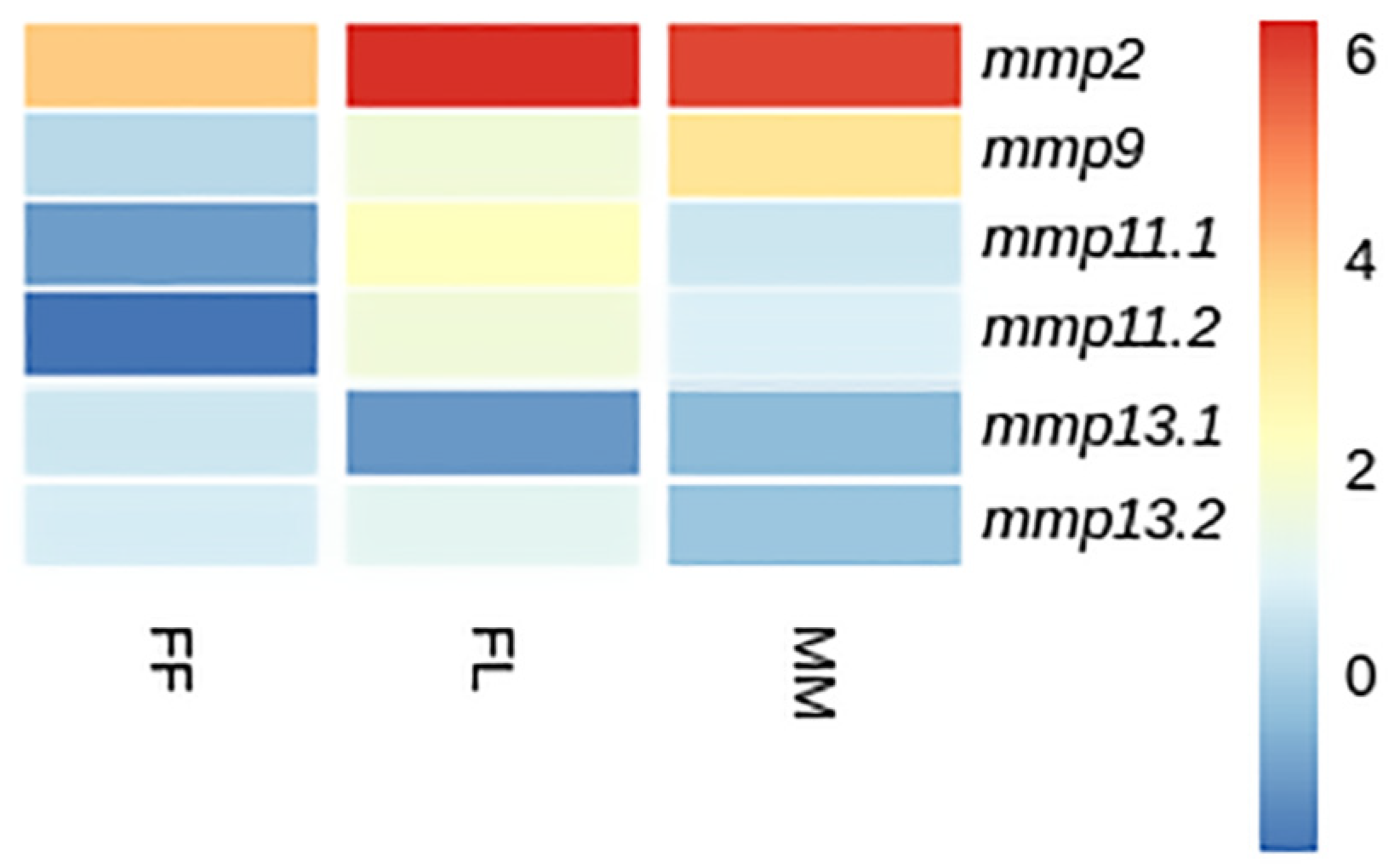
3.4.1. Developmental Expression Patterns
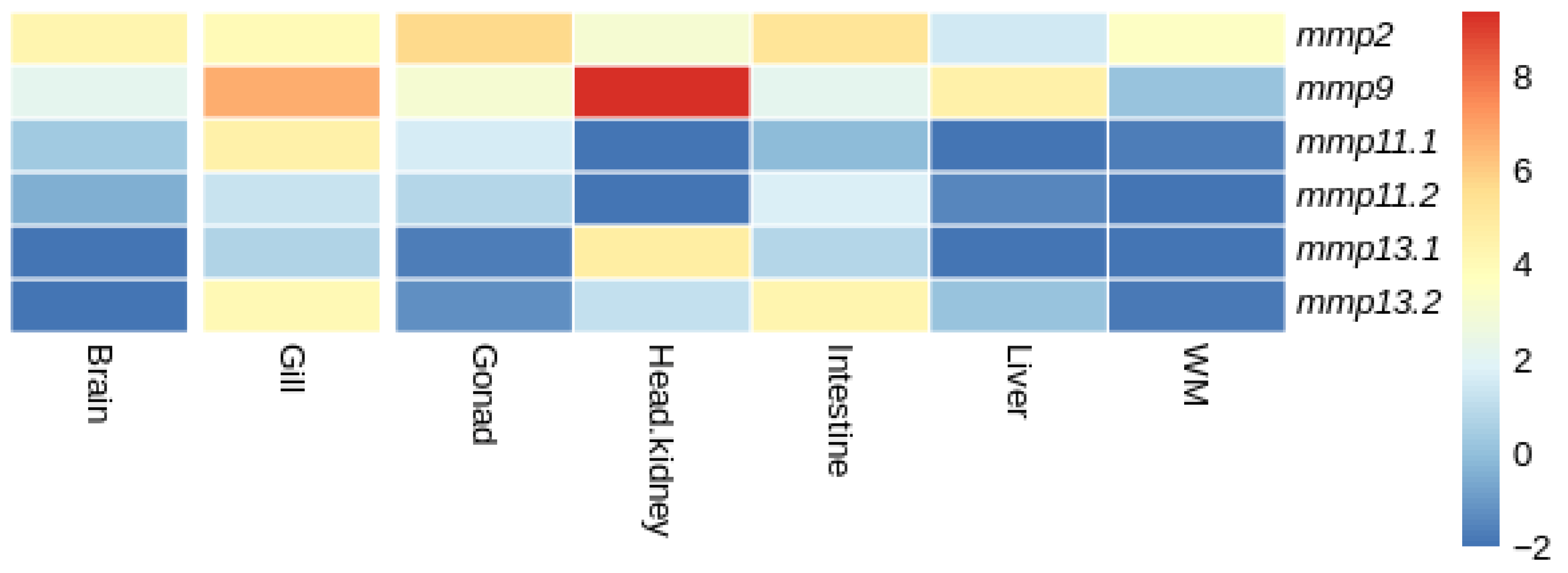
3.4.2. Tissue Expression Patterns
4. Discussion
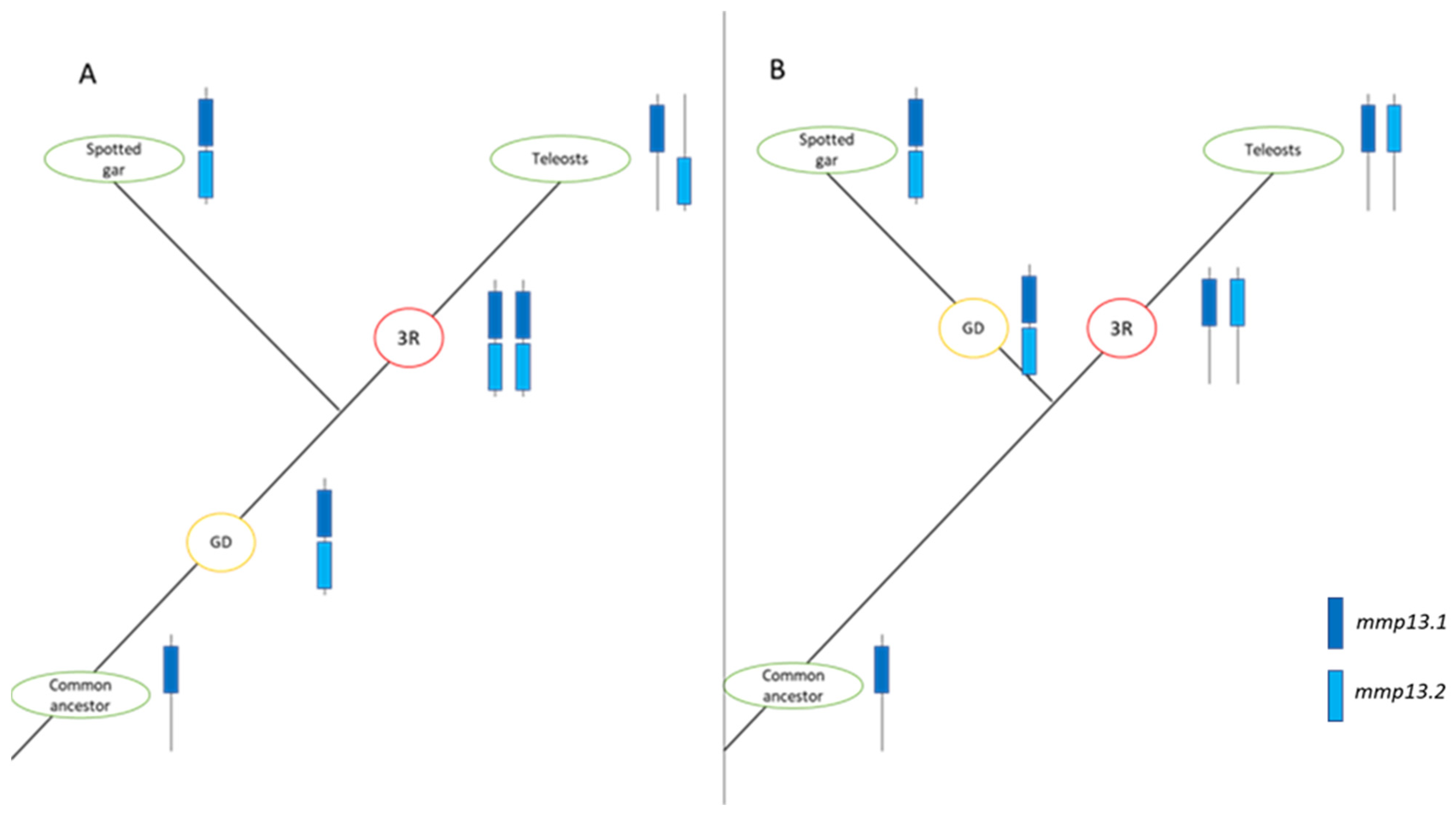
4.1. Evolutionary Path of Matrix Metalloproteases in Teleosts
4.2. Conservation of Structure
4.3. Expression Analysis
4.4. Fate of Paralogs
4.5. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sherman, V.R.; Yang, W.; Meyers, M.A. The Materials Science of Collagen. J. Mech. Behav. Biomed. Mater. 2015, 52, 22–50. [Google Scholar] [CrossRef]
- Moorehead, C.; Prudnikova, K.; Marcolongo, M. The Regulatory Effects of Proteoglycans on Collagen Fibrillogenesis and Morphology Investigated Using Biomimetic Proteoglycans. J. Struct. Biol. 2019, 206, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Sternlicht, M.D.; Werb, Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001, 17, 463–516. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.E.; Vuong, T.T.; Rønning, S.B.; Kolset, S.O. Matrix Metalloproteinases in Fish Biology and Matrix Turnover. Matrix Biol. 2015, 44–46, 86–93. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Matrix Metalloproteinases (MMPs): Chemical–Biological Functions and (Q)SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef]
- Huxley-Jones, J.; Clarke, T.K.; Beck, C.; Toubaris, G.; Robertson, D.L.; Boot-Handford, R.P. The Evolution of the Vertebrate Metzincins; Insights from Ciona Intestinalis and Danio Rerio. BMC Evol. Biol. 2007, 7, 63. [Google Scholar] [CrossRef]
- Fanjul-Fernández, M.; Folgueras, A.R.; Cabrera, S.; López-Otín, C. Matrix Metalloproteinases: Evolution, Gene Regulation and Functional Analysis in Mouse Models. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 3–19. [Google Scholar] [CrossRef]
- Marino-Puertas, L.; Goulas, T.; Gomis-Rüth, F.X. Matrix Metalloproteinases Outside Vertebrates. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2026–2035. [Google Scholar] [CrossRef]
- Robertson, F.M.; Gundappa, M.K.; Grammes, F.; Hvidsten, T.R.; Redmond, A.K.; Lien, S.; Martin, S.A.M.; Holland, P.W.H.; Sandve, S.R.; Macqueen, D.J. Lineage-Specific Rediploidization Is a Mechanism to Explain Time-Lags between Genome Duplication and Evolutionary Diversification. Genome Biol. 2017, 18, 111. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Van de Peer, Y.; Braasch, I.; Meyer, A. Comparative Genomics Provides Evidence for an Ancient Genome Duplication Event in Fish. Philos. Trans. R. Soc. B Biol. Sci. 2001, 356, 1661–1679. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. Turning a Hobby into a Job: How Duplicated Genes Find New Functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Evolution by Gene Duplication; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar] [CrossRef]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Van de Peer, Y. Genome Duplication, a Trait Shared by 22,000 Species of Ray-Finned Fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Van De Peer, Y. From 2R to 3R: Evidence for a Fish-Specific Genome Duplication (FSGD). BioEssays 2005, 27, 937–945. [Google Scholar] [CrossRef]
- Szklarczyk, R.; Huynen, M.A.; Snel, B. Complex Fate of Paralogs. BMC Evol. Biol. 2008, 8, 337. [Google Scholar] [CrossRef]
- Edger, P.P.; Pires, J.C. Gene and Genome Duplications: The Impact of Dosage-Sensitivity on the Fate of Nuclear Genes. Chromosom. Res. 2009, 17, 699–717. [Google Scholar] [CrossRef]
- Glasauer, S.M.K.K.; Neuhauss, S.C.F.F. Whole-Genome Duplication in Teleost Fishes and Its Evolutionary Consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Drobek, M. Paralogous Genes Involved in Embryonic Development: Lessons from the Eye and Other Tissues. Genes 2022, 13, 2082. [Google Scholar] [CrossRef]
- Young, D.; Das, N.; Anowai, A.; Dufour, A. Matrix Metalloproteases as Influencers of the Cells’ Social Media. Int. J. Mol. Sci. 2019, 20, 3847. [Google Scholar] [CrossRef]
- Pauletto, M.; Manousaki, T.; Ferraresso, S.; Babbucci, M.; Tsakogiannis, A.; Louro, B.; Vitulo, N.; Quoc, V.H.; Carraro, R.; Bertotto, D.; et al. Genomic Analysis of Sparus Aurata Reveals the Evolutionary Dynamics of Sex-Biased Genes in a Sequential Hermaphrodite Fish. Commun. Biol. 2018, 1, 119. [Google Scholar] [CrossRef] [PubMed]
- Tine, M.; Kuhl, H.; Gagnaire, P.A.; Louro, B.; Desmarais, E.; Martins, R.S.T.; Hecht, J.; Knaust, F.; Belkhir, K.; Klages, S.; et al. European Sea Bass Genome and Its Variation Provide Insights into Adaptation to Euryhalinity and Speciation. Nat. Commun. 2014, 5, 5770. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Ridwan Amode, M.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate Detection of Unreliable Alignment Regions Accounting for the Uncertainty of Multiple Parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Tsipourlianos, A.; Cardoso, J.C.R.; Angelakopoulos, R.; Kotoula, A.; Power, D.M.; Mamuris, Z.; Moutou, K.A. Regulatory Subfunctionalization Drives OXPHOS Evolution in Teleosts. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A Hub for Data Retrieval across Taxonomic Space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An Illustrator for the Presentation and Visualization of Biological Sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Papandroulakis, N.; Divanach, P.; Anastasiadis, P.; Kentouri, M. The Pseudo-Green Water Technique for Intensive Rearing of Sea Bream (Sparus aurata) Larvae. Aquac. Int. 2001, 9, 205–216. [Google Scholar] [CrossRef]
- Kourkouta, C.; Tsipourlianos, A.; Power, D.M.; Moutou, K.A.; Koumoundouros, G. Variability of Key-Performance-Indicators in Commercial Gilthead Seabream Hatcheries. Sci. Rep. 2022, 12, 17896. [Google Scholar] [CrossRef]
- Stamperna, K.; Giannoulis, T.; Cañon-Beltrán, K.; Dovolou, E.; Kalemkeridou, M.; Nanas, I.; Rizos, D.; Moutou, K.A.; Mamuris, Z.; Amiridis, G.S. Oviductal Epithelial Cells Transcriptome and Extracellular Vesicles Characterization during Thermoneutral and Heat Stress Conditions in Dairy Cows. Theriogenology 2022, 187, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Amălinei, C.; Căruntu, I.; Anca Bălan, R. Biology of Metalloproteinases. Rom. J. Morphol. Embryol. 2007, 48, 323–334. [Google Scholar]
- Gout, J.F.; Hao, Y.; Johri, P.; Arnaiz, O.; Doak, T.G.; Bhullar, S.; Couloux, A.; Guérin, F.; Malinsky, S.; Potekhin, A.; et al. Dynamics of Gene Loss Following Ancient Whole-Genome Duplication in the Cryptic Paramecium Complex. Mol. Biol. Evol. 2023, 40, msad107. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Ma, L.; Wang, G.; Li, M.; Zhang, Z. Old Genes Experience Stronger Translational Selection than Young Genes. Gene 2016, 590, 29–34. [Google Scholar] [CrossRef]
- Xu, X.Y.; Shen, Y.B.; Fu, J.J.; Liu, F.; Guo, S.Z.; Yang, X.M.; Li, J. Le Matrix Metalloproteinase 2 of Grass Carp Ctenopharyngodon Idella (CiMMP2) Is Involved in the Immune Response against Bacterial Infection. Fish Shellfish Immunol. 2012, 33, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Ke, F.; Wang, Y.; Hong, J.; Xu, C.; Chen, H.; Zhou, S.B. Characterization of MMP-9 Gene from a Normalized CDNA Library of Kidney Tissue of Yellow Catfish (Pelteobagrus fulvidraco). Fish Shellfish Immunol. 2015, 45, 260–267. [Google Scholar] [CrossRef]
- Aase-Remedios, M.E.; Ferrier, D.E.K. Improved Understanding of the Role of Gene and Genome Duplications in Chordate Evolution With New Genome and Transcriptome Sequences. Front. Ecol. Evol. 2021, 9, 703163. [Google Scholar] [CrossRef]
