Effects of Chronic Heat Stress on Growth, Apoptosis, Antioxidant Enzymes, Transcriptomic Profiles, and Immune-Related Genes of Hong Kong Catfish (Clarias fuscus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design and Management
2.3. Morphological Statistics
2.4. TUNEL Staining of Liver Tissue for Apoptosis Detection
2.5. Serum and Liver Indexes Test
2.6. RNA Extraction, Library Construction, Sequencing and Assembly
2.7. Screening of Differentially Expressed Genes (DEGs) and Functional Enrichment Analysis
2.8. Real-Time Fluorescence Quantitative PCR (qPCR) Validation
2.9. Statistical Analysis
3. Results
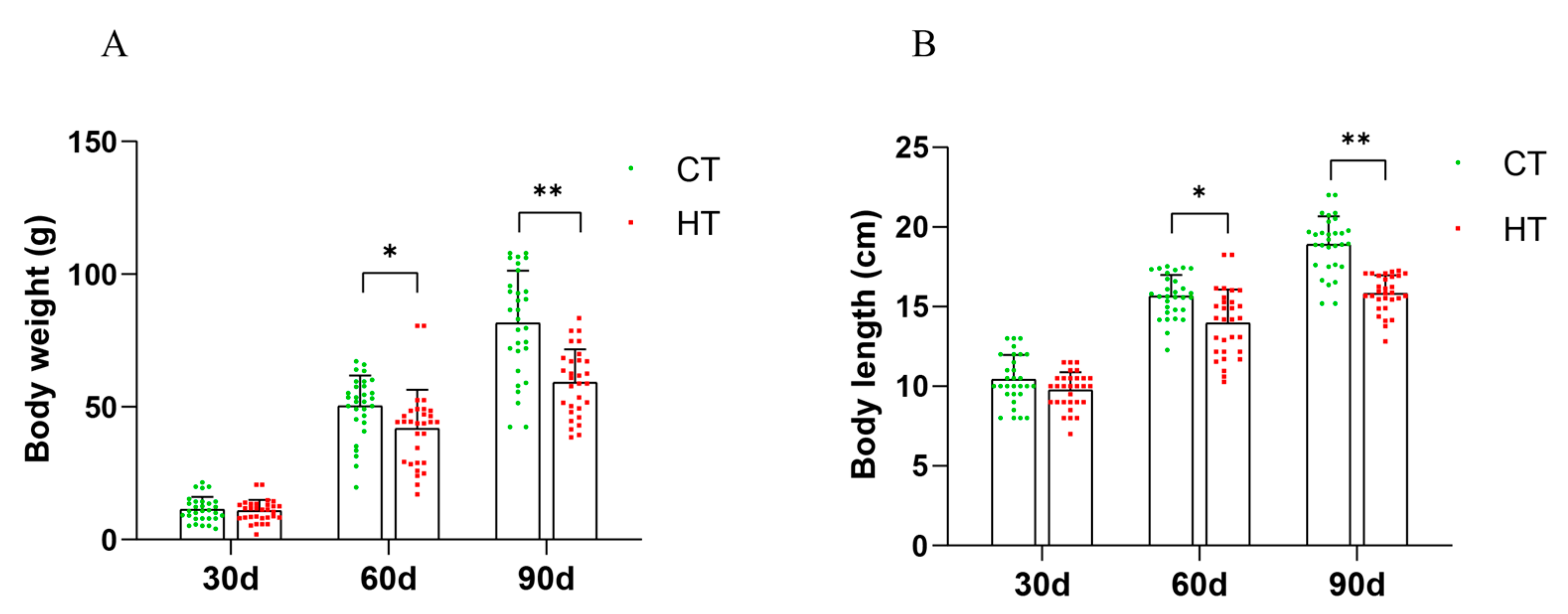
3.1. Morphological Statistics
3.2. Hepatic Tissue Analysis via TUNEL Assay
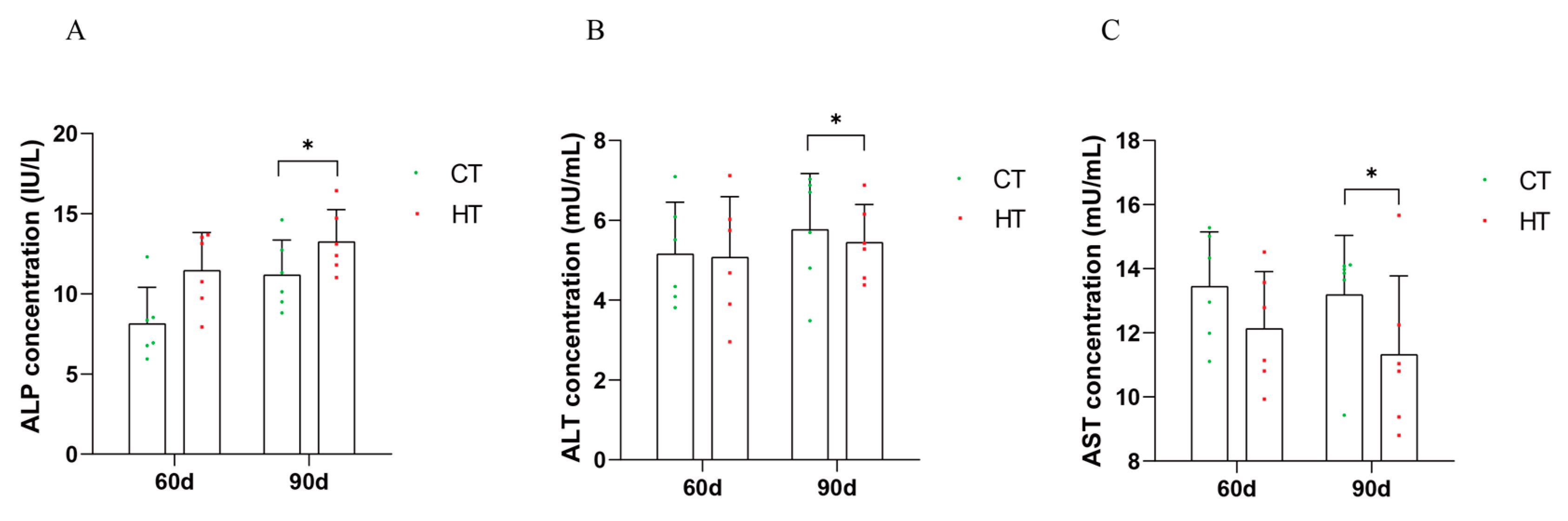
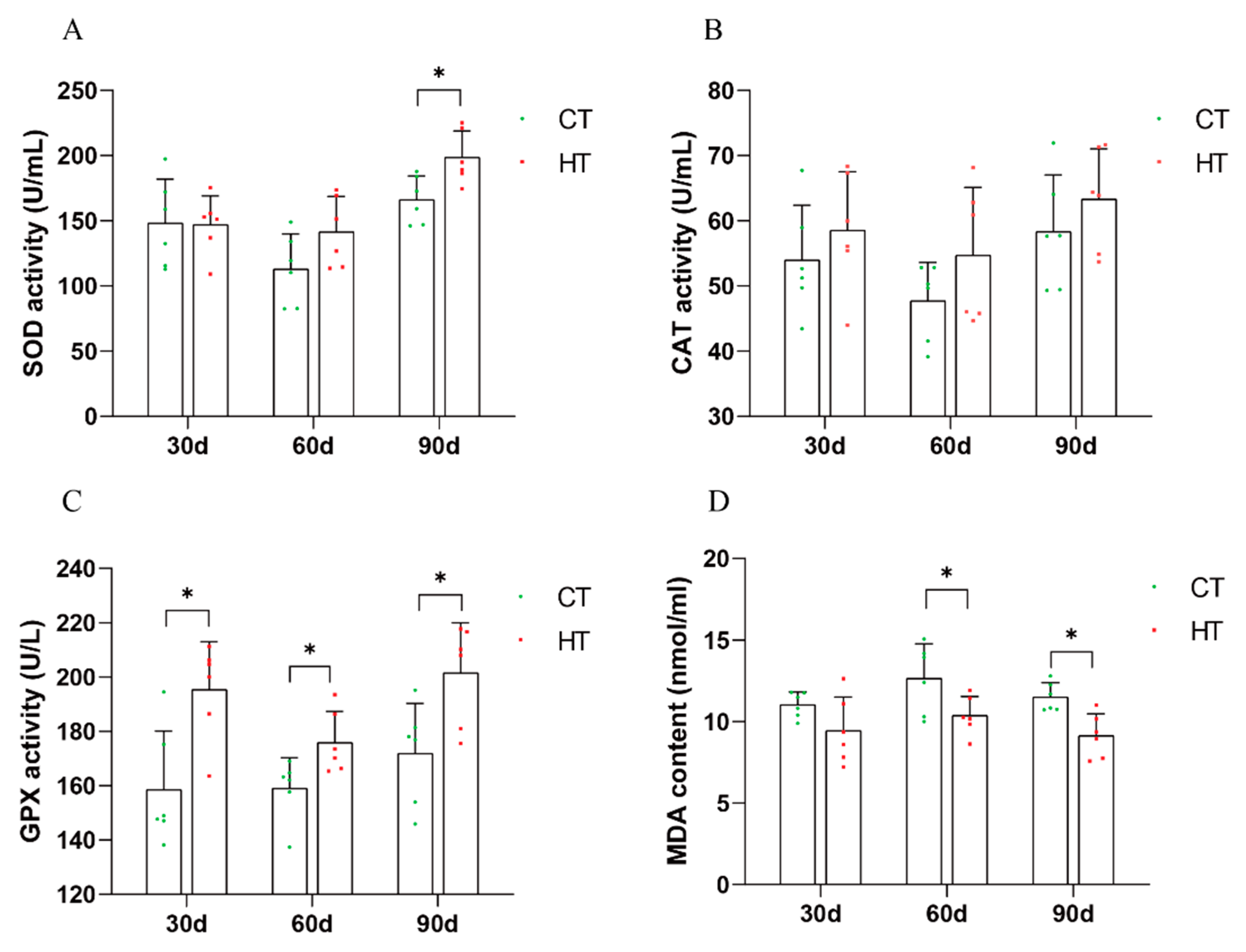
3.3. Effect of Chronic High Temperature on Biochemical Indexes in Serum and Liver of C. fuscus
3.4. Transcriptome Sequencing Analysis
3.4.1. Sequencing of Reads in C. fuscus Liver
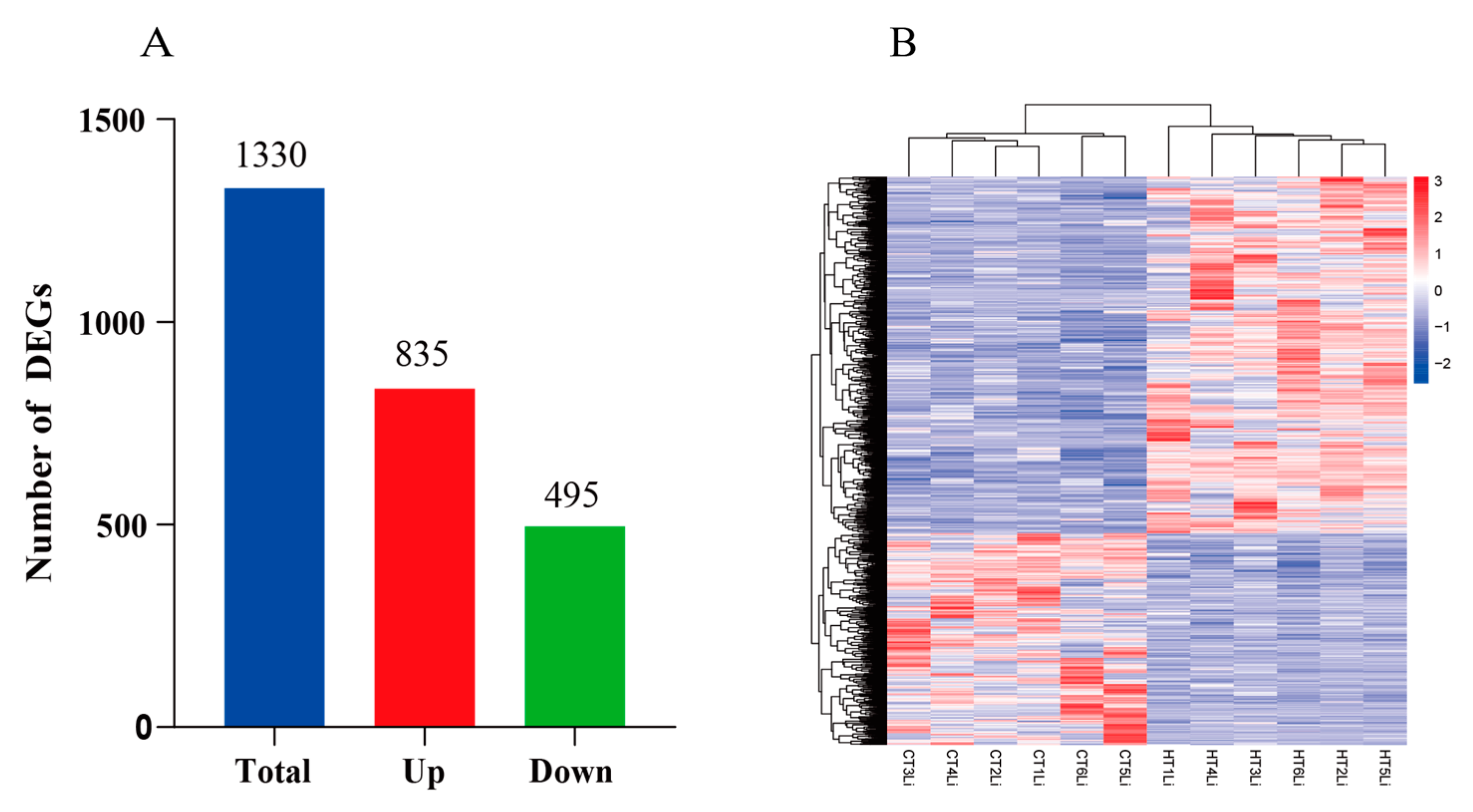
3.4.2. Analysis of Differentially Expressed Genes (DEGs)
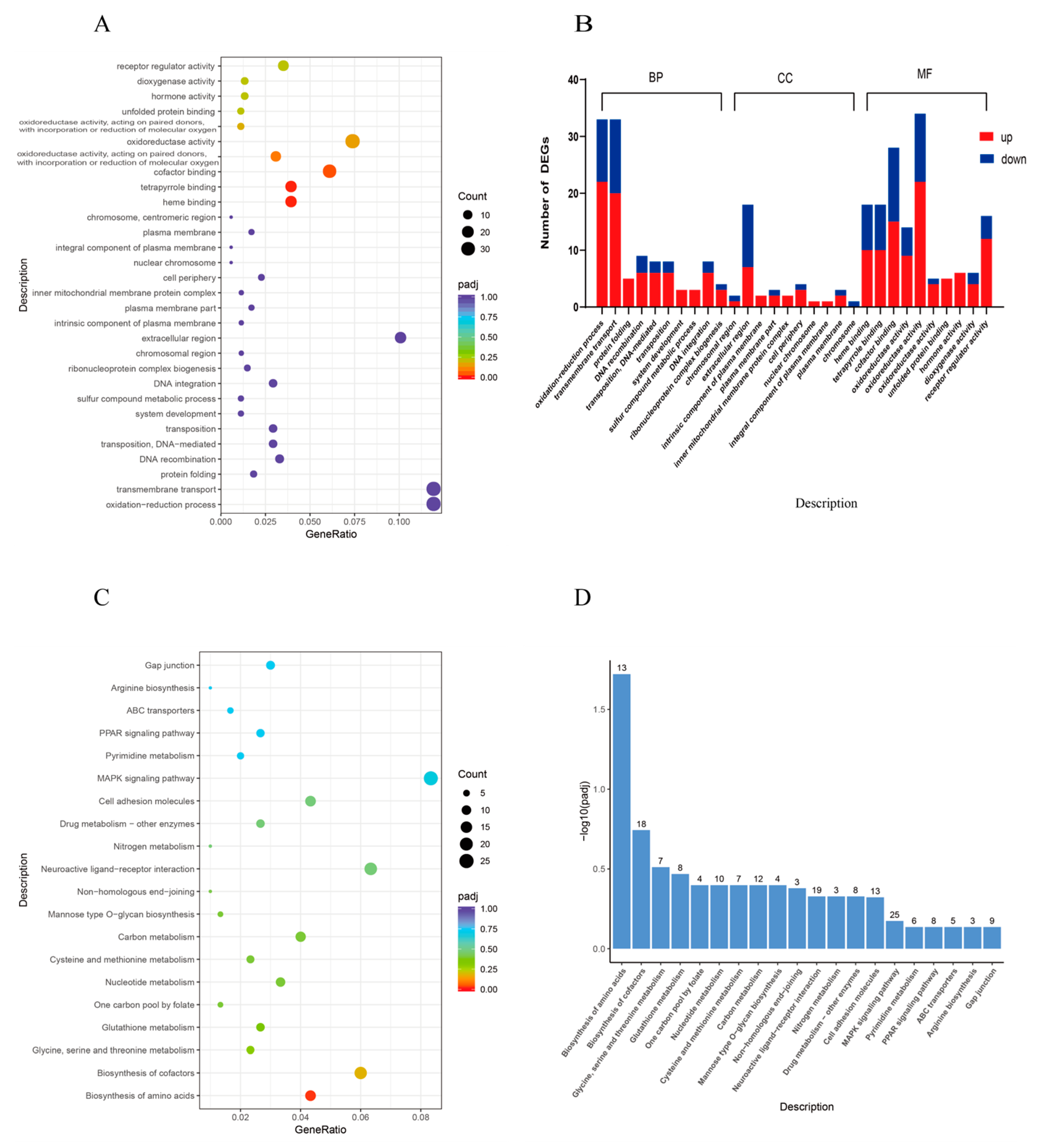
3.4.3. Functional Classification of DEGs
3.5. qRT-PCR Validation of Transcriptome Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, T.; Gu, W.; Liu, E.; Wang, B.; Wang, G.; Dong, F.; Guo, F.; Jiao, W.; Sun, Y.; Wang, X.; et al. miR-301b-5p and Its Target Gene Nfatc2ip Regulate Inflammatory Responses in the Liver of Rainbow Trout (Oncorhynchus mykiss) under High Temperature Stress. Ecotoxicol. Environ. Saf. 2022, 242, 113915. [Google Scholar] [CrossRef] [PubMed]
- Braz-Mota, S.; Fé, L.; Delunardo, F.A.; Sadauskas-Henrique, H.; Almeida-Val, V.; Val, A. Exposure to Waterborne Copper and High Temperature Induces the Formation of Reactive Oxygen Species and Causes Mortality in the Amazonian Fish Hoplosternum Littorale. Hydrobiologia 2017, 789, 157–166. [Google Scholar] [CrossRef]
- Neuheimer, A.B.; Thresher, R.E.; Lyle, J.M.; Semmens, J.M. Tolerance Limit for Fish Growth Exceeded by Warming Waters. Nat. Clim. Chang. 2011, 1, 110–113. [Google Scholar] [CrossRef]
