Transcriptomic Association Analysis of the Metabolic Mechanism of Sulfamethoxazole in Channel Catfish (Ictalurus punctatus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Maintenance and Treatment of Channel Catfish
2.2. RNA Isolation, RNA-seq Library Construction and Sequencing
2.3. Differential Expression of Genes Analysis (DEGs)
2.4. GO Functional Enrichment Analysis for DEGs
2.5. Pathway Analysis of DEGs Enrichment
2.6. Quantitative Real-Time Reverse
3. Results
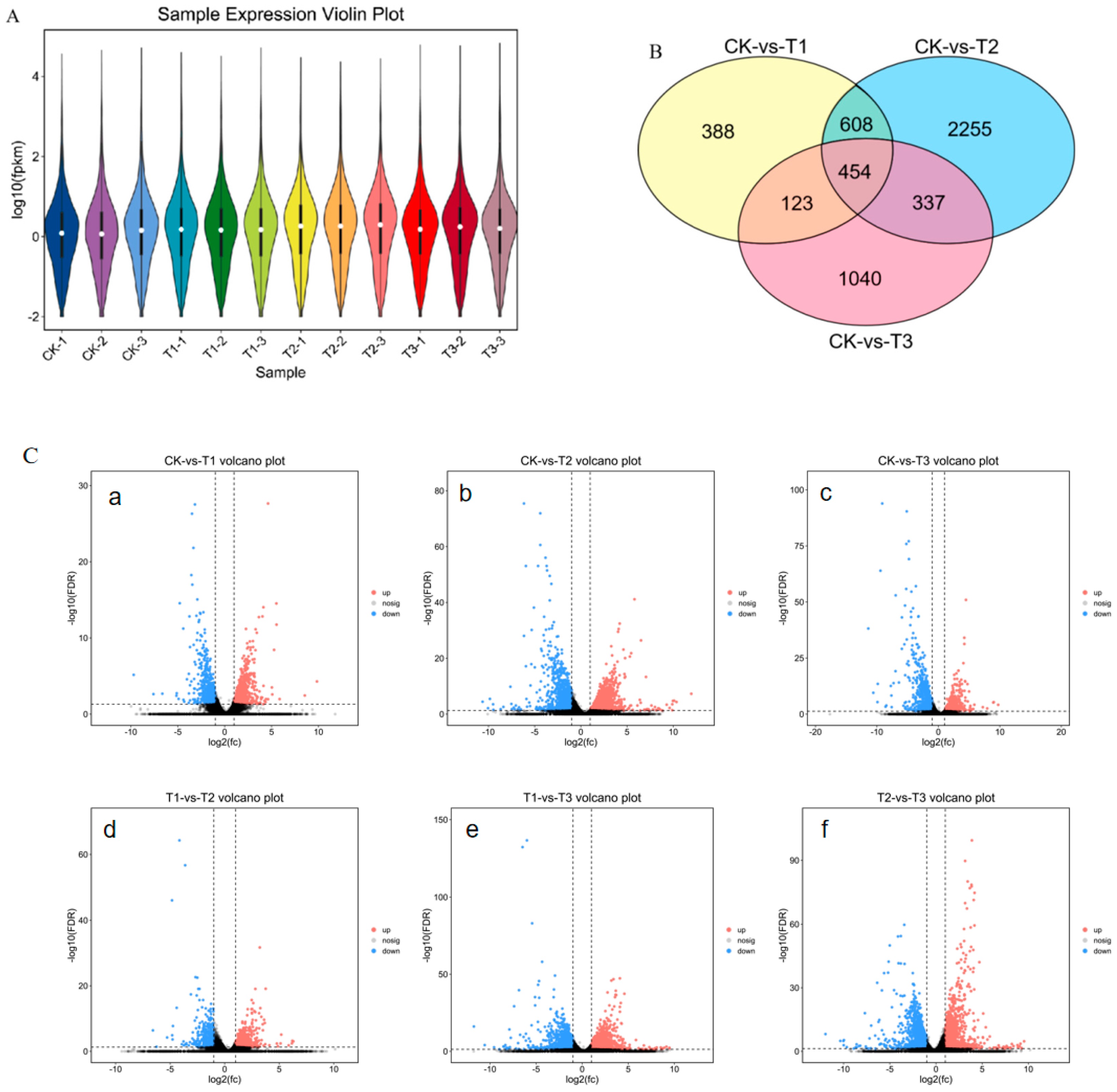
3.1. Illumina Sequencing and Quality Assembly
3.2. Comparative Analysis with the Reference Genome
3.3. Analysis of DEGs
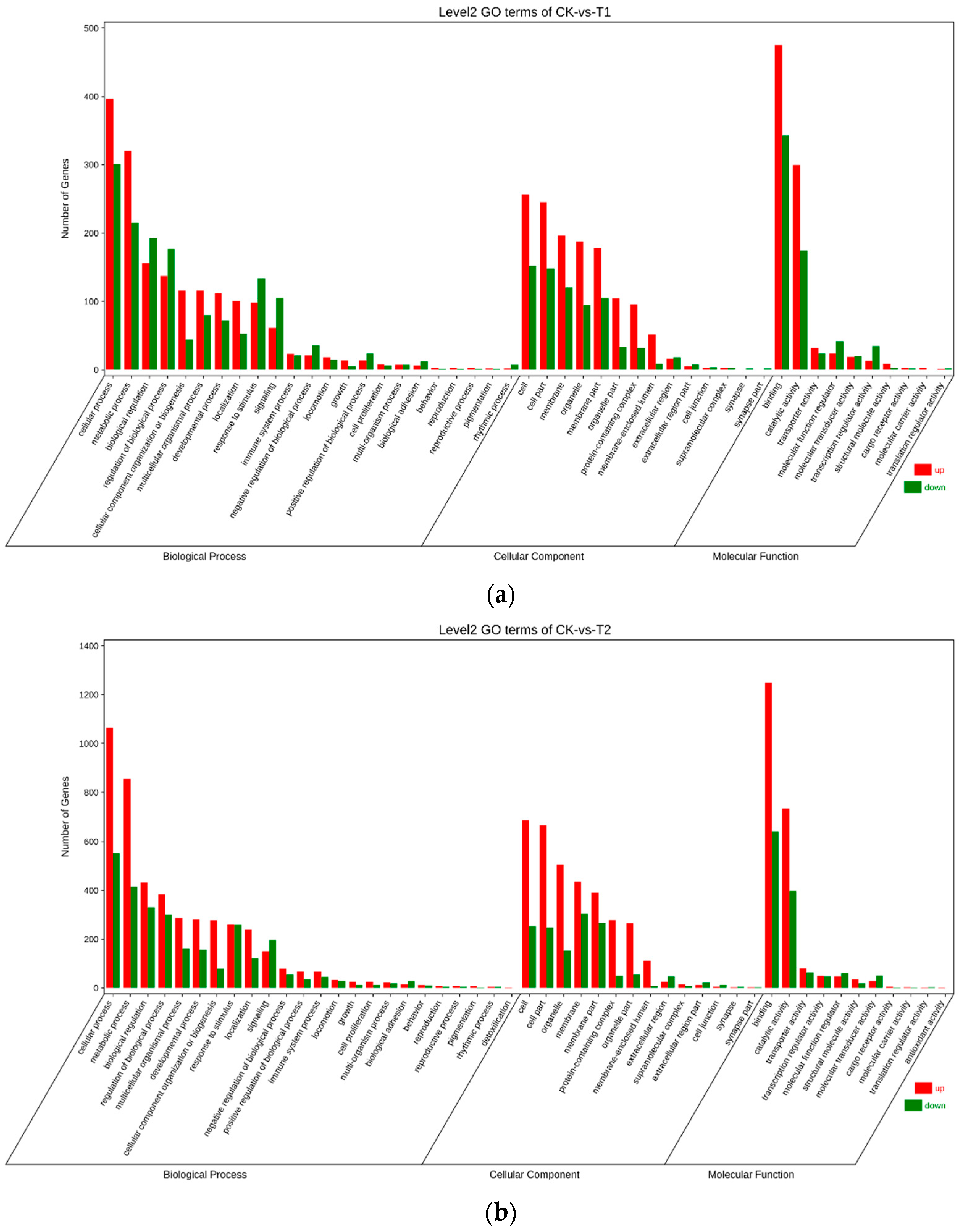
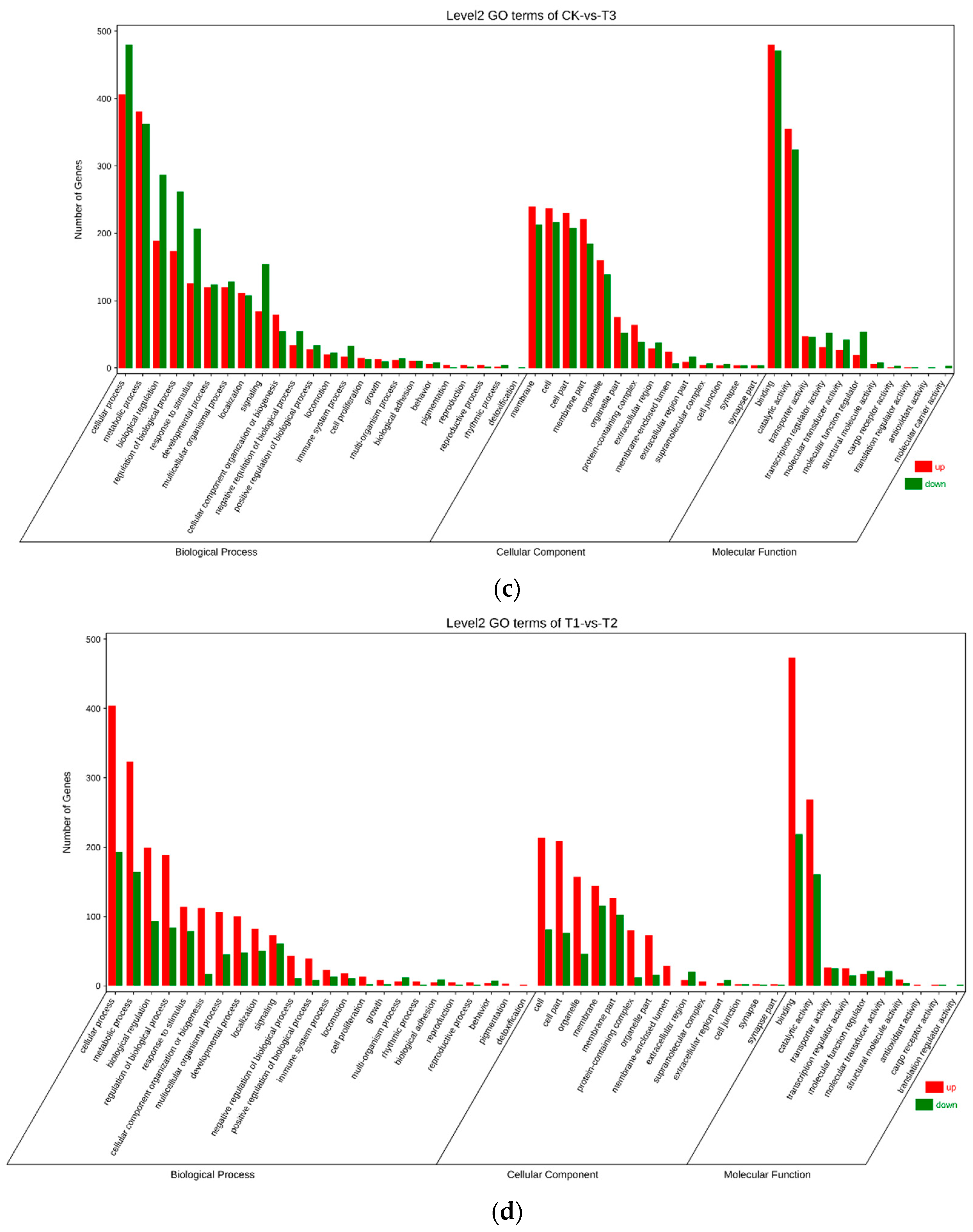
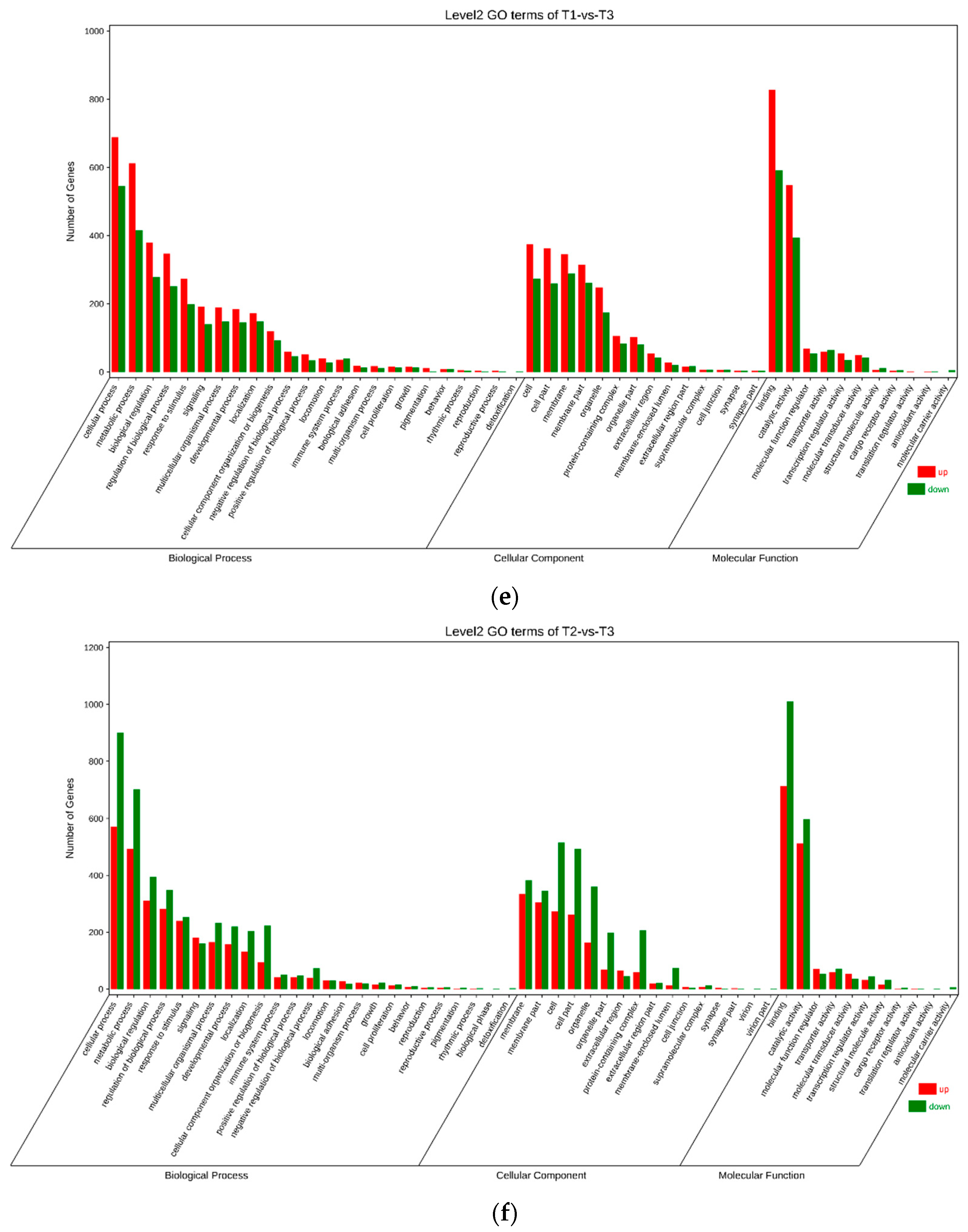
3.4. Gene Ontology (GO) Annotation of Differentially Expressed Genes (DEGs)
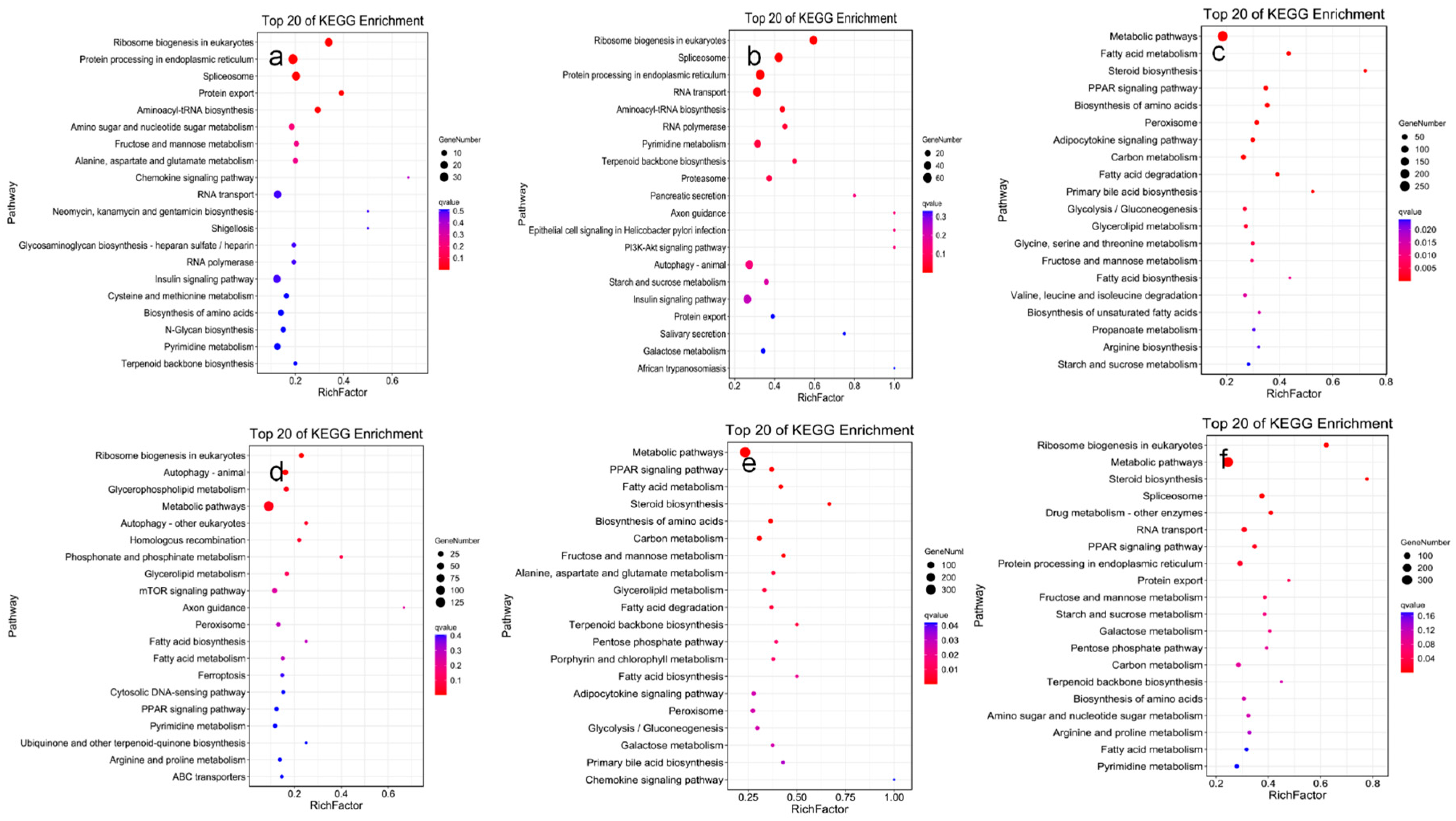
3.5. KEGG Pathway Analysis of DEGs
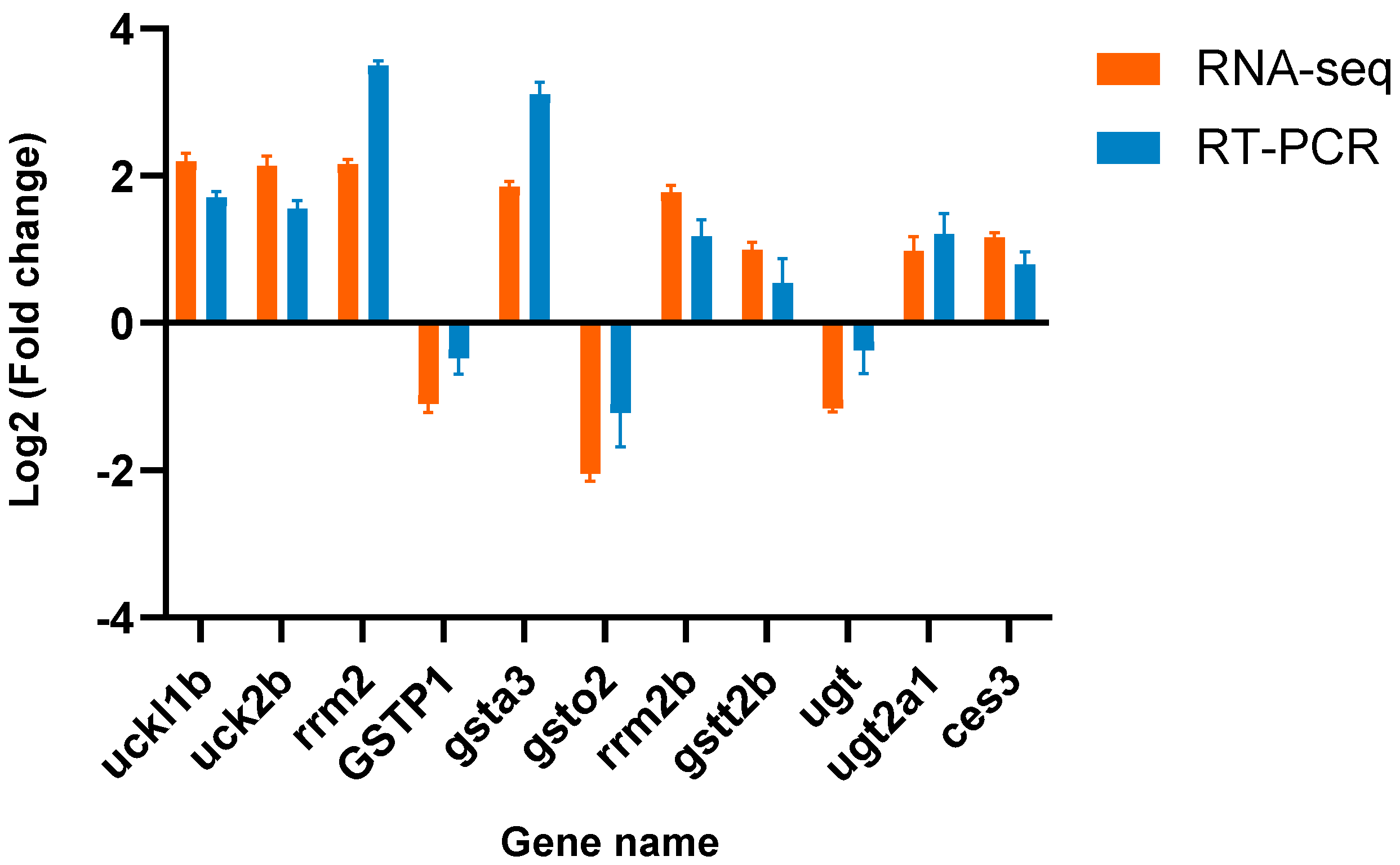
3.6. Verification of the Differential Expression of DEGs
4. Discussion
4.1. Glucose and Lipid Metabolism
4.2. Antioxidant
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhong, L.; Song, C.; Chen, X.; Deng, W.; Xiao, Y.; Wang, M.; Qin, Q.; Luan, S.; Kong, J.; Bian, W. Channel catfish in China: Historical aspects, current status, and problems. Aquaculture 2016, 465, 367–373. [Google Scholar] [CrossRef]
- Hawke, J.P.; Khoo, L.H. 14 Infectious diseases. In Developments in Aquaculture and Fisheries Science; Elsevier: Amsterdam, The Netherlands, 2004; Volume 34. [Google Scholar]
- A’yunin, Q.; Andayani, S.; Yuwanita, R. Histopathological Analysis of Pangasius sp. Infected by Edwardsiella tarda Causes Edwardsiellosis Disease; IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2020; Volume 441. [Google Scholar]