- Catchen, J.M.; Conery, J.S.; Postlethwait, J.H. Automated Identification of Conserved Synteny after Whole-Genome Duplication. Genome Res. 2009, 19, 1497–1505. [Google Scholar] [CrossRef]
- Luo, J.; Chai, J.; Chai, J.; Wen, Y.; Wen, Y.; Wen, Y.; Tao, M.; Lin, G.; Liu, X.; Ren, L.; et al. From Asymmetrical to Balanced Genomic Diversification during Rediploidization: Subgenomic Evolution in Allotetraploid Fish. Sci. Adv. 2020, 6, 7677–7704. [Google Scholar] [CrossRef]
- Woods, I.G.; Kelly, P.D.; Chu, F.; Ngo-Hazelett, P.; Yan, Y.-L.; Huang, H.; Postlethwait, J.H.; Talbot, W.S. A Comparative Map of the Zebrafish Genome. Genome Res. 2000, 10, 1903–1914. [Google Scholar] [CrossRef]
- Small, C.D.; El-Khoury, M.; Deslongchamps, G.; Benfey, T.J.; Crawford, B.D. Matrix Metalloproteinase 13 Activity Is Required for Normal and Hypoxia-Induced Precocious Hatching in Zebrafish Embryos. J. Dev. Biol. 2020, 8, 3. [Google Scholar] [CrossRef]
- Woolfe, A.; Elgar, G. Comparative Genomics Using Fugu Reveals Insights into Regulatory Subfunctionalization. Genome Biol. 2007, 8, R53. [Google Scholar] [CrossRef]
- Wyatt, R.A.; Keow, J.Y.; Harris, N.D.; Haché, C.A.; Li, D.H.; Crawford, B.D. The Zebrafish Embryo: A Powerful Model System for Investigating Matrix Remodeling. Zebrafish 2009, 6, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Macqueen, D.J.; Johnston, I.A. A Well-Constrained Estimate for the Timing of the Salmonid Whole Genome Duplication Reveals Major Decoupling from Species Diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Thorgaard, G.H. Tetraploidy and the Evolution of Salmonid Fishes. In Evolutionary Genetics of Fishes; Springer: Boston, MA, USA, 1984; pp. 1–53. [Google Scholar] [CrossRef]
- Grone, B.P.; Maruska, K.P. Divergent Evolution of Two Corticotropin-Releasing Hormone (CRH) Genes in Teleost Fishes. Front. Neurosci. 2015, 9, 365. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hu, J.; Qu, C.; Wang, L.; Huang, G.; Niu, P.; Zhong, Z.; Hong, F.; Wang, G.; Postlethwait, J.H.; et al. Molecular Evolution and Functional Divergence of Zebrafish (Danio rerio) Cryptochrome Genes. Sci. Rep. 2015, 5, 8113. [Google Scholar] [CrossRef]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The Spotted Gar Genome Illuminates Vertebrate Evolution and Facilitates Human-Teleost Comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef]
- Foley, C.J.; Kuliopulos, A. Mouse Matrix Metalloprotease-1a (Mmp1a) Gives New Insight Into MMP Function. J. Cell. Physiol. 2014, 229, 1875. [Google Scholar] [CrossRef]