- Gradil, A.M.; Wright, G.M.; Speare, D.J.; Wadowska, D.W.; Purcell, S.; Fast, M.D. The Effects of Temperature and Body Size on Immunological Development and Responsiveness in Juvenile Shortnose Sturgeon (Acipenser brevirostrum). Fish Shellfish Immunol. 2014, 40, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Qu, Y.-K.; Wang, A.-M.; Yu, Y.-B.; Yang, W.-P.; Lv, F.; Nie, Q. Effects of Carotenoids on the Growth Performance, Biochemical Parameters, Immune Responses and Disease Resistance of Yellow Catfish (Pelteobagrus fulvidraco) under High-Temperature Stress. Aquaculture 2019, 503, 293–303. [Google Scholar] [CrossRef]
- Liu, Z.-M.; Zhu, X.-L.; Lu, J.; Cai, W.-J.; Ye, Y.-P.; Lv, Y.-P. Effect of High Temperature Stress on Heat Shock Protein Expression and Antioxidant Enzyme Activity of Two Morphs of the Mud Crab Scylla Paramamosain. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2018, 223, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Rebl, A.; Korytář, T.; Borchel, A.; Bochert, R.; Strzelczyk, J.E.; Goldammer, T.; Verleih, M. The Synergistic Interaction of Thermal Stress Coupled with Overstocking Strongly Modulates the Transcriptomic Activity and Immune Capacity of Rainbow Trout (Oncorhynchus mykiss). Sci. Rep. 2020, 10, 14913. [Google Scholar] [CrossRef]
- Verma, A.K.; Pal, A.K.; Manush, S.M.; Das, T.; Dalvi, R.S.; Chandrachoodan, P.P.; Ravi, P.M.; Apte, S.K. Persistent Sub-Lethal Chlorine Exposure Augments Temperature Induced Immunosuppression in Cyprinus Carpio Advanced Fingerlings. Fish Shellfish Immunol. 2007, 22, 547–555. [Google Scholar] [CrossRef]
- Bowden, T.J. Modulation of the Immune System of Fish by Their Environment. Fish Shellfish Immunol. 2008, 25, 373–383. [Google Scholar] [CrossRef]
- Lee, S.; Ji, K.; Choi, K. Effects of Water Temperature on Perchlorate Toxicity to the Thyroid and Reproductive System of Oryzias Latipes. Ecotoxicol. Environ. Saf. 2014, 108, 311–317. [Google Scholar] [CrossRef]
- Logan, C.A.; Somero, G.N. Effects of Thermal Acclimation on Transcriptional Responses to Acute Heat Stress in the Eurythermal Fish Gillichthys Mirabilis (Cooper). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1373–R1383. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.O.; Eweedah, N.M.; El-Sharawy, M.E.; Awad, S.S.; Van Doan, H.; Paray, B.A. Dietary White Button Mushroom Improved the Growth, Immunity, Antioxidative Status and Resistance against Heat Stress in Nile Tilapia (Oreochromis niloticus). Aquaculture 2020, 523, 735229. [Google Scholar] [CrossRef]