- Shoemaker, C.A.; Martins, M.L.; Xu, D.H.; Klesius, P.H. Effect of Ichthyophthirius multifiliis parasitism on the survival, hematology and bacterial load in channel catfish previously exposed to Edwardsiella ictaluri. Parasitol. Res. 2012, 111, 2223–2228. [Google Scholar] [CrossRef]
- Xu, D.H.; Pridgeon, J.W.; Klesius, P.H.; Shoemaker, C.A. Parasitism by protozoan Ichthyophthirius multifiliis enhanced invasion of Aeromonas hydrophila in tissues of channel catfish. Vet. Parasitol. 2012, 184, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Shengshan, Z.; Liang, P. The Harm and Problem Analysis of Drug Abuse in Aquaculture. Chin. J. Fish. 2015, 28, 47–54. [Google Scholar]
- Roberts, M. Drug Resistance in Environments Associated with Aquaculture; University of Washington Water Center: Seattle, WA, USA, 2007. [Google Scholar]
- Masters, P.A.; O’Bryan, T.A.; Zurlo, J.; Miller, D.Q.; Joshi, N. Trimethoprim-Sulfamethoxazole Revisited. Arch. Intern. Med. 2003, 163, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Wang, Y.; Zou, X.; Hu, K.; Sun, B.; Fang, W.; Fu, G.; Yang, X. Pharmacokinetics of sulfamethoxazole and trimethoprim in Pacific white shrimp, Litopenaeus vannamei, after oral administration of single-dose and multiple-dose. Environ. Toxicol. Pharmacol. 2017, 52, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Prasannamedha, G.; Kumar, P.S. A review on contamination and removal of sulfamethoxazole from aqueous solution using cleaner techniques: Present and future perspective. J. Clean. Prod. 2020, 250, 119553. [Google Scholar] [CrossRef]
- Yan, Z.; Lu, G.; Ye, Q.; Liu, J. Long-term effects of antibiotics, norfloxacin, and sulfamethoxazole, in a partial life-cycle study with zebrafish (Danio rerio): Effects on growth, development, and reproduction. Environ. Sci. Pollut. Res. 2016, 23, 18222–18228. [Google Scholar] [CrossRef] [PubMed]