- Ribeiro, A.J.M.; Tyzack, J.D.; Borkakoti, N.; Holliday, G.L.; Thornton, J.M. A Global Analysis of Function and Conservation of Catalytic Residues in Enzymes. J. Biol. Chem. 2020, 295, 314. [Google Scholar] [CrossRef] [PubMed]
- Valasatava, Y.; Rosato, A.; Furnham, N.; Thornton, J.M.; Andreini, C. To What Extent Do Structural Changes in Catalytic Metal Sites Affect Enzyme Function? J. Inorg. Biochem. 2018, 179, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Wargelius, A.; Fjelldal, P.G.; Grini, A.; Gil-Martens, L.; Kvamme, B.O.; Hansen, T. MMP-13 (Matrix MetalloProteinase 13) Expression Might Be an Indicator for Increased ECM Remodeling and Early Signs of Vertebral Compression in Farmed Atlantic Salmon (Salmo salar L.). J. Appl. Ichthyol. 2010, 26, 366–371. [Google Scholar] [CrossRef]
- de Vrieze, E.; Sharif, F.; Metz, J.R.; Flik, G.; Richardson, M.K. Matrix Metalloproteinases in Osteoclasts of Ontogenetic and Regenerating Zebrafish Scales. Bone 2011, 48, 704–712. [Google Scholar] [CrossRef]
- Liu, R.; Imangali, N.; Ethiraj, L.P.; Carney, T.J.; Winkler, C. Transcriptome Profiling of Osteoblasts in a Medaka (Oryzias latipes) Osteoporosis Model Identifies Mmp13b as Crucial for Osteoclast Activation. Front. Cell Dev. Biol. 2022, 10, 775512. [Google Scholar] [CrossRef]
- Mazzoni, T.S.; Nostro, F.L.L.; Antoneli, F.N.; Quagio-Grassiotto, I. Action of the Metalloproteinases in Gonadal Remodeling during Sex Reversal in the Sequential Hermaphroditism of the Teleostei Fish Synbranchus marmoratus (Synbranchiformes: Synbranchidae). Cells 2018, 7, 34. [Google Scholar] [CrossRef]
- Castillo-Briceño, P.; Sepulcre, M.P.; Chaves-Pozo, E.; Meseguer, J.; García-Ayala, A.; Mulero, V. Collagen Regulates the Activation of Professional Phagocytes of the Teleost Fish Gilthead Seabream. Mol. Immunol. 2009, 46, 1409–1415. [Google Scholar] [CrossRef]
- Mundell, N.A.; Jessen, J.R.; Mundell, N.A.; Jessen, J.R.; Desimone, D.W. Extracellular Matrix Remodeling in Zebrafish Development. In Extracellular Matrix in Development; Springer: Berlin/Heidelberg, Germany, 2013; pp. 187–218. [Google Scholar] [CrossRef]
- Bai, S.; Thummel, R.; Godwin, A.R.; Nagase, H.; Itoh, Y.; Li, L.; Evans, R.; McDermott, J.; Seiki, M.; Sarras, M.P. Matrix Metalloproteinase Expression and Function during Fin Regeneration in Zebrafish: Analysis of MT1-MMP, MMP2 and TIMP2. Matrix Biol. 2005, 24, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ceballos-Francisco, D.; Guardiola, F.A.; Huang, D.; Esteban, M.Á. Skin Wound Healing in Gilthead Seabream (Sparus aurata L.) Fed Diets Supplemented with Arginine. Fish Shellfish Immunol. 2020, 104, 347–358. [Google Scholar] [CrossRef]