- Kim, J.-H.; Kim, S.K.; Hur, Y.B. Temperature-Mediated Changes in Stress Responses, Acetylcholinesterase, and Immune Responses of Juvenile Olive Flounder Paralichthys Olivaceus in a Bio-Floc Environment. Aquaculture 2019, 506, 453–458. [Google Scholar] [CrossRef]
- Cardoso, P.G.; Resende-de-Oliveira, R.; Rocha, E. Combined Effects of Increased Temperature and Levonorgestrel Exposure on Zebrafish Female Liver, Using Stereology and Immunohistochemistry against Catalase, CYP1A, HSP90 and Vitellogenin. Environ. Pollut. 2019, 252, 1059–1067. [Google Scholar] [CrossRef]
- Livingstone, D.R. Contaminant-Stimulated Reactive Oxygen Species Production and Oxidative Damage in Aquatic Organisms. Mar. Pollut. Bull. 2001, 42, 656–666. [Google Scholar] [CrossRef]
- Lopes, P.A.; Pinheiro, T.; Santos, M.C.; da Luz Mathias, M.; Collares-Pereira, M.J.; Viegas-Crespo, A.M. Response of Antioxidant Enzymes in Freshwater Fish Populations (Leuciscus Alburnoides Complex) to Inorganic Pollutants Exposure. Sci. Total Environ. 2001, 280, 153–163. [Google Scholar] [CrossRef]
- Basu, N.; Todgham, A.E.; Ackerman, P.A.; Bibeau, M.R.; Nakano, K.; Schulte, P.M.; Iwama, G.K. Heat Shock Protein Genes and Their Functional Significance in Fish. Gene 2002, 295, 173–183. [Google Scholar] [CrossRef]
- Regoli, F.; Giuliani, M.E. Oxidative Pathways of Chemical Toxicity and Oxidative Stress Biomarkers in Marine Organisms. Mar. Environ. Res. 2014, 93, 106–117. [Google Scholar] [CrossRef]
- Zheng, J.-L.; Zeng, L.; Shen, B.; Xu, M.-Y.; Zhu, A.-Y.; Wu, C.-W. Antioxidant Defenses at Transcriptional and Enzymatic Levels and Gene Expression of Nrf2-Keap1 Signaling Molecules in Response to Acute Zinc Exposure in the Spleen of the Large Yellow Croaker Pseudosciaena Crocea. Fish Shellfish Immunol. 2016, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-H.; Guo, Z.-X.; Luo, S.-W.; Wang, A.-L. Effects of High Temperature on Biochemical Parameters, Oxidative Stress, DNA Damage and Apoptosis of Pufferfish (Takifugu obscurus). Ecotoxicol. Environ. Saf. 2018, 150, 190–198. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Pan, C.; Liu, E.; Zhao, X.; Ling, Q. Alterations to Transcriptomic Profile, Histopathology, and Oxidative Stress in Liver of Pikeperch (Sander lucioperca) under Heat Stress. Fish Shellfish Immunol. 2019, 95, 659–669. [Google Scholar] [CrossRef]