- Chalermchaikit, T.; Dilokkiat, S.; Srisagha, S.; Lertworapreecha, N. Residues of Oxytetracycline, Sulfamethoxazole, Sulfamethoxazole+Trimethoprim and Enrofloxacin after withdrawal time in white shrimp (Penaeus vannamei) detected by Microbial Inhibition Disc Assay versus Screening Test Kit “SAM-Test”. Songklanakarin J. Sci. Technol. 2005, 27 (Suppl. S1), 283–290. [Google Scholar]
- Dutta, H.M.; Adhikari, S.; Singh, N.K.; Roy, P.K.; Munshi, J.S.D. Histopathological changes induced by malathion in the liver of a freshwater catfish, Heteropneustes fossilis (Bloch). Bull. Environ. Contam. Toxicol. 1993, 51, 895. [Google Scholar] [CrossRef]
- Martinsen, B.; Horsberg, T.E.; Varma, K.J.; Sams, R. Single dose pharmacokinetic study of florfenicol in Atlantic salmon (Salmo salar) in seawater at 11 °C. Aquaculture 1993, 112, 1–11. [Google Scholar] [CrossRef]
- Terzi, E.; Corum, O.; Bilen, S.; Kenanoglu, O.N.; Atik, O.; Uney, K. Pharmacokinetics of danofloxacin in rainbow trout after different routes of administration. Aquaculture 2020, 520, 734984. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. FASTP: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Jiang, H.; Cao, D.; Liu, L.; Hu, S.; Wang, Q. Comparative transcriptome analysis of the accessory sex gland and testis from the Chinese mitten crab (Eriocheir sinensis). PLoS ONE 2013, 8, e53915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Henkel, C.V.; Dirks, R.P.; de Wijze, D.L.; Minegishi, Y.; Aoyama, J.; Jansen, H.J.; Turner, B.; Knudsen, B.; Bundgaard, M.; Hvam, K.L.; et al. First draft genome sequence of the Japanese eel. Anguilla Jpn. Gene 2012, 511, 195–201. [Google Scholar]
