Functional and Genetic Insights into the Role of the NR4A1 Gene in the Litter Size of the Shaanbei White Cashmere Goat
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis of Regulation and Conservation of the NR4A1 Gene
2.2. Tissue Sample Collection, Total RNA Extraction, and cDNA Synthesis
2.3. Reverse Transcriptase Quantitative Real-Time PCR (RT-qPCR)
2.4. Animal Selection, Ear Tissue Sample Collection, and Genomic DNA Extraction
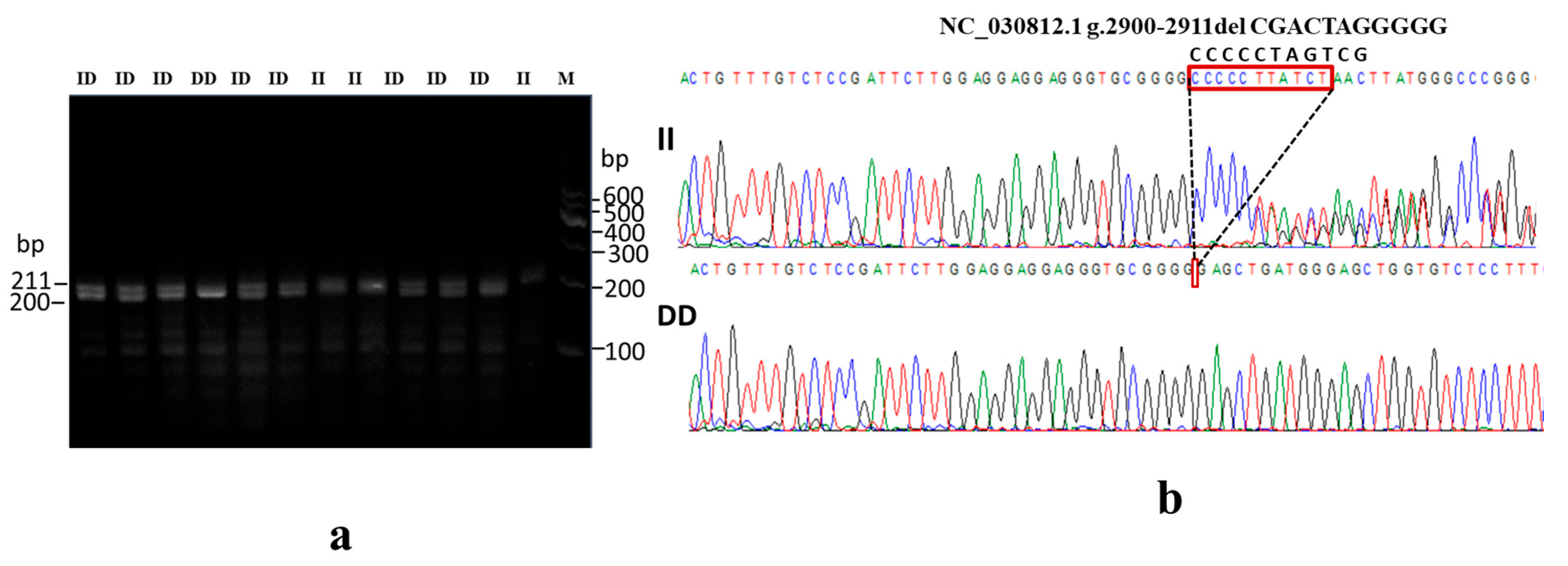
2.5. Insertion–Deletion Identification and Primer Design
2.6. PCR Amplification, Polymorphism Detection, and Genotyping
2.7. SNP Loci Identification for the NR4A1 Gene Through Resequencing
2.8. Population Genetic Parameter Estimation
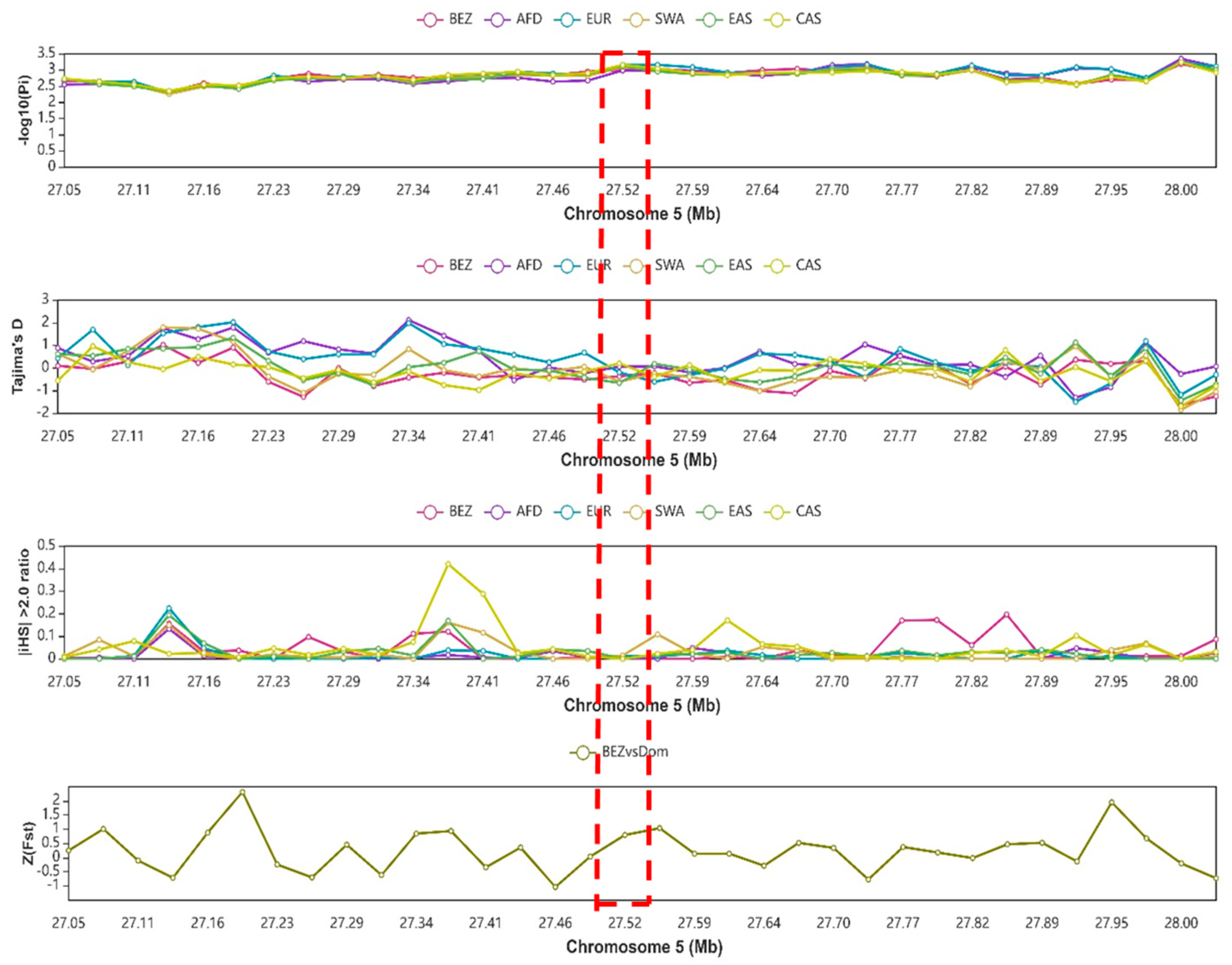
2.9. Analysis of Selection Signal at the Region of the NR4A1 Gene
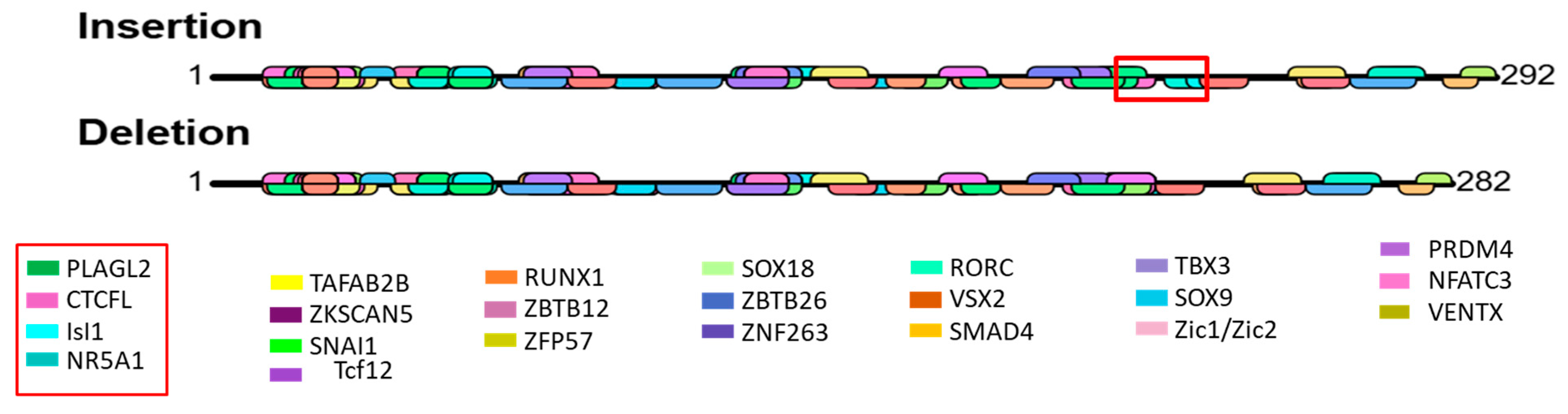
2.10. Identification of Transcription Factor Binding Site
2.11. Data Analysis
3. Results
3.1. NR4A1 Gene Interaction Network
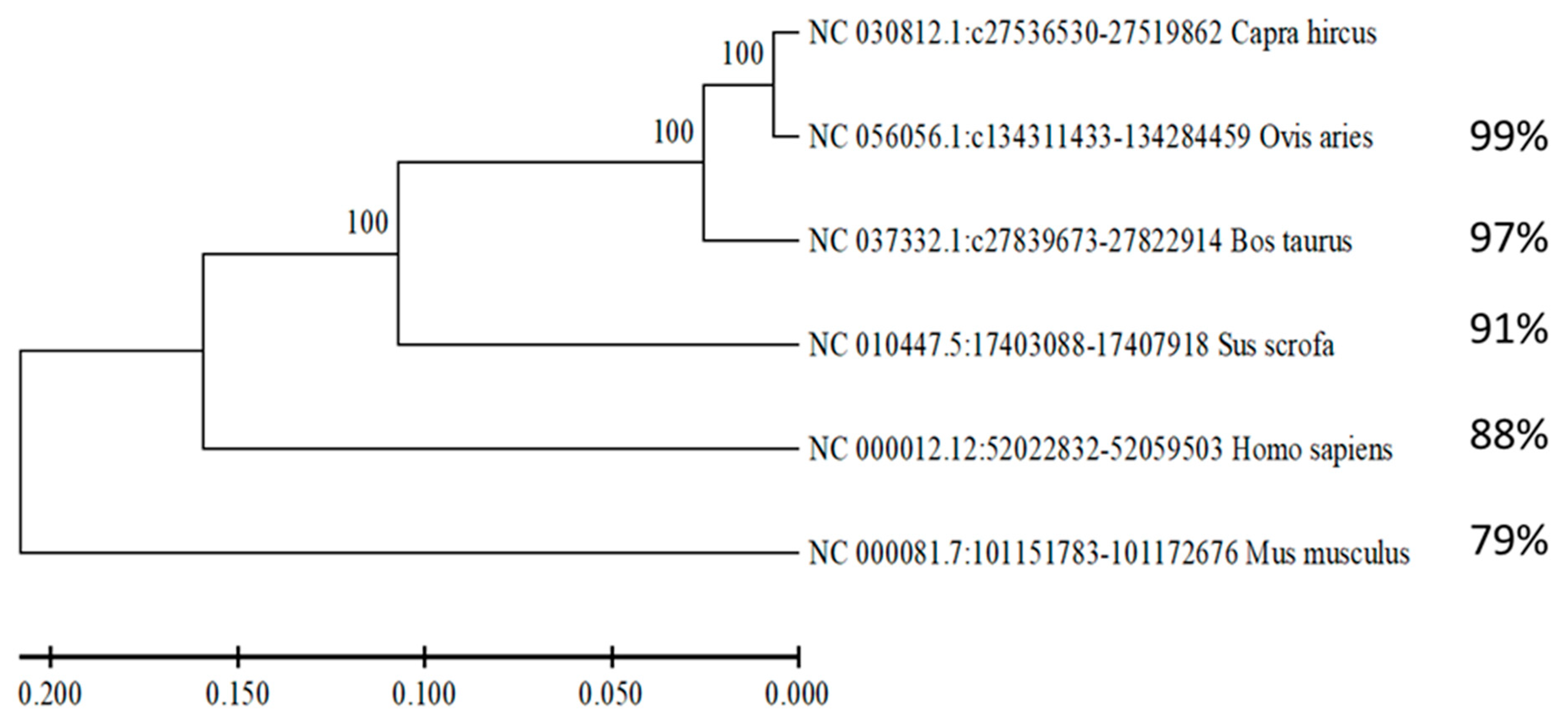
3.2. NR4A1 Gene Conservation Analysis Across Taxonomic Order
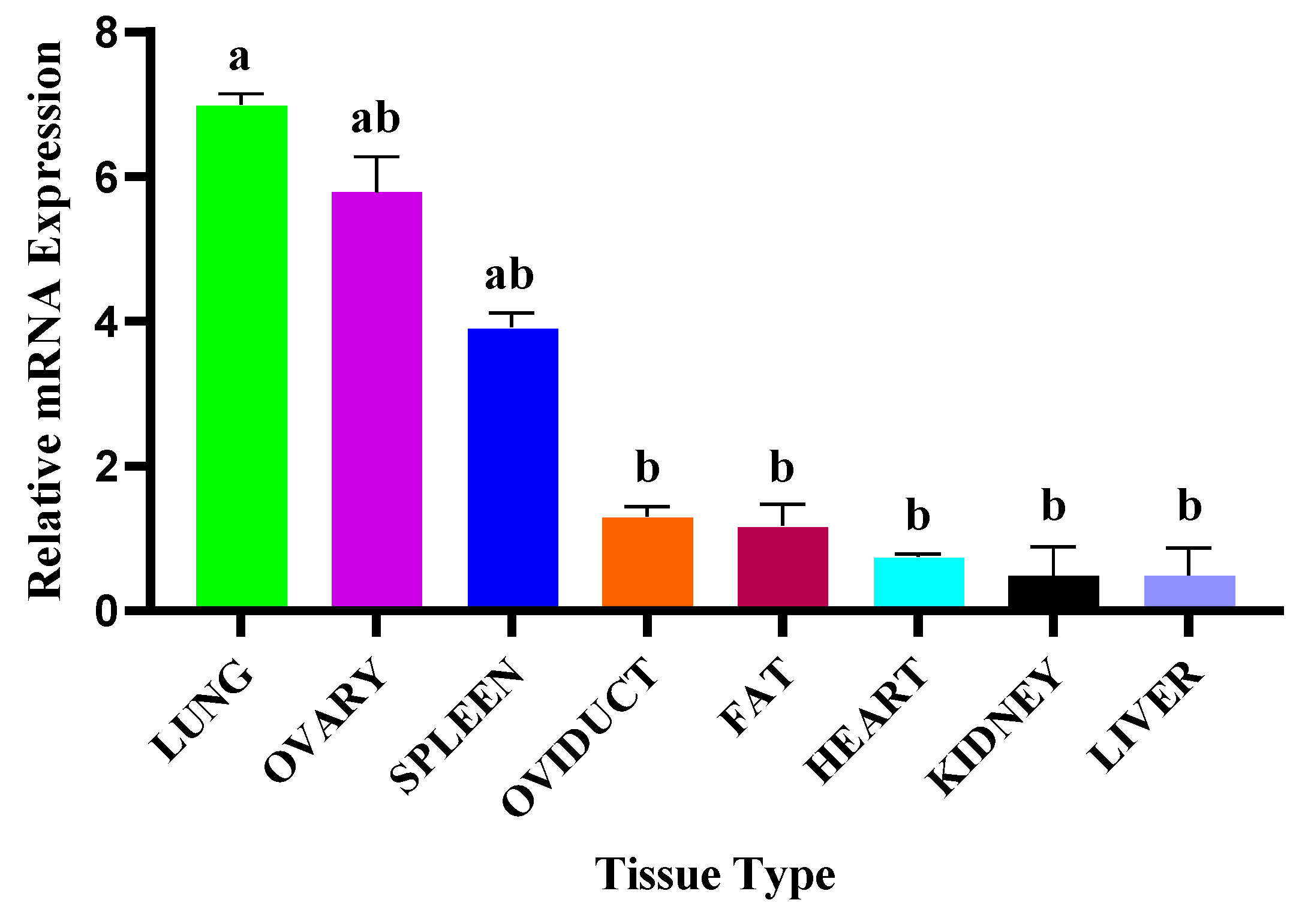
3.3. NR4A1 Gene mRNA Transcriptional Profiles
3.4. Genotyping and Population Genetic Parameter Analysis
3.5. Association of NR4A1 Gene InDel Variants with Goat Litter Size
3.6. Association of the NR4A1 Gene SNP Loci with Goat Litter Size
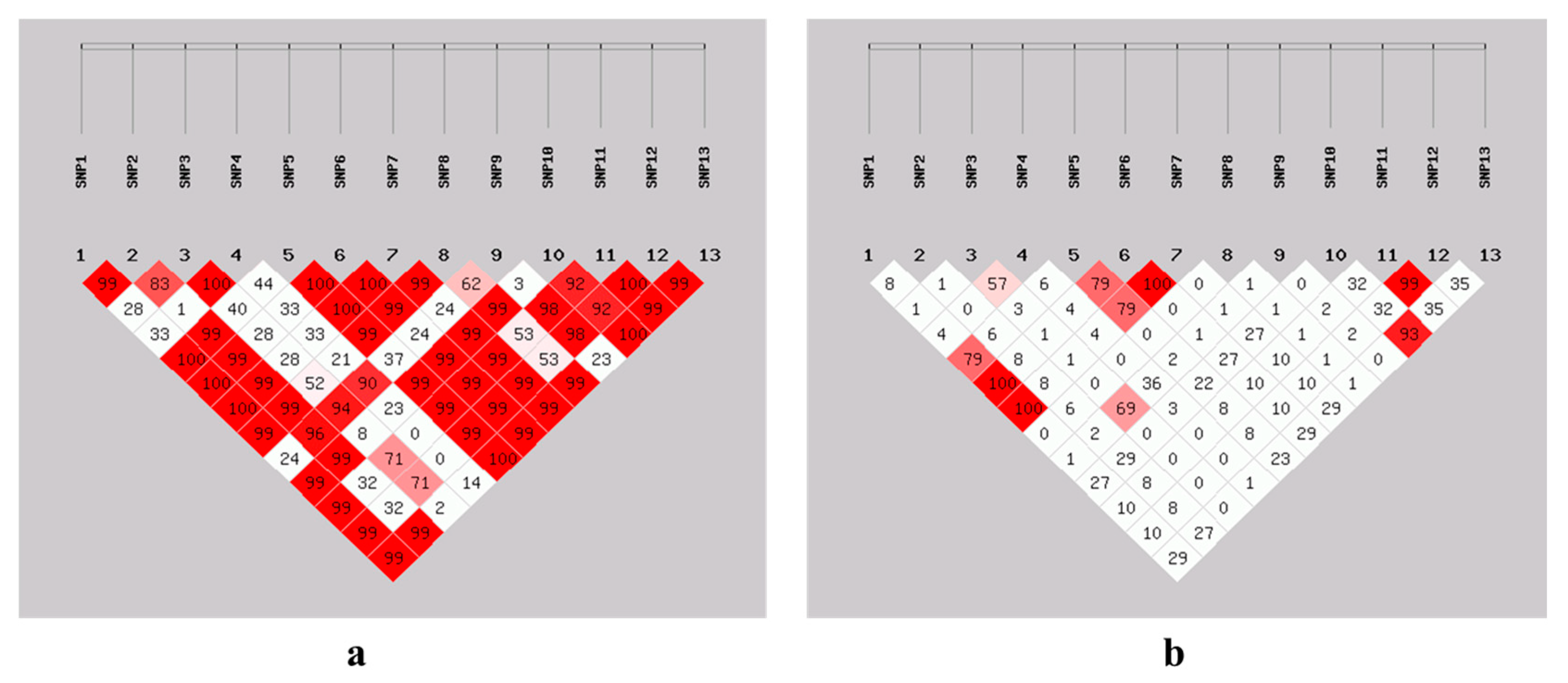
3.7. Linkage Disequilibrium Estimation Among SNPs
3.8. Analysis of Selection Signals on the NR4A1 Gene’s Genomic Region
3.9. Prediction of Transcription Factor Binding Site
4. Discussion
4.1. Function, Evolutionary Conservation, and Expression of Goat NR4A1
4.2. Effect of NR4A1 Gene Genetic Variability and Selection Signal
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SBWC | Shaanbei White Cashmere goat |
| NR4A1 | Nuclear receptor subfamily 4 group A member 1 |
| InDel | Insertion–deletion |
| SNP | Single nucleotide polymorphism |
| MAS | Marker-assisted selection |
| MSA | Multiple Sequence Alignment |
| GGVD | Goat Genomic Variation Database |
| MAF | Minor allele frequencies |
| Ne | Effective allele number |
| PIC | Polymorphism information content |
| He | Heterozygosity |
| Ho | Homozygosity |
| HWE | Hardy–Weinberg equilibrium |
| W&C | Weir and Cockerham |
| BEZ | Bezoar goat |
| SWA | Southwest Asian goat |
| AFD | African dairy goat |
| EUR | European goat |
| EAS | East Asian goat |
| iHS | Integrated haplotype score |
| XPEHH | Cross-population extended haplotype homozygosity |
| Fst | Z-transformed |
| CLR | Composite likelihood ratio |
| TFBS | Transcription factor binding site |
| LD | Linkage disequilibrium |
| AIC | Akakie Information Criteria |
References
- Zhong, T.; Hou, D.; Zhao, Q.; Zhan, S.; Wang, L.; Li, L.; Zhang, H.; Zhao, W.; Yang, S.; Niu, L. Comparative Whole-Genome Resequencing to Uncover Selection Signatures Linked to Litter Size in Hu Sheep and Five Other Breeds. BMC Genom. 2024, 25, 480. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Han, Y.; Chen, Y.; Liu, X.; Liang, H.; Wang, C.; Khan, M.Z. Potential Candidate Genes Associated with Litter Size in Goats: A Review. Animals 2025, 15, 82. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ye, J.; Liu, J.; Li, Z.; Deng, M.; Guo, Y.; Liu, G.; Sun, B.; Li, Y.; Liu, D. Whole-Genome Sequencing Identified Candidate Genes Associated with High and Low Litter Size in Chuanzhong Black Goats. Front. Vet. Sci. 2024, 11, 1420164. [Google Scholar] [CrossRef] [PubMed]