- Hubbard, A.H.; Zhang, X.; Jastrebski, S.; Singh, A.; Schmidt, C. Understanding the Liver under Heat Stress with Statistical Learning: An Integrated Metabolomics and Transcriptomics Computational Approach. BMC Genom. 2019, 20, 502. [Google Scholar] [CrossRef]
- Xu, D.; Zhou, S.; Yang, H. Carbohydrate and Amino Acids Metabolic Response to Heat Stress in the Intestine of the Sea Cucumber Apostichopus Japonicus. Aquac. Res. 2017, 48, 5883–5891. [Google Scholar] [CrossRef]
- Dettleff, P.; Zuloaga, R.; Fuentes, M.; Gonzalez, P.; Aedo, J.; Estrada, J.M.; Molina, A.; Valdés, J.A. High-Temperature Stress Effect on the Red Cusk-Eel (Geypterus chilensis) Liver: Transcriptional Modulation and Oxidative Stress Damage. Biology 2022, 11, 990. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L.; Li, C.; Liu, E.; Zhu, H.; Ling, Q. Heat Stress-Induced Endoplasmic Reticulum Stress Promotes Liver Apoptosis in Largemouth Bass (Micropterus salmoides). Aquaculture 2022, 546, 737401. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, T.; He, Y.; Zhan, W.; Xie, Q.; Lou, B. Integration of Transcriptome and Proteome Analyses Reveals the Regulation Mechanisms of Larimichthys Polyactis Liver Exposed to Heat Stress. Fish Shellfish Immunol. 2023, 135, 108704. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, K.M.; Hinch, S.G.; Sierocinski, T.; Pavlidis, P.; Miller, K.M. Transcriptomic Responses to High Water Temperature in Two Species of Pacific Salmon. Evol. Appl. 2014, 7, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Luo, L. Genome-Wide Identification of Hsp70/110 Genes in Rainbow Trout and Their Regulated Expression in Response to Heat Stress. PeerJ 2020, 8, e10022. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, Y.; Meng, W.; Zhong, X.; Shan, Y.; Gao, T. Comparative Transcriptomic Analysis Brings New Insights into the Response to Acute Temperature Acclimation in Burbot (Lota Lota Lota). Aquac. Rep. 2021, 20, 100657. [Google Scholar] [CrossRef]