- Schmidt, J.G.; Andersen, E.W.; Ersbøll, B.K.; Nielsen, M.E. Muscle Wound Healing in Rainbow Trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2016, 48, 273–284. [Google Scholar] [CrossRef]
- Kandhwal, M.; Behl, T.; Singh, S.; Sharma, N.; Arora, S.; Bhatia, S.; Al-Harrasi, A.; Sachdeva, M.; Bungau, S. Role of Matrix Metalloproteinase in Wound Healing. Am. J. Transl. Res. 2022, 14, 4391. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y. Role of Matrix Metalloproteinases in Skeletal Muscle. Cell Adhes. Migr. 2009, 3, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Murawala, H.; Patel, S.; Ranadive, I.; Desai, I.; Balakrishnan, S. Variation in Expression and Activity Pattern of Mmp2 and Mmp9 on Different Time Scales in the Regenerating Caudal Fin of Poecilia latipinna. J. Fish Biol. 2018, 92, 1604–1619. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, J.; Akimenko, M.A. Inhibition of Mmp13a during Zebrafish Fin Regeneration Disrupts Fin Growth, Osteoblasts Differentiation, and Laminin Organization. Dev. Dyn. 2020, 249, 187–198. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, H.; Zhao, G.; Yokoyama, T.; Vega, H.; Huang, Y.; Sood, R.; Bishop, K.; Maduro, V.; Accardi, J.; et al. ATP6V1H Deficiency Impairs Bone Development through Activation of MMP9 and MMP13. PLoS Genet. 2017, 13, e1006481. [Google Scholar] [CrossRef]
- Kessels, M.Y.; Huitema, L.A.F.; Boeren, S.; Kranenbarg, S.; Schulte-Merker, S.; Van Leeuwen, J.L.; De Vries, S.C. Proteomics Analysis of the Zebrafish Skeletal Extracellular Matrix. PLoS ONE 2014, 9, e90568. [Google Scholar] [CrossRef]
- Vincenti, M.P.; Brinckerhoff, C.E. Transcriptional Regulation of Collagenase (MMP-1, MMP-13) Genes in Arthritis: Integration of Complex Signaling Pathways for the Recruitment of Gene-Specific Transcription Factors. Arthritis Res. 2002, 4, 157–164. [Google Scholar] [CrossRef]
- Castillo-Briceño, P.; Arizcun-Arizcun, M.; Meseguer, J.; Mulero, V.; García-Ayala, A. Correlated Expression Profile of Extracellular Matrix-Related Molecules during the Inflammatory Response of the Teleost Fish Gilthead Seabream. Dev. Comp. Immunol. 2010, 34, 1051–1058. [Google Scholar] [CrossRef]