- El-Shorbagy, H.M.; Abdel-Aal, E.S.; Mohamed, S.A.; El-Ghor, A.A. Association of PRLR, IGF1, and LEP Genes Polymorphism with Milk Production and Litter Size in Egyptian Zaraibi Goat. Trop. Anim. Health Prod. 2022, 54, 321. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Xu, H.Z.; Zhao, Z.Q.; Narisu; Mao, J.W.; Guan, D.L.; Xie, C. Polymorphisms of Osteopontin Gene and Their Association with Placental Efficiency and Prolificacy in Goats. J. Appl. Anim. Res. 2015, 43, 272–278. [Google Scholar] [CrossRef]
- Zonaed Siddiki, A.M.A.M.; Miah, G.; Islam, M.S.; Kumkum, M.; Rumi, M.H.; Baten, A.; Hossain, M.A. Goat Genomic Resources: The Search for Genes Associated with Its Economic Traits. Int. J. Genomics 2020, 5940205. [Google Scholar] [CrossRef]
- Hui, Y.; Zhang, Y.; Wang, K.; Pan, C.; Chen, H.; Qu, L.; Song, X.; Lan, X. Goat DNMT3B: An Indel Mutation Detection, Association Analysis with Litter Size and mRNA Expression in Gonads. Theriogenology 2020, 147, 108–115. [Google Scholar] [CrossRef]
- Wang, K.; Liu, X.; Qi, T.; Hui, Y.; Yan, H.; Qu, L.; Lan, X.; Pan, C. Whole-Genome Sequencing to Identify Candidate Genes for Litter Size and to Uncover the Variant Function in Goats (Capra hircus). Genomics 2021, 113, 142–150. [Google Scholar] [CrossRef]
- Bai, Y.; Bo, D.; Bi, Y.; Areb, E.; Zhu, H.; Pan, C.; Lan, X. Analysis of Goat PPP6C mRNA Profile, Detection of Genetic Variations, and Their Associations with Litter Size. Anim. Reprod. Sci. 2024, 268, 107544. [Google Scholar] [CrossRef]
- Wijayanti, D.; Zhang, S.; Bai, Y.; Pan, C.; Chen, H.; Qu, L.; Guo, Z.; Lan, X. Investigation on mRNA Expression and Genetic Variation within Goat SMAD2 Gene and Its Association with Litter Size. Anim. Biotechnol. 2023, 34, 2111–2119. [Google Scholar] [CrossRef]
- Wijayanti, D.; Zhang, S.; Yang, Y.; Bai, Y.; Akhatayeva, Z.; Pan, C.; Zhu, H.; Qu, L.; Lan, X. Goat SMAD Family Member 1 (SMAD1): mRNA Expression, Genetic Variants, and Their Associations with Litter Size. Theriogenology 2022, 193, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Wijayanti, D.; Bai, Y.; Hanif, Q.; Chen, H.; Zhu, H.; Qu, L.; Guo, Z.; Lan, X. Goat CLSTN2 Gene: Tissue Expression Profile, Genetic Variation, and Its Associations with Litter Size. Anim. Biotechnol. 2023, 34, 2674–2683. [Google Scholar] [CrossRef] [PubMed]
- Buthelezi, N.L.; Mtileni, B.; Nephawe, K.A.; Idowu, P.A.; Modiba, M.C.; Mpedi, H.; Mpofu, T.J. The Impact of Parity, Litter Size and Birth Weight Variations within a Litter on Piglet Pre-Weaning Performance. Czech J. Anim. Sci. 2024, 69, 255–268. [Google Scholar] [CrossRef]
- Rutherford, K.; Baxter, E.; D’Eath, R.; Turner, S.; Arnott, G.; Roehe, R.; Ask, B.; SandØe, P.; Moustsen, V.; Thorup, F.; et al. The Welfare Implications of Large Litter Size in the Domestic Pig I: Biological Factors. Anim. Welf. 2013, 22, 199–218. [Google Scholar] [CrossRef]
- Ju, M.; Wang, X.; Li, X.; Zhang, M.; Shi, L.; Hu, P.; Zhang, B.; Han, X.; Wang, K.; Li, X.; et al. Effects of Litter Size and Parity on Farrowing Duration of Landrace × Yorkshire Sows. Animals 2021, 12, 94. [Google Scholar] [CrossRef]