- Ibaraki, H.; Wu, X.; Uji, S.; Yokoi, H.; Sakai, Y.; Suzuki, T. Transcriptome analysis of vertebral bone in the flounder, Paralichthys olivaceus (Teleostei, Pleuronectiformes), using Illumina sequencing. Mar. Genom. 2015, 24, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.M.; Yang, X.L.; Xie, X.Y.; Yang, Z.Y.; Hu, K.; Wu, Y.J.; Jiang, Y.Y.; Liu, T.F.; Fang, W.H.; Huang, X.Y. Comparative transcriptome analysis of Anguilla japonicaAnguilla japonica livers following exposure to methylene blue. Aquac. Res. 2018, 49, 1232–1241. [Google Scholar] [CrossRef]
- Zhu, F.J.; Hu, K.; Yang, Z.Y.; Yang, X.L. Comparative transcriptome analysis of the hepatopancreas of Eriocheir sinensis following oral gavage with enrofloxacin. Can. J. Fish. Aquat. Sci. 2017, 74, 435–444. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, Y.; Mo, J.; Sun, H.; Li, Q. Sulfamethoxazole-Altered Transcriptomein Green Alga Raphidocelis subcapitata Suggests Inhibition of Translation and DNA Damage Repair. Front. Microbiol. 2021, 12, 541451. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Zheng, M.; Wang, G.; Zhao, H. Exposed to Sulfamethoxazole induced hepatic lipid metabolism disorder and intestinal microbiota changes on zebrafish (Danio rerio). Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2022, 253, 109245. [Google Scholar] [CrossRef]
- Jedlitschky, G.; Cassidy, A.J.; Sales, M.; Pratt, N.; Burchell, B. Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem. J. 1999, 340, 837–843. [Google Scholar] [CrossRef]
- Sneitz, N.; Zhang, X.; Laajanen, K.; Yee, K.K.; Dalton, P.; Ding, X.; Finel, M. Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenetics Genom. 2009, 19, 923–934. [Google Scholar] [CrossRef]
- Sten, T.; Bichlmaier, I.; Kuuranne, T.; Leinonen, A.; Yli-Kauhaluoma, J.; Finel, M. UDP-Glucuronosyltransferases (UGTs) 2B7 and UGT2B17 Display Converse Specificity in Testosterone and Epitestosterone Glucuronidation, whereas UGT2A1 Conjugates Both Androgens Similarly. Drug Metab. Dispos. 