Identification of MAPK10 as a Candidate Gene for High Milk Production in Water Buffaloes Through a Genome-Wide Association Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Welfare Statement
2.2. Phenotypes and Animal Resources
2.3. Sample Collection, Sequencing and Data Storage
2.4. Alignments and Variant Identification
2.5. Variation Filtering
2.6. Population Structure Analysis
2.7. Genome-Wide Association Mapping
2.8. Candidate-Associated Gene Pathway Enrichment
3. Results
3.1. Phenotypic Value Statistics of the Traits
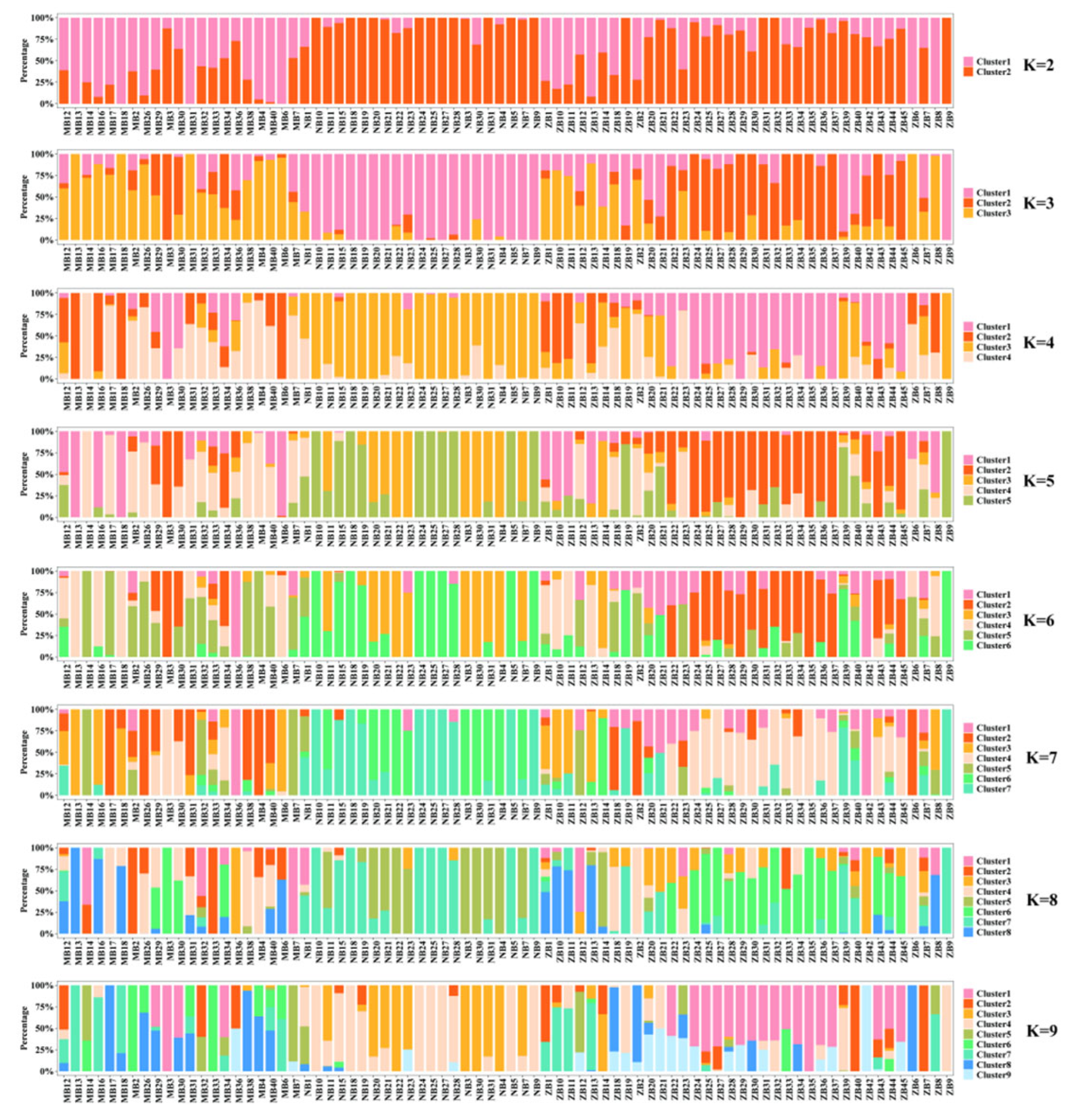
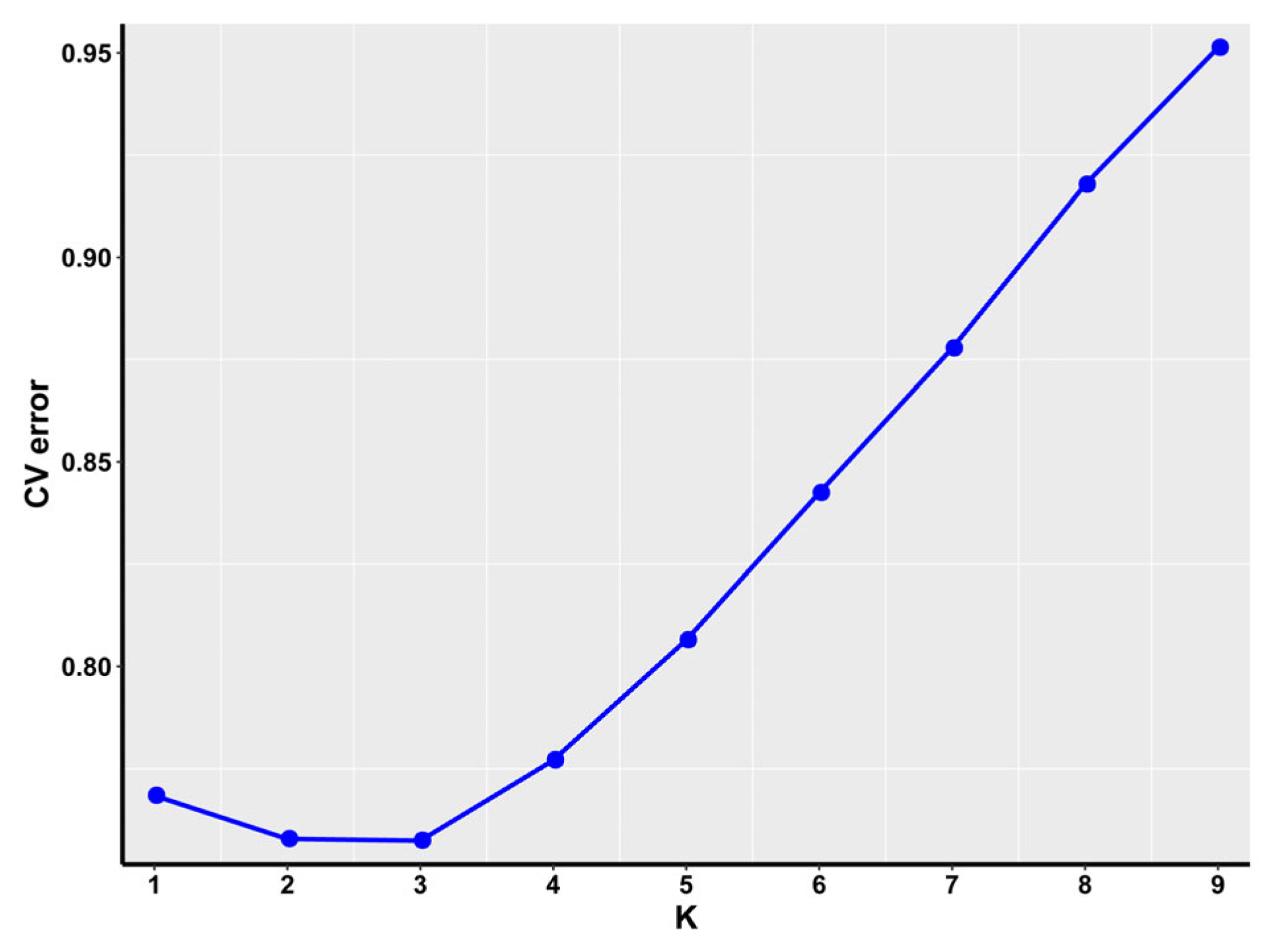
3.2. Population Structure
3.3. Results of the Genome-Wide Associations
3.4. Kyoto Encyclopedia of Genes and Genome Pathway Analysis of Candidate Genes
3.5. Significant Association of Milk Protein Content with SNP Validation
4. Discussion
4.1. Molecular Genetic Structure
4.2. Genome-Wide Association Analysis of Reproductive-Related Traits
4.3. The Role of MAPK10 in Enhancing Buffalo Milk Production: Current Findings and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, Y.X.; Sun, H.; Shaukat, A.; Deng, T.X.; Abdel-Shafy, H.; Che, Z.X.; Zhou, Y.; Hu, C.M.; Li, H.Z.; Wu, Q.P.; et al. Novel Insight Into the Role of ACSL1 Gene in Milk Production Traits in Buffalo. Front. Genet. 2022, 13, 896910. [Google Scholar] [CrossRef]
- Barłowska, J.; Szwajkowska, M.; Litwińczuk, Z.; Król, J. Nutritional Value and Technological Suitability of Milk from Various Animal Species Used for Dairy Production. Compr. Rev. Food Sci. Food Saf. 2011, 10, 291–302. [Google Scholar] [CrossRef]
- Pisanu, S.; Cacciotto, C.; Pagnozzi, D.; Puggioni, G.M.G.; Uzzau, S.; Ciaramella, P.; Guccione, J.; Penati, M.; Pollera, C.; Moroni, P.; et al. Proteomic changes in the milk of water buffaloes (water buffalo) with subclinical mastitis due to intramammary infection by Staphylococcus aureus and by non-aureus staphylococci. Sci. Rep. 2019, 9, 15850. [Google Scholar] [CrossRef]