- Boonkum, W.; Permthongchoochai, S.; Chankitisakul, V.; Duangjinda, M. Genetic Strategies for Enhancing Litter Size and Birth Weight Uniformity in Piglets. Front. Vet. Sci. 2025, 12, 1512701. [Google Scholar] [CrossRef]
- Parra-Vargas, M.; Ramon-Krauel, M.; Lerin, C.; Jimenez-Chillaron, J.C. Size Does Matter: Litter Size Strongly Determines Adult Metabolism in Rodents. Cell Metab. 2020, 32, 334–340. [Google Scholar] [CrossRef]
- Crean, D.; Murphy, E.P. Targeting NR4A Nuclear Receptors to Control Stromal Cell Inflammation, Metabolism, Angiogenesis, and Tumorigenesis. Front. Cell Dev. Biol. 2021, 9, 1–10. [Google Scholar] [CrossRef]
- Pearen, M.A.; Muscat, G.E.O. Orphan Nuclear Receptors and the Regulation of Nutrient Metabolism: Understanding Obesity. Physiology 2012, 27, 156–166. [Google Scholar] [CrossRef]
- Cruz-Nieves, R.P.; Ramírez-Rosales, G.E.; González-Ramírez, J.; Sánchez-Muñoz, F.; Ruiz-Hernández, A. Analysing the Gene Expression Profiles of the Orphan Nuclear Receptors NR4A1, NR4A2 and NR4A3 in Premalignant Lesions of the Cervix and Cervicitis. Eur. J. Obstet. Gynecol. Reprod. Biol. X 2024, 24, 100355. [Google Scholar] [CrossRef]
- Safe, S.; Jin, U.-H.; Morpurgo, B.; Abdayyeh, A.; Singh, M.; Tjalkens, R.B.; Steroid, J.; Mol, B.; Author, B. Nuclear Receptor 4A (NR4A) Family-Orphans No More. J. Steroid. Biochem. Mol. Biol. 2016, 157, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Zheng, J.; Cui, S.; Wang, J.; Wang, Y.; Li, Y.; Zhu, J.; Lin, Y. Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Promotes the Adipogenesis of Intramuscular Preadipocytes through PI3K/AKT Pathway in Goats. Animals 2024, 14, 2051. [Google Scholar] [CrossRef] [PubMed]
- Parreira, J.R.; Hernández-Castellano, L.E.; Argüello, A.; Capote, J.; Castro, N.; De Sousa Araújo, S.; De Almeida, A.M. Understanding Seasonal Weight Loss Tolerance in Dairy Goats: A Transcriptomics Approach. BMC Genom. 2020, 21, 629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-K.; Li, J.; Liu, J.; Guo, B.; Leung, A.; Zhang, G.; Zhang, B.-T. Icaritin Requires Phosphatidylinositol 3 Kinase (PI3K)/Akt Signaling to Counteract Skeletal Muscle Atrophy Following Mechanical Unloading. Sci. Rep. 2016, 6, 20300. [Google Scholar] [CrossRef]
- Kasch, J.; Kanzleiter, I.; Saussenthaler, S.; Schürmann, A.; Keijer, J.; van Schothorst, E.; Klaus, S.; Schumann, S. Insulin Sensitivity Linked Skeletal Muscle Nr4a1 DNA Methylation Is Programmed by the Maternal Diet and Modulated by Voluntary Exercise in Mice. J. Nutr. Biochem. 2018, 57, 86–92. [Google Scholar] [CrossRef]
- Chao, L.C.; Wroblewski, K.; Zhang, Z.; Pei, L.; Vergnes, L.; Ilkayeva, O.R.; Ding, S.Y.; Reue, K.; Watt, M.J.; Newgard, C.B.; et al. Insulin Resistance and Altered Systemic Glucose Metabolism in Mice Lacking Nur77. Diabetes 2009, 58, 2788–2796. [Google Scholar] [CrossRef]
- Ran, X.; Hu, F.; Mao, N.; Ruan, Y.; Yi, F.; Niu, X.; Huang, S.; Li, S.; You, L.; Zhang, F.; et al. Differences in Gene Expression and Variable Splicing Events of Ovaries between Large and Small Litter Size in Chinese Xiang Pigs. Porc. Health Manag. 2021, 7, 52. [Google Scholar] [CrossRef]