- Tian, C.-X.; Lin, X.-H.; Zhou, D.-Y.; Chen, Y.; Shen, Y.-J.; Ye, M.-H.; Duan, C.-Y.; Zhang, Y.-L.; Yang, B.-L.; Deng, S.-P.; et al. A Chromosome-Level Genome Assembly of Hong Kong Catfish (Clarias fuscus) Uncovers a Sex-Determining Region. BMC Genom. 2023, 24, 291. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Sun, F.; Zhang, J.; Feng, J.; Liu, H.; Rajendran, K.V.; Sun, L.; Zhang, Y.; Jiang, Y.; et al. RNA-Seq Reveals Expression Signatures of Genes Involved in Oxygen Transport, Protein Synthesis, Folding, and Degradation in Response to Heat Stress in Catfish. Physiol. Genom. 2013, 45, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Khieokhajonkhet, A.; Sangphrom, S.; Aeksiri, N.; Tatsapong, P.; Wuthijaree, K.; Kaneko, G. Effects of Long-Term Exposure to High Temperature on Growth Performance, Chemical Composition, Hematological and Histological Changes, and Physiological Responses in Hybrid Catfish [♂Clarias Gariepinus (Burchell, 1822) ×♀C. Macrocephalus (Günther, 1864)]. J. Therm. Biol. 2022, 105, 103226. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yang, Y.; Sarath Babu, V.; Chen, J.; Li, F.; Yang, M.; Li, N.; Li, J.; Lin, L.; Qin, Z. The Antibacterial Activity of Erythrocytes from Clarias Fuscus Associated with Phagocytosis and Respiratory Burst Generation. Fish Shellfish Immunol. 2021, 119, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Lisachov, A.; Nguyen, D.H.M.; Panthum, T.; Ahmad, S.F.; Singchat, W.; Ponjarat, J.; Jaisamut, K.; Srisapoome, P.; Duengkae, P.; Hatachote, S.; et al. Emerging Importance of Bighead Catfish (Clarias macrocephalus) and North African Catfish (C. Gariepinus) as a Bioresource and Their Genomic Perspective. Aquaculture 2023, 573, 739585. [Google Scholar] [CrossRef]
- Lin, X.; Tan, J.; Shen, Y.; Yang, B.; Zhang, Y.; Liao, Y.; Wang, P.; Zhou, D.; Li, G.; Tian, C. A High-Density Genetic Linkage Map and QTL Mapping for Sex in Clarias Fuscus. Aquaculture 2022, 561, 738723. [Google Scholar] [CrossRef]
- Liver Transcriptome Analysis Revealing Response to High-Temperature Stress in Glyptosternum Maculatum (Sisoridae: Siluriformes). Aquac. Rep. 2023, 29, 101538. [CrossRef]
- Wang, Z.; Hu, J.; Mei, Z.; Zhang, Y.; Liu, Q.; Yang, D. Mannan-Oligosaccharide Induces Trained Immunity Activation and Alleviates Pathological Liver Injury in Turbot (Scophthalmus maximus). Aquaculture 2024, 578, 740097. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Dupré, R.K.; Wood, S.C. Behavioral Temperature Regulation by Aquatic Ectotherms during Hypoxia. Can. J. Zool. 1988, 66, 2649–2652. [Google Scholar] [CrossRef]