- Deng, T.; Liang, A.; Liang, S.; Ma, X.; Lu, X.; Duan, A.; Pang, C.; Hua, G.; Liu, S.; Campanile, G.; et al. Integrative Analysis of Transcriptome and GWAS Data to Identify the Hub Genes Associated With Milk Yield Trait in Buffalo. Front. Genet. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Srirattana, K.; Hufana-Duran, D.; Atabay, E.P.; Duran, P.G.; Atabay, E.C.; Lu, K.; Liang, Y.; Chaikhun-Marcou, T.; Theerakittayakorn, K.; Parnpai, R. Current status of assisted reproductive technologies in buffaloes. Anim. Sci. J. 2022, 93, e13767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yao, Z.; Li, X.; Zhang, Z.; Liu, X.; Yang, P.; Chen, N.; Xia, X.; Lyu, S.; Shi, Q.; et al. Assessing genomic diversity and signatures of selection in Pinan cattle using whole-genome sequencing data. BMC Genom. 2022, 23, 460. [Google Scholar] [CrossRef]
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Z.; Zhang, Z.; Li, H.; Liu, D.; Zhang, Q.; Bradbury, P.J.; Buckler, E.S.; Zhang, Z. Expanding the BLUP alphabet for genomic prediction adaptable to the genetic architectures of complex traits. Heredity 2018, 121, 648–662. [Google Scholar] [CrossRef]
- Ning, C.; Wang, D.; Kang, H.; Mrode, R.; Zhou, L.; Xu, S.; Liu, J.F. A rapid epistatic mixed-model association analysis by linear retransformations of genomic estimated values. Bioinformatics 2018, 34, 1817–1825. [Google Scholar] [CrossRef]
- Breen, E.J.; MacLeod, I.M.; Ho, P.N.; Haile-Mariam, M.; Pryce, J.E.; Thomas, C.D.; Daetwyler, H.D.; Goddard, M.E. BayesR3 enables fast MCMC blocked processing for largescale multi-trait genomic prediction and QTN mapping analysis. Commun. Biol. 2022, 5, 661. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.; Tu, Y.; Lv, X. Accelerating BWA-MEM Read Mapping on GPUs. In Proceedings of the ICS ’23: Proceedings of the 37th International Conference on Supercomputing, Orlando, FL, USA, 21–23 June 2023; pp. 155–166. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Bathke, J.; Lühken, G. OVarFlow: A resource optimized GATK 4 based Open source Variant calling workFlow. BMC Bioinform. 2021, 22, 402. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Tingzhu, Y.; Pasandideh, M.; Liang, A.; Hua, G.; Farmanullah; Schreurs, N.M.; Raza, S.H.A.; Salzano, A.; Campanile, G.; et al. Genetic Association of PPARGC1A Gene Single Nucleotide Polymorphism with Milk Production Traits in Italian Mediterranean Buffalo. BioMed Res. Int. 2021, 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Haldar, T.; Ghosh, S. Effect of population stratification on false positive rates of population-based association analyses of quantitative traits. Ann. Hum. Genet. 2012, 76, 237–245. [Google Scholar] [CrossRef]
- Persyn, E.; Redon, R.; Bellanger, L.; Dina, C. The impact of a fine-scale population stratification on rare variant association test results. PLoS ONE 2018, 13, e0207677. [Google Scholar] [CrossRef]
- Elhaik, E. Principal Component Analyses (PCA)-based findings in population genetic studies are highly biased and must be reevaluated. Sci. Rep. 2022, 12, 14683. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yao, S.; Meng, Q.; Liu, X.; Han, H.; Kan, C.; Liu, G. A novel signaling transduction pathway of melatonin on lactose synthesis in cows via melatonin receptor 1 (MT1) and prolactin receptor (PRLR). PeerJ 2023, 11, e15932. [Google Scholar] [CrossRef]
- Qian, L.; Lopez, V.; Seo, Y.A.; Kelleher, S.L. Prolactin regulates ZNT2 expression through the JAK2/STAT5 signaling pathway in mammary cells. Am. J. Physiol.-Cell Physiol. 2009, 297, C369–C377. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, N.; Zhu, S.; Lu, S.; Jiang, H.; Zhou, H. Estradiol reduced 5-HT reuptake by downregulating the gene expression of Plasma Membrane Monoamine Transporter (PMAT, Slc29a4) through estrogen receptor β and the MAPK/ERK signaling pathway. Eur. J. Pharmacol. 2022, 924, 174939. [Google Scholar] [CrossRef]
- Maya-Bernal, J.L.; Ramírez-Santiago, G. Spatio-temporal dynamics of a cell signal pathway with negative feedbacks: The MAPK/ERK pathway. Eur. Phys. J. E 2016, 39, 28. [Google Scholar] [CrossRef]