- Yu, H.; Li, X.; Zhao, J.; Wang, W.; Wei, Q.; Mao, D. NR4A1-Mediated Regulation of Lipid Droplets in Progesterone Synthesis in Goat Luteal Cells. Biol. Reprod. 2024, 111, 640–654. [Google Scholar] [CrossRef]
- Cui, L.; Bao, H.; Zhu, W.; Tang, Y.; Luo, Q.; Si, Y.; Fu, Q.; Jiang, Z. hUMSCs Transplantation Regulates AMPK/NR4A1 Signaling Axis to Inhibit Ovarian Fibrosis in POI Rats. Stem Cell Rev. Rep. 2023, 19, 1449–1465. [Google Scholar] [CrossRef]
- Kumchoo, T.; Mekchay, S. Association of NR4A1 and GNB2L1 Genes with Reproductive Traits in Commercial Pig Breeds. Genet. Mol. Res. 2015, 14, 16276–16284. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, J.; Pei, X.; Xia, Y.; Li, H.; Wang, W.; Mao, D. Dual Role of NR4A1 in Porcine Ovarian Granulosa Cell Differentiation and Granulosa-Lutein Cell Regression in Vitro. Theriogenology 2023, 198, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, B.; Yu, K.; Song, J.; Wang, L.; Zhang, X.; Li, Y. Nur77 Improves Ovarian Function in Reproductive Aging Mice by Activating Mitophagy and Inhibiting Apoptosis. Reprod. Biol. Endocrinol. 2024, 22, 86. [Google Scholar] [CrossRef]
- Kang, Z.; Bai, Y.; Lan, X.; Zhao, H. Goat AKAP12: Indel Mutation Detection, Association Analysis with Litter Size and Alternative Splicing Variant Expression. Front. Genet. 2021, 12, 648256. [Google Scholar] [CrossRef] [PubMed]
- Xin, D.; Bai, Y.; Bi, Y.; He, L.; Kang, Y.; Pan, C.; Zhu, H.; Chen, H.; Qu, L.; Lan, X. Insertion/Deletion Variants within the IGF2BP2 Gene Identified in Reported Genome-Wide Selective Sweep Analysis Reveal a Correlation with Goat Litter Size. J. Zhejiang Univ. Sci. B 2021, 22, 757–766. [Google Scholar] [CrossRef]
- Tang, Q.; Zhang, Y.; Yang, Y.; Hu, H.; Lan, X.; Pan, C. The KMT2A Gene: mRNA Differential Expression in the Ovary and a Novel 13-Nt Nucleotide Sequence Variant Associated with Litter Size in Cashmere Goats. Domest. Anim. Endocrinol. 2021, 74, 106538. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Lan, X.; Zhu, H.; Qu, L.; Pan, C. Polymorphism within the GATA Binding Protein 4 Gene Is Significantly Associated with Goat Litter Size. Anim. Biotechnol. 2023, 34, 4291–4300. [Google Scholar] [CrossRef]
- Cui, Y.; Yan, H.; Wang, K.; Xu, H.; Zhang, X.; Zhu, H.; Liu, J.; Qu, L.; Lan, X.; Pan, C. Insertion/Deletion within the KDM6A Gene Is Significantly Associated with Litter Size in Goat. Front. Genet. 2018, 9, 91. [Google Scholar] [CrossRef]
- Lin, G.; He, Q.; Cai, J.; Yang, X.; Wang, Y. Detection of Single Nucleotide Polymorphisms Based on Triple-Helix Molecular Switch Combined with Invader Assay. Microchem. J. 2024, 199, 109954. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, E.; Kang, Z.; Bi, Y.; Liu, H.; Xu, H.; Wang, Z.; Lei, C.; Chen, H.; Lan, X. CircRNA Profiling Reveals an Abundant circBDP1 That Regulates Bovine Fat Development by Sponging miR-181b/miR-204 Targeting Sirt1/TRARG1. J. Agric. Food Chem. 2022, 70, 14312–14328. [Google Scholar] [CrossRef]
- Bi, Y.; Zhang, S.; Li, J.; He, L.; Kang, Y.; Chen, H.; Lan, X.; Pan, C. The mRNA Expression Profile of the Goat Prion Protein Testis-Specific (PRNT) Gene and Its Associations with Litter Size. Theriogenology 2021, 165, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yan, H.; Xu, H.; Yang, Q.; Zhang, S.; Pan, C.; Chen, H.; Zhu, H.; Liu, J.; Qu, L.; et al. A Novel Indel within Goat Casein Alpha S1 Gene Is Significantly Associated with Litter Size. Gene 2018, 671, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Aljanabi, S.M.; Martinez, I. Universal and Rapid Salt-Extraction of High Quality Genomic DNA for PCR-Based Techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar]
- Yue, C.; Bai, W.L.; Zheng, Y.Y.; Hui, T.Y.; Sun, J.M.; Guo, D.; Guo, S.L.; Wang, Z.Y. Correlation Analysis of Candidate Gene SNP for High-Yield in Liaoning Cashmere Goats with Litter Size and Cashmere Performance. Anim. Biotechnol. 2021, 32, 43–50. [Google Scholar] [CrossRef]
- Fu, W.; Wang, R.; Yu, J.; Hu, D.; Cai, Y.; Shao, J.; Jiang, Y. GGVD: A Goat Genome Variation Database for Tracking the Dynamic Evolutionary Process of Selective Signatures and Ancient Introgressions. J. Genet. Genom. 2021, 48, 248–256. [Google Scholar] [CrossRef]
- Akhmet, N.; Zhu, L.; Song, J.; Akhatayeva, Z.; Zhang, Q.; Su, P.; Li, R.; Pan, C.; Lan, X. Exploring the Sheep MAST4 Gene Variants and Their Associations with Litter Size. Animals 2024, 14, 591. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Aydinli, M.; Liang, C.; Dandekar, T. Motif and Conserved Module Analysis in DNA (Promoters, Enhancers) and RNA (lncRNA, mRNA) Using AlModules. Sci. Rep. 2022, 12, 17588. [Google Scholar] [CrossRef]
- Shi, Y.Y.; He, L. SHEsis, a Powerful Software Platform for Analyses of Linkage Disequilibrium, Haplotype Construction, and Genetic Association at Polymorphism Loci. Cell Res. 2005, 15, 97–98, Erratum in Cell Res. 2006, 16, 851. [Google Scholar] [CrossRef]
- Myers, A.J.; Gibbs, J.R.; Webster, J.A.; Rohrer, K.; Zhao, A.; Marlowe, L.; Kaleem, M.; Leung, D.; Bryden, L.; Nath, P.; et al. A Survey of Genetic Human Cortical Gene Expression. Nat. Genet. 2007, 39, 1494–1499. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, R.; Kasimanickam, V.; Grende, K. Endometrial Expression of Various Genes (ISGs, PPARs, RXRs and MUC1) on Day 16 Post-Ovulation in Repeat Breeder Cows, with or without Subclinical Endometritis. Theriogenology 2020, 142, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Punetha, M.; Chouhan, V.S.; Sonwane, A.; Singh, G.; Bag, S.; Green, J.A.; Whitworth, K.; Sarkar, M. Early Growth Response Gene Mediates in VEGF and FGF Signaling as Dissected by CRISPR in Corpus Luteum of Water Buffalo. Sci. Rep. 2020, 10, 6849. [Google Scholar] [CrossRef] [PubMed]
- Kalous, J.; Aleshkina, D.; Anger, M. A Role of PI3K/Akt Signaling in Oocyte Maturation and Early Embryo Development. Cells 2023, 12, 1830. [Google Scholar] [CrossRef]
- Prince, L.R.; Prosseda, S.D.; Higgins, K.; Carlring, J.; Prestwich, E.C.; Ogryzko, N.V.; Rahman, A.; Basran, A.; Falciani, F.; Taylor, P.; et al. NR4A Orphan Nuclear Receptor Family Members, NR4A2 and NR4A3, Regulate Neutrophil Number and Survival. Blood 2017, 130, 1014–1025. [Google Scholar] [CrossRef]
- Przygrodzka, E.; Sokołowska, G.; Myszczynski, K.; Krawczynski, K.; Kaczmarek, M.M. Clustered microRNAs: The Molecular Mechanism Supporting the Maintenance of Luteal Function during Early Pregnancy. FASEB J. 2020, 34, 6582–6597. [Google Scholar] [CrossRef]