2009, 37, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Perreault, M.; Gauthier-Landry, L.; Trottier, J.; Verreault, M.; Caron, P.; Finel, M.; Barbier, O. The Human UDP-glucuronosyltransferase UGT2A1 and UGT2A2 enzymes are highly active in bile acid glucuronidation. Drug Metab. Dispos. 2013, 41, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Leaver, M.J.; Wright, J.; Hodgson, P.; Boukouvala, E.; George, S.G. Piscine UDP-glucuronosyltransferase 1B. Aquat. Toxicol. 2007, 84, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Sanghani, S.P.; Sanghani, P.C.; Schiel, M.A.; Bosron, W.F. Human carboxylesterases: An update on CES1, CES2 and CES3. Protein Pept. Lett. 2009, 16, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Mardones, J.; Gallardo-Escárate, C. Deltamethrin (AlphaMax) reveals modulation of genes related to oxidative stress in the ectoparasite Caligus rogercresseyi: Implications on delousing drug effectiveness. Aquaculture 2014, 433, 421–429. [Google Scholar] [CrossRef]
- Gaaied, S.; Oliveira, M.; Domingues, I.; Banni, M. 2,4-Dichlorophenoxyacetic acid herbicide effects on zebrafish larvae: Development, neurotransmission and behavior as sensitive endpoints. Environ. Sci. Pollut. Res. 2020, 27, 3686–3696. [Google Scholar] [CrossRef]
- Schmuck, E.M.; Board, P.G.; Whitbread, A.K.; Tetlow, N.; Cavanaugh, J.A.; Blackburn, A.C.; Masoumi, A. Characterization of the monomethylarsonate reductase and dehydroascorbate reductase activities of Omega class glutathione transferase variants: Implications for arsenic metabolism and the age-at-onset of Alzheimer’s and Parkinson’s diseases. Pharmacogenetics Genom. 2005, 15, 493–501. [Google Scholar] [CrossRef]
- Dong, M.; Zhu, L.; Shao, B.; Zhu, S.; Wang, J.; Xie, H.; Wang, J.; Wang, F. The effects of endosulfan on cytochrome P450 enzymes and glutathione S-transferases in zebrafish (Danio rerio) livers. Ecotoxicol. Environ. Saf. 2013, 92, 1–9. [Google Scholar] [CrossRef]