- Zhao, H.; Ke, H.; Zhang, L.; Zhao, Z.; Lai, J.; Zhou, J.; Huang, Z.; Li, H.; Du, J.; Li, Q. Integrated Analysis about the Effects of Heat Stress on Physiological Responses and Energy Metabolism in Gymnocypris Chilianensis. Sci. Total Environ. 2022, 806, 151252. [Google Scholar] [CrossRef] [PubMed]
- Baum, D.; Laughton, R.; Armstrong, J.D.; Metcalfe, N.B. The Effect of Temperature on Growth and Early Maturation in a Wild Population of Atlantic Salmon Parr. J. Fish Biol. 2005, 67, 1370–1380. [Google Scholar] [CrossRef]
- Chang, Y. Effects of Cooling Temperature Stress on Hematology and Serum Chemistry Values of Cyprinus Carpio. J. Fish. China 2006, 30, 701–706. [Google Scholar]
- Hoang, T.; Ho, H.C.; Le, N.P.T.; Bui, T.H.H. Effects of High Temperature on Survival and Feed Consumption of Banana Shrimp Penaeus Merguiensis. Aquaculture 2020, 522, 735152. [Google Scholar] [CrossRef]
- Li, A.J.; Leung, P.T.Y.; Bao, V.W.W.; Lui, G.C.S.; Leung, K.M.Y. Temperature-Dependent Physiological and Biochemical Responses of the Marine Medaka Oryzias Melastigma with Consideration of Both Low and High Thermal Extremes. J. Therm. Biol. 2015, 54, 98–105. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Yang, F.-F.; Liao, S.-A.; Miao, Y.-T.; Ye, C.-X.; Wang, A.-L.; Tan, J.-W.; Chen, X.-Y. High Temperature Induces Apoptosis and Oxidative Stress in Pufferfish (Takifugu obscurus) Blood Cells. J. Therm. Biol. 2015, 53, 172–179. [Google Scholar] [CrossRef]
- Islam, M.J.; Kunzmann, A.; Slater, M.J. Extreme Winter Cold-Induced Osmoregulatory, Metabolic, and Physiological Responses in European Seabass (Dicentrarchus labrax) Acclimatized at Different Salinities. Sci. Total Environ. 2021, 771, 145202. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Ye, C.-X.; Guo, Z.-X.; Wang, A.-L. Immune and Physiological Responses of Pufferfish (Takifugu obscurus) under Cold Stress. Fish Shellfish Immunol. 2017, 64, 137–145. [Google Scholar] [CrossRef]