- Mucha, S.; Tortereau, F.; Doeschl-Wilson, A.; Rupp, R.; Conington, J. Animal Board Invited Review: Meta-Analysis of Genetic Parameters for Resilience and Efficiency Traits in Goats and Sheep. Animal 2022, 16, 100456. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Q.; Pan, C.-Y.; Lan, X.-Y. The Free Fatty Acid Receptor 2 (FFA2): Mechanisms of Action, Biased Signaling, and Clinical Prospects. Pharmacol. Ther. 2025, 272, 108878. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, R.; Pan, C.; Chen, H.; Qu, L.; Wu, L.; Guo, Z.; Zhu, H.; Lan, X. Genetic Variations and mRNA Expression of Goat DNAH1 and Their Associations with Litter Size. Cells 2022, 11, 1371. [Google Scholar] [CrossRef]
- Abuzahra, M.; Abu Eid, L.; Effendi, M.H.; Mustofa, I.; Lamid, M.; Rehman, S. Polymorphism Studies and Candidate Genes Associated with Litter Size Traits in Indonesian Goats. a Systematic Review. F1000Research 2023, 12, 61. [Google Scholar] [CrossRef]
- Saranya, S.K.; Thomas, M.; Aravindakshan, T.V.; Venkatachalapathy, R.T.; Sukumaran, J.; Kanakkaparambil, R. Diplotype-Based Assessment of GNRHR Gene Polymorphisms and Their Role in Determining Litter Size at Birth in the Malabari Goat Population of South India. Small Rumin. Res. 2024, 233, 107229. [Google Scholar] [CrossRef]
- Bai, Y.; Li, J.; Zhu, H.; Liu, J.; Dong, S.; Li, L.; Qu, L.; Chen, H.; Song, X.; Lan, X. Deletion Mutation within the Goat PPP3CA Gene Identified by GWAS Significantly Affects Litter Size. Reprod. Fertil. Dev. 2021, 33, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Jiang, E.; Kang, Z.; Wang, X.; Liu, Y.; Liu, X.; Wang, Z.; Li, X.; Lan, X. Detection of Insertions/Deletions (InDels) within the Goat Runx2 Gene and Their Association with Litter Size and Growth Traits. Anim. Biotechnol. 2021, 32, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Wang, J.X.; Liu, Q.Y.; Chu, M.X.; Di, R.; He, J.N.; Cao, G.L.; Fang, L.; Feng, T.; Li, N. Analysis on DNA Sequence of TSHB Gene and Its Association with Reproductive Seasonality in Goats. Mol. Biol. Rep. 2013, 40, 1893–1904. [Google Scholar] [CrossRef]
- Bi, Y.; Li, J.; Wang, X.; He, L.; Lan, K.; Qu, L.; Lan, X.; Song, X.; Pan, C. Two Novel Rare Strongly Linked Missense SNPs (P27R and A85G) Within the GDF9 Gene Were Significantly Associated With Litter Size in Shaanbei White Cashmere (SBWC) Goats. Front. Vet. Sci. 2020, 7, 406. [Google Scholar] [CrossRef]
- Yuan, C.; Guo, T.T.; Liu, J.B.; Yue, Y.J.; Yang, B.H. Conservation and Utilization of Indigenous Goats and Breeding of New Breeds in China. In Sustainable Goat Production in Adverse Environments; Springer: Cham, Switzerland, 2017; Volume 1, pp. 457–472. [Google Scholar] [CrossRef]
- Georgakopoulos-Soares, I.; Deng, C.; Agarwal, V.; Chan, C.S.Y.; Zhao, J.; Inoue, F.; Ahituv, N. Transcription Factor Binding Site Orientation and Order Are Major Drivers of Gene Regulatory Activity. Nat. Commun. 2023, 14, 2333. [Google Scholar] [CrossRef]
- Muley, V.Y. Prediction and Analysis of Transcription Factor Binding Sites: Practical Examples and Case Studies Using R Programming. In Reverse Engineering of Regulatory Networks; Springer: Berlin/Heidelberg, Germany, 2024; pp. 199–225. [Google Scholar]


| Primers Name | Primer Sequence (5′->3′) | Sizes (bp) | Tm (°C) | Region |
|---|---|---|---|---|
| P1 | F: TCTGCCTTTGGGACAGCAAG | 235 | 60.54 | upstream |