| Gene Symbol | Description | Primer Name | Nucleotide Sequence (5′-3′) | Expected Product (bp) |
|---|---|---|---|---|
| uckl1b | uridine-cytidine kinase-like 1 | uckl1b-F uckl1b-R | GCCTGTGTAAGGTGCTGTGT GCTGCTGCTGCTGCTCTATG | 173 |
| uck2b | uridine-cytidine kinase 2-B | uck2b-F uck2b-R | CATCCTCAACGGCGGCTTCA TCAGTGCGGTCTGCTGGACT | 100 |
| rrm2 | ribonucleotide reductase regulatory subunit M2 | rrm2-F rrm2-R | TCCACCGACAACACCTTCCG GGCAGTGGTCACAGGAGCAT | 105 |
| GSTP1 | glutathione S-transferase pi 1 | GSTP1-F GSTP1-R | GCTCTGCGGATCATGCTGTC TGCGTCCCAAATGCCTCAAA | 178 |
| gsta3 | glutathione S-transferase 3 | gsta3-F gsta3-R | AGCCAAGGAACGCACTCTTC GAGCACCTCCTCCAGCATGA | 127 |
| gsto2 | glutathione S-transferase omega 1 | gsto2-F gsto2-R | CCGTGGTTTGAGAGGGTGGA TGACTGCCTTCACTGCTGGA | 112 |
| rrm2b | ribonucleoside-diphosphate reductase subunit M2 B-like | rrm2b-F rrm2b-R | ACCATGCGAACCGACTCCAA CGACTGTCCAGAACGATGCC | 175 |
| gstt2b | glutathione S-transferase theta 2B | gstt2b-F gstt2b-R | ACCTGTCCTGGCAGCATTCA ACGGCAGCATCCATCTTGTC | 118 |
| ugt | UDP-glucuronosyltransferase | ugt-F ugt-R | GCCGTGTTCTGGACCGAGTT GCAGGAGGAAGGCGATGACA | 118 |
| ugt2a1 | UDP-glucuronosyltransferase 2C1-like | ugt2a1-F ugt2a1-R | TGGCGATACACAGGCGAGAA GTCAGGCTGATCCGCAAACA | 195 |
| ces3 | carboxylesterase 5A-like | ces3-F ces3-R | TGTTCGCACAGGCTCACCAA GCTTCTGAGGCAGCTCCACA | 155 |
| Sample | Raw Data (bp) | Clean Data (bp) | Q20 (%) | Q30 (%) | N (%) | GC (%) |
|---|---|---|---|---|---|---|
| CK-1 | 8,528,218,500 | 8,455,531,252 | 8,312,672,702 (98.31%) | 8,034,203,258 (95.02%) | 88,081 (0.00%) | 3,943,852,525 (46.64%) |
| CK-2 | 8,098,164,900 | 8,022,273,318 | 7,873,934,629 (98.15%) | 7,590,887,887 (94.62%) | 85,023 (0.00%) | 3,885,940,274 (48.44%) |