- Shahjahan, M.; Islam, M.J.; Hossain, M.T.; Mishu, M.A.; Hasan, J.; Brown, C. Blood Biomarkers as Diagnostic Tools: An Overview of Climate-Driven Stress Responses in Fish. Sci. Total Environ. 2022, 843, 156910. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Roosta, Z.; Hajimoradloo, A.; Vakili, F. The Effects of Lactobacillus Acidophilus as Feed Supplement on Skin Mucosal Immune Parameters, Intestinal Microbiota, Stress Resistance and Growth Performance of Black Swordtail (Xiphophorus helleri). Fish Shellfish Immunol. 2015, 42, 533–538. [Google Scholar] [CrossRef]
- Dalvi, R.S.; Das, T.; Debnath, D.; Yengkokpam, S.; Baruah, K.; Tiwari, L.R.; Pal, A.K. Metabolic and Cellular Stress Responses of Catfish, Horabagrus Brachysoma (Günther) Acclimated to Increasing Temperatures. J. Therm. Biol. 2017, 65, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ren, M.; Liang, H.; Ge, X.; Xu, H.; Wu, L. Transcriptome Analysis of the Effect of High-Temperature on Nutrient Metabolism in Juvenile Grass Carp (Ctenopharyngodon idellus). Gene 2022, 809, 146035. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, Y.; Fang, L.; Guan, C.; Xu, Y. Heat-Shock Protein 90 Alleviates Oxidative Stress and Reduces Apoptosis in Liver of Seriola Aureovittata (Yellowtail kingfish) under High-Temperature Stress. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2023, 270, 110927. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Yeager-Armstead, M.; Murray, E. Protective and Antioxidant Role of Selenium on Arsenic Trioxide-Induced Oxidative Stress and Genotoxicity in the Fish Hepatoma Cell Line PLHC-1. Environ. Toxicol. Chem. 2012, 31, 2861–2869. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-L.; Zuo, D.; Wang, L.-M.; Sun, T.; Wang, Q.; Zhao, Y.-L. Effects of White Spot Syndrome Virus Infection on Immuno-Enzyme Activities and Ultrastructure in Gills of Cherax Quadricarinatus. Fish Shellfish Immunol. 2012, 32, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; ZhiQiang, J.; FanPing, M.; Ying, L.; ZhenYu, W. The effects of sharply changes in temperature on survival and indices of physiology and biochemistry in Pacific cod Gadus macrocephalus. Fish. Sci. 2012, 31, 463–466. [Google Scholar]
- Lin, Y.-H.; Shiau, S.-Y. Dietary Selenium Requirements of Juvenile Grouper, Epinephelus Malabaricus. Aquaculture 2005, 250, 356–363. [Google Scholar] [CrossRef]
- Kim, B.-S.; Jung, S.J.; Choi, Y.J.; Kim, N.N.; Choi, C.Y.; Kim, J.-W. Effects of Different Light Wavelengths from LEDs on Oxidative Stress and Apoptosis in Olive Flounder (Paralichthys olivaceus) at High Water Temperatures. Fish Shellfish Immunol. 2016, 55, 460–468. [Google Scholar] [CrossRef]