| R: GCCTTGAGCCCTATTCACCC | 60.47 | |||
| P2 | F: GGGTGAGATGTGGAGAGCAG | 199 | 59.82 | upstream |
| R: TAGGGGGATTTGCTCTGTGG | 59.08 | |||
| P3 | F: AGCCCCATCTCTGGACATACT | 200 | 60.06 | intron |
| R: AATGGGAGCGTTGTCTGGG | 60 | |||
| P4 | F: CACCAGGAAGAGGTCCCAAC | 368 | 59.96 | intron |
| R: GGGTCAACAGGAGAGGAGGA | 60.25 | |||
| P5 | F: CCTCGCCACACCTTGCATTT | 233 | 61.53 | upstream |
| R: CACATTCCTCTCCCCACCTC | 59.46 | |||
| P6 | F: AGGCTGTGTGTTTGTCCCAG | 197 | 60.47 | intron |
| R: GGGTTCGGCCATATCCTCAG | 59.96 | |||
| P7 | F: GGTTGTAAGAAGGCGCAGAG | 486 | 58.92 | upstream |
| R: GCCCTTCCAACCAATAGCAC | 59.18 | |||
| P8 | F: ACACCTTTACCCGAGAGAGC | 223 | 59.1 | intron |
| R: ACTCTCTGGACCCTGAACAC | 58.66 | |||
| NR4A1-qPCR | F: ACAGACAGCCTGAAAGGACG | 93 | 59.97 | exon 1–2 |
| R: GACCAGGGAGGTGAGTAGGT | 59.96 | |||
| GAPDH-qPCR | F: AAAGTGGACATCGTTGCCAT | 116 | 58.09 | exon 2 |
| R: CCGTTCTCTGCCTTGACTGT | 59.97 |
| Genotype Frequencies | Allele Frequencies | Genetic Variation Metrics | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| II | ID | DD | I | D | Ho | He | Ne | PIC | HWE | W&C |
| 0.216 | 0.561 | 0.224 | 0.496 | 0.504 | 0.500 | 0.499 | 1.999 | 0.374 | p < 0.05 | −0.1211 |
| n = 245 | n =637 | n = 254 | ||||||||
| N = 1136 | ||||||||||
| SNPs | Genotypes | Genomic Location | Variant ID | Allele Frequencies | Ho | He | Ne | PIC | HWE |
|---|---|---|---|---|---|---|---|---|---|
| SNP1 | TT (115) | NC 030812.1: g. 27520435T>C | Novel | 0.96 (T) | 0.96 | 0.04 | 1.04 | 0.03 | p > 0.05 |
| TC (5) | 0.04 (C) | ||||||||
| SNP2 | GG (76) | NC 030812.1: g. 27521198G>C | rs668853994 | 0.80 (G) | 0.68 | 0.32 | 1.48 | 0.27 | p > 0.05 |
| GC (39) | 0.20 (C) | ||||||||
| CC (5) | |||||||||
| SNP3 | CC (101) | NC 030812.1: g. 27521415C>G | rs657635772 | 0.92 (C) | 0.85 | 0.15 | 1.18 | 0.14 | p > 0.05 |
| CG (18) | |||||||||
| GG (1) | 0.08 (G) | ||||||||
| SNP4 | CC (108) | NC 030812.1: g. 27522352C>T | Novel | 0.95 (C) | 0.91 | 0.09 | 1.10 | 0.09 | p > 0.05 |
| CT (12) | 0.05 (T) | ||||||||
| SNP5 | CC (116) | NC 030812.1: g. 27526206C>T | Novel | 0.98 (C) | 0.97 | 0.03 | 1.03 | 0.03 | p > 0.05 |
| CT (4) | 0.02 (T) | ||||||||
| SNP6 | AA (115) | NC 030812.1: g. 27527050A>G | rs683100351 | 0.98 (A) | 0.96 | 0.04 | 1.04 | 0.04 | p > 0.05 |
| AG (5) | 0.02 (G) | ||||||||
| SNP7 | CC (115) | NC 030812.1: g. 27527625C>T | rs677241829 | 0.98 (C) | 0.96 | 0.04 | 1.04 | 0.04 | p > 0.05 |
| CT (5) | 0.02 (T) | ||||||||
| SNP8 | CC (81) | NC 030812.1: g. 27528181C>A | Novel | 0.79 (C) | 0.68 | 0.32 | 1.47 | 0.27 | p < 0.05 |
| CA (27) | 0.21 (A) | ||||||||
| AA (10) | |||||||||
| SNP9 | GG (96) | NC 030812.1: g. 27531784G>C | rs661365536 | 0.90 (G) | 0.81 | 0.19 | 1.23 | 0.17 | p > 0.05 |
| GC (23) | 0.10 (C) | ||||||||
| CC (1) | |||||||||
| SNP10 | GG (103) | NC 030812.1: g. 27532725G>A | Novel | 0.93 (G) | 0.87 | 0.13 | 1.15 | 0.12 | p > 0.05 |
| GA (17) | 0.07 (A) | ||||||||
| SNP11 | CC (3) | NC 030812.1: g. 27533799C>G | rs669578807 | 0.17 (C) | 0.72 | 0.27 | 1.38 | 0.24 | p > 0.05 |
| CG (34) | 0.83 (G) | ||||||||
| GG (83) | |||||||||
| SNP12 | AA (3) | NC 030812.1: g. 27533803A>G | rs658171703 | 0.17 (A) | 0.72 | 0.27 | 1.38 | 0.24 | p > 0.05 |
| AG (34) | |||||||||
| GG (83) | 0.83 (G) | ||||||||