- Kuzmin, E.; Vandersluis, B.; Ba, A.N.N.; Wang, W.; Koch, E.N.; Usaj, M.; Khmelinskii, A.; Usaj, M.M.; van Leeuwen, J.; Kraus, O.; et al. Exploring Whole-Genome Duplicate Gene Retention with Complex Genetic Interaction Analysis. Science 2020, 368, eaaz5667. [Google Scholar] [CrossRef] [PubMed]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic Salmon Genome Provides Insights into Rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Hou, G.Y.; Kong, X.F.; Li, C.Y.; Zeng, J.M.; Li, H.D.; Xiao, G.B.; Li, X.M.; Sun, X.W. The Fate of Recent Duplicated Genes Following a Fourth-Round Whole Genome Duplication in a Tetraploid Fish, Common Carp (Cyprinus carpio). Sci. Rep. 2015, 5, srep08199. [Google Scholar] [CrossRef]
- Duarte, J.M.; Cui, L.; Wall, P.K.; Zhang, Q.; Zhang, X.; Leebens-Mack, J.; Ma, H.; Altman, N.; DePamphilis, C.W. Expression Pattern Shifts Following Duplication Indicative of Subfunctionalization and Neofunctionalization in Regulatory Genes of Arabidopsis. Mol. Biol. Evol. 2006, 23, 469–478. [Google Scholar] [CrossRef]
- Maouche, A.; Curran, E.; Goupil, A.S.; Sambroni, E.; Bellaiche, J.; Le Gac, F.; Lareyre, J.J. New Insights into the Evolution, Hormonal Regulation, and Spatiotemporal Expression Profiles of Genes Involved in the Gfra1/Gdnf and Kit/Kitlg Regulatory Pathways in Rainbow Trout Testis. Fish Physiol. Biochem. 2018, 44, 1599–1616. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.R.; Berg, J.J.; Blinka, S.; Rausher, M.D.; Baum, D.A. Precise Spatio-Temporal Regulation of the Anthocyanin Biosynthetic Pathway Leads to Petal Spot Formation in Clarkia gracilis (Onagraceae). New Phytol. 2013, 197, 958–969. [Google Scholar] [CrossRef]
- Shew, C.J.; Carmona-Mora, P.; Soto, D.C.; Mastoras, M.; Roberts, E.; Rosas, J.; Jagannathan, D.; Kaya, G.; O’Geen, H.; Dennis, M.Y. Diverse Molecular Mechanisms Contribute to Differential Expression of Human Duplicated Genes. Mol. Biol. Evol. 2021, 38, 3060–3077. [Google Scholar] [CrossRef]
- MacCarthy, T.; Bergman, A. The Limits of Subfunctionalization. BMC Evol. Biol. 2007, 7, 213. [Google Scholar] [CrossRef]





| Species | Representative of | Genome |
|---|---|---|
| Sparus aurata (Gilthead sea bream) | Teleosts | GCA_900880675.1 |
| Dicentrarchus labrax (European sea bass) | Teleosts | GCA_000689215.1 |
| Seriola dumerilii (Greater amberjack) | Teleosts | GCA_002260705.1 |
| Gasterosteus aculeatus (Stickleback) | Teleosts | BROAD S1 * |
| Takifugu rubripes (Fugu) | Teleosts | GCA_901000725.2 |
| Tetraodon nigroviridis (Tetraodon) | Teleosts | TETRAODON 8.0 * |
| Oreochromis niloticus (Nile tilapia) | Teleosts | GCA_001858045.3 |
| Oryzias latipes (Japanese medaka) | Teleosts | GCA_002234675.1 |