| CK-3 | 7,866,796,200 | 7,790,326,860 | 7,647,431,709 (98.17%) | 7,375,843,021 (94.68%) | 80,343 (0.00%) | 3,689,368,216 (47.36%) |
| T1-1 | 8,295,867,300 | 8,217,440,673 | 8,051,283,794 (97.98%) | 7,743,396,884 (94.23%) | 86,100 (0.00%) | 3,870,550,118 (47.10%) |
| T1-2 | 7,531,641,600 | 7,457,377,413 | 7,295,029,054 (97.82%) | 6,998,206,939 (93.84%) | 77,000 (0.00%) | 3,534,572,880 (47.40%) |
| T1-3 | 7,386,847,200 | 7,322,938,214 | 7,188,711,492 (98.17%) | 6,930,035,630 (94.63%) | 76,341 (0.00%) | 3,486,858,742 (47.62%) |
| T2-1 | 7,918,878,000 | 7,834,621,505 | 7,681,087,705 (98.04%) | 7,395,471,402 (94.39%) | 80,981 (0.00%) | 3,668,850,749 (46.83%) |
| T2-2 | 7,269,383,100 | 7,204,736,382 | 7,067,332,104 (98.09%) | 6,806,621,229 (94.47%) | 73,881 (0.00%) | 3,398,786,165 (47.17%) |
| T2-3 | 8,150,140,800 | 8,070,530,230 | 7,925,943,852 (98.21%) | 7,650,521,989 (94.80%) | 84,886 (0.00%) | 3,763,940,093 (46.64%) |
| T3-1 | 6,986,844,300 | 6,926,075,339 | 6,798,229,961 (98.15%) | 6,544,811,236 (94.50%) | 71,170 (0.00%) | 3,232,926,751 (46.68%) |
| T3-2 | 7,957,478,400 | 7,900,931,726 | 7,767,533,892 (98.31%) | 7,497,502,331 (94.89%) | 82,047 (0.00%) | 3,703,797,826 (46.88%) |
| T3-3 | 7,329,309,900 | 7,265,578,461 | 7,146,030,274 (98.35%) | 6,900,631,072 (94.98%) | 75,379 (0.00%) | 3,418,766,468 (47.05%) |
| Sample | Total | Unmapped (%) | Unique_Mapped (%) | Multiple_Mapped (%) | Total_Mapped (%) |
|---|---|---|---|---|---|
| CK-1 | 56,593,710 | 4,177,297 (7.38%) | 49,626,790 (87.69%) | 2,789,623 (4.93%) | 52,416,413 (92.62%) |
| CK-2 | 53,663,248 | 4,566,882 (8.51%) | 46,317,822 (86.31%) | 2,778,544 (5.18%) | 49,096,366 (91.49%) |
| CK-3 | 52,108,754 | 4,102,178 (7.87%) | 45,449,379 (87.22%) | 2,557,197 (4.91%) | 48,006,576 (92.13%) |
| T1-1 | 54,955,458 | 4,414,562 (8.03%) | 48,089,747 (87.51%) | 2,451,149 (4.46%) | 50,540,896 (91.97%) |