- Haftcheshmeh, S.M.; Abedi, M.; Mashayekhi, K.; Mousavi, M.J.; Navashenaq, J.G.; Mohammadi, A.; Momtazi-Borojeni, A.A. Berberine as a natural modulator of inflammatory signaling pathways in the immune system: Focus on NF-κB, JAK/STAT, and MAPK signaling pathways. Phytother. Res. 2022, 36, 1216–1230. [Google Scholar] [CrossRef]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Non-Coding RNA 2021, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Hynes, A.C.; Sreenan, J.; MKane, M.T. The phosphatidylinositol signalling system in elongating bovine blastocysts; formation of phosphoinositides, inositol phosphates and stimulation by growth factors. Reprod. Fertil. Dev. 2002, 14, 515–523. [Google Scholar] [CrossRef] [PubMed]





| Methods | SNP | Chr | Pos | p | Candidate Genes |
|---|---|---|---|---|---|
| cBLUP | 1 | NC_037551.1 | 16,156,790 | 3.8 × 10−8 | MAPK10 |
| cBLUP | 2 | NC_037561.1 | 28,948,029 | 1.8 × 10−9 | ZNF84, ZNF26, ZNF605 |
| BayesR | 1 | NC_037551.1 | 28,948,029 | 2.8 × 10−8 | ZNF84, ZNF26, ZNF605 |
| GMATs | 1 | NC_037561.1 | 16,156,790 | 1.4 × 10−9 | MAPK10 |
| GMATs | 2 | NC_037561.1 | 28,948,029 | 1.5 × 10−8 | ZNF84, ZNF26, ZNF605 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Chen, H.; Cao, D.; Yang, X. Identification of MAPK10 as a Candidate Gene for High Milk Production in Water Buffaloes Through a Genome-Wide Association Study. Animals 2025, 15, 2567. https://doi.org/10.3390/ani15172567
Li W, Chen H, Cao D, Yang X. Identification of MAPK10 as a Candidate Gene for High Milk Production in Water Buffaloes Through a Genome-Wide Association Study. Animals. 2025; 15(17):2567. https://doi.org/10.3390/ani15172567
Chicago/Turabian StyleLi, Wangchang, Huan Chen, Duming Cao, and Xiaogan Yang. 2025. "Identification of MAPK10 as a Candidate Gene for High Milk Production in Water Buffaloes Through a Genome-Wide Association Study" Animals 15, no. 17: 2567. https://doi.org/10.3390/ani15172567
APA StyleLi, W., Chen, H., Cao, D., & Yang, X. (2025). Identification of MAPK10 as a Candidate Gene for High Milk Production in Water Buffaloes Through a Genome-Wide Association Study. Animals, 15(17), 2567. https://doi.org/10.3390/ani15172567