| Poecilia formosa (Amazon molly) | Teleosts | GCA_000485575.1 |
| Xiphophorus maculatus (Monterrey platyfish) | Teleosts | GCA_001444195.1 |
| Danio rerio (Zebrafish) | Teleosts | GCA_000002035.4 |
| Salmo salar (Atlantic salmon) | Teleosts | GCA_905237065.2 |
| Oncorhynchus mykiss (Rainbow trout) | Teleosts | GCA_013265735.3 |
| Lepisosteus oculatus (Spotted gar) | Actinopterygii | GCA_000242695.1 |
| Latimeria chalumnae (Coelacanth) | Sarcopterygii | GCA_000225785.1 |
| Homo sapiens (Human) | Sarcopterygii | GCA_000001405.28 |
| Mus musculus (Common mouse) | Sarcopterygii | GCA_000001635.9 |
| Gallus gallus (Chicken) | Sarcopterygii | GCA_000002315.5 |
| Xenopus tropicalis (Tropical clawed frog) | Sarcopterygii | GCA_000004195.3 |
| Callorhinchus milii (Elephant shark) | Chondrichthyes | GCA_000165045.2 |
| Gene | Gilthead Sea Bream (S. aurata) | European Sea Bass (D. labrax) |
|---|---|---|
| mmp2 | ENSSAUG00010014278 | ENSDLAG00005001814 |
| mmp9 | ENSSAUG00010026521 | ENSDLAG00005018682 |
| mmp11.1 | ENSSAUG00010019504 | ENSDLAG00005011760 |
| mmp11.2 | ENSSAUG00010022437 | ENSDLAG00005009945 |
| mmp13.1 | ENSSAUG00010014101 | ENSDLAG00005008130 |
| mmp13.2 | ENSSAUG00010010684 | ENSDLAG00005008348 |
| Gene Name | Gilthead Sea Bream | European Sea Bass |
|---|---|---|
| mmp11 | Dispersed | Dispersed |
| mmp13 | WGD/segmental | WGD/segmental |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelakopoulos, R.; Tsipourlianos, A.; Maravelakis, I.D.; Giannoulis, T.; Mamuris, Z.; Moutou, K.A. Evolutionary Dynamics of Matrix Metalloproteases with Collagenolytic Activity in Teleosts. Animals 2025, 15, 3270. https://doi.org/10.3390/ani15223270
Angelakopoulos R, Tsipourlianos A, Maravelakis ID, Giannoulis T, Mamuris Z, Moutou KA. Evolutionary Dynamics of Matrix Metalloproteases with Collagenolytic Activity in Teleosts. Animals. 2025; 15(22):3270. https://doi.org/10.3390/ani15223270
Chicago/Turabian StyleAngelakopoulos, Rafael, Andreas Tsipourlianos, Ioannis Damianos Maravelakis, Themistoklis Giannoulis, Zissis Mamuris, and Katerina A. Moutou. 2025. "Evolutionary Dynamics of Matrix Metalloproteases with Collagenolytic Activity in Teleosts" Animals 15, no. 22: 3270. https://doi.org/10.3390/ani15223270
APA StyleAngelakopoulos, R., Tsipourlianos, A., Maravelakis, I. D., Giannoulis, T., Mamuris, Z., & Moutou, K. A. (2025). Evolutionary Dynamics of Matrix Metalloproteases with Collagenolytic Activity in Teleosts. Animals, 15(22), 3270. https://doi.org/10.3390/ani15223270