| SNP13 | GG (104) | NC 030812.1: g. 27534820G>A | Novel | 0.93 (G) | 0.88 | 0.12 | 1.14 | 0.12 | p > 0.05 |
| GA (16) | 0.07 (A) |
| Ancestral Goat Group | Number of Samples | Reference Allele Frequency | Alternate Allele Frequency |
|---|---|---|---|
| Bezoar | 24 | 0.771 | 0.229 |
| Africa | 54 | 0.713 | 0.287 |
| Africa Dairy | 15 | 0.767 | 0.233 |
| Europe | 28 | 0.786 | 0.214 |
| Southwest Asia | 34 | 0.735 | 0.265 |
| East Asia | 57 | 0.868 | 0.132 |
| Coefficient Table | Goodness of Fit | |||||||
|---|---|---|---|---|---|---|---|---|
| Predictor | Estimate (β) | SE | z-Value | p-Value | Odd Ratio | χ2 Test | AIC | p-Value |
| Intercept | −2.5735 | 0.2445 | −10.524 | <2 × 10−16 | 0.076 | 40.484 | 751.14 | 1.62 × 10−9 |
| Genotype ID | +0.1320 | 0.2848 | +0.463 | 0.643 | 1.141 | |||
| Genotype II | +1.3801 | 0.2875 | +4.800 | 1.58 × 10−6 | 3.975 | |||
| SNP | Genotypes | N | Mean ± SEM | p-Value |
|---|---|---|---|---|
| SNP1 | TT | 115 | 1.65 a ± 0.07 | 8.24 × 10−17 |
| TC | 5 | 1.00 b ± 0.00 | ||
| SNP2 | GG | 76 | 1.75 a ± 0.08 | 0.032 |
| GC | 39 | 1.38 c ± 0.11 | ||
| CC | 5 | 1.60 b ± 0.24 | ||
| SNP3 | CC | 101 | 1.70 a ± 0.07 | 0.003 |
| CG | 18 | 1.22 b ± 0.13 | ||
| GG | 1 | 1.00 | ||
| SNP4 | CC | 108 | 1.69 a ± 0.07 | 5 × 10−6 |
| CT | 12 | 1.08 b ± 0.08 | ||
| SNP5 | CC | 116 | 1.65 a ± 0.07 | 9.20 × 10−17 |
| CT | 4 | 1.00 b ± 0.00 | ||
| SNP6 | AA | 115 | 1.65 a ± 0.07 | 8.24 × 10−17 |
| AG | 5 | 1.00 b ± 0.00 | ||
| SNP7 | CC | 115 | 1.65 a ± 0.07 | 8.24 × 10−17 |
| CT | 5 | 1.00 b ± 0.00 | ||
| SNP8 | CC | 81 | 1.52 c ± 0.08 | 0.034 |
| CA | 27 | 1.70 b ± 0.12 | ||
| AA | 10 | 2.10 a ± 0.23 | ||
| SNP9 | GG | 96 | 1.70 a ± 0.07 | 0.033 |
| GC | 23 | 1.35 b ± 0.13 | ||
| CC | 1 | 1.00 | ||
| SNP10 | GG | 103 | 1.69 a ± 0.07 | 0.001 |
| GA | 17 | 1.24 b ± 0.11 | ||
| SNP11 | CC | 3 | 1.00 c ± 0.00 | 1.95 × 10−11 |
| CG | 34 | 1.56 b ± 0.10 | ||
| GG | 83 | 1.67 a ± 0.08 | ||
| SNP12 | AA | 3 | 1.00 c ± 0.00 | 1.95 × 10−11 |
| AG | 34 | 1.56 b ± 0.10 | ||
| GG | 83 | 1.67 a ± 0.08 | ||
| SNP13 | GG | 104 | 1.68 a ± 0.07 | 0.003 |
| GA | 16 | 1.25 b ± 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Areb, E.; Bi, Y.; Bai, Y.; Zhu, Q.; Ma, L.; Pan, C.; Chen, X.; Lan, X. Functional and Genetic Insights into the Role of the NR4A1 Gene in the Litter Size of the Shaanbei White Cashmere Goat. Animals 2025, 15, 1729. https://doi.org/10.3390/ani15121729
Areb E, Bi Y, Bai Y, Zhu Q, Ma L, Pan C, Chen X, Lan X. Functional and Genetic Insights into the Role of the NR4A1 Gene in the Litter Size of the Shaanbei White Cashmere Goat. Animals. 2025; 15(12):1729. https://doi.org/10.3390/ani15121729
Chicago/Turabian StyleAreb, Ebadu, Yutian Bi, Yangyang Bai, Qihui Zhu, Lingyuan Ma, Chuanying Pan, Xiaolei Chen, and Xianyong Lan. 2025. "Functional and Genetic Insights into the Role of the NR4A1 Gene in the Litter Size of the Shaanbei White Cashmere Goat" Animals 15, no. 12: 1729. https://doi.org/10.3390/ani15121729
APA StyleAreb, E., Bi, Y., Bai, Y., Zhu, Q., Ma, L., Pan, C., Chen, X., & Lan, X. (2025). Functional and Genetic Insights into the Role of the NR4A1 Gene in the Litter Size of the Shaanbei White Cashmere Goat. Animals, 15(12), 1729. https://doi.org/10.3390/ani15121729