| T1-2 | 49,875,330 | 4,236,696 (8.49%) | 43,303,053 (86.82%) | 2,335,581 (4.68%) | 45,638,634 (91.51%) |
| T1-3 | 48,971,260 | 3,965,702 (8.10%) | 42,764,114 (87.32%) | 2,241,444 (4.58%) | 45,005,558 (91.90%) |
| T2-1 | 52,463,010 | 4,222,162 (8.05%) | 45,801,883 (87.30%) | 2,438,965 (4.65%) | 48,240,848 (91.95%) |
| T2-2 | 48,161,994 | 4,038,301 (8.38%) | 42,007,026 (87.22%) | 2,116,667 (4.39%) | 44,123,693 (91.62%) |
| T2-3 | 54,071,356 | 4,310,287 (7.97%) | 47,335,777 (87.54%) | 2,425,292 (4.49%) | 49,761,069 (92.03%) |
| T3-1 | 46,375,682 | 3,417,071 (7.37%) | 40,595,071 (87.54%) | 2,363,540 (5.10%) | 42,958,611 (92.63%) |
| T3-2 | 52,855,970 | 3,919,932 (7.42%) | 46,517,939 (88.01%) | 2,4180,99 (4.57%) | 48,936,038 (92.58%) |
| T3-3 | 48,693,526 | 3,443,780 (7.07%) | 42,888,480 (88.08%) | 2,361,266 (4.85%) | 45,249,746 (92.93%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Sun, R.; Zhang, L.; Liu, Y.; Ai, X. Transcriptomic Association Analysis of the Metabolic Mechanism of Sulfamethoxazole in Channel Catfish (Ictalurus punctatus). Animals 2024, 14, 1059. https://doi.org/10.3390/ani14071059
Du X, Sun R, Zhang L, Liu Y, Ai X. Transcriptomic Association Analysis of the Metabolic Mechanism of Sulfamethoxazole in Channel Catfish (Ictalurus punctatus). Animals. 2024; 14(7):1059. https://doi.org/10.3390/ani14071059
Chicago/Turabian StyleDu, Xiangxuan, Ruyu Sun, Lei Zhang, Yongtao Liu, and Xiaohui Ai. 2024. "Transcriptomic Association Analysis of the Metabolic Mechanism of Sulfamethoxazole in Channel Catfish (Ictalurus punctatus)" Animals 14, no. 7: 1059. https://doi.org/10.3390/ani14071059
APA StyleDu, X., Sun, R., Zhang, L., Liu, Y., & Ai, X. (2024). Transcriptomic Association Analysis of the Metabolic Mechanism of Sulfamethoxazole in Channel Catfish (Ictalurus punctatus). Animals, 14(7), 1059. https://doi.org/10.3390/ani14071059