- Teles, M.; Oliveira, M.; Jerez-Cepa, I.; Franco-Martínez, L.; Tvarijonaviciute, A.; Tort, L.; Mancera, J.M. Transport and Recovery of Gilthead Sea Bream (Sparus aurata L.) Sedated With Clove Oil and MS222: Effects on Oxidative Stress Status. Front. Physiol. 2019, 10, 523. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, D.; Zhao, C.; Wang, W.; Zhang, X.; Liu, X.; Liu, Y.; Xiao, Z.; Xu, S.; Xiao, Y.; et al. Histological and Enzymatic Responses of Japanese Flounder (Paralichthys olivaceus) and Its Hybrids (P. olivaceus ♀ × P. dentatus ♂) to Chronic Heat Stress. Fish Physiol. Biochem. 2014, 40, 1031–1041. [Google Scholar] [CrossRef]
- Dettleff, P.; Zuloaga, R.; Fuentes, M.; Gonzalez, P.; Aedo, J.; Estrada, J.M.; Molina, A.; Valdés, J.A. Physiological and Molecular Responses to Thermal Stress in Red Cusk-Eel (Genypterus chilensis) Juveniles Reveals Atrophy and Oxidative Damage in Skeletal Muscle. J. Therm. Biol. 2020, 94, 102750. [Google Scholar] [CrossRef]
- Roychowdhury, P.; Aftabuddin, M.; Pati, M.K. Thermal Stress–Induced Oxidative Damages in the Liver and Associated Death in Fish, Labeo Rohita. Fish Physiol. Biochem. 2021, 47, 21–32. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, E.; Li, C.; Pan, C.; Zhao, X.; Wang, Y.; Ling, Q. Effects of Heat Stress on Histopathology, Antioxidant Enzymes, and Transcriptomic Profiles in Gills of Pikeperch Sander Lucioperca. Aquaculture 2020, 534, 736277. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Wang, G.; Chen, Y.; Guo, J.; Pan, C.; Liu, E.; Ling, Q. Physicochemical Changes in Liver and Hsc70 Expression in Pikeperch Sander Lucioperca under Heat Stress. Ecotoxicol. Environ. Saf. 2019, 181, 130–137. [Google Scholar] [CrossRef]
- Wang, Y.; Peng, X.; Yang, Z.; Zhao, W.; Xu, W.; Hao, J.; Wu, W.; Shen, X.L.; Luo, Y.; Huang, K. iTRAQ Mitoproteome Analysis Reveals Mechanisms of Programmed Cell Death in Arabidopsis Thaliana Induced by Ochratoxin A. Toxins 2017, 9, 167. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Z.; Liu, Z.; Wang, X.; Yu, H.; Peng, C.; Hou, X.; Lu, W.; Xing, Q.; Hu, J.; et al. Metabonomic Analysis Provides New Insights into the Response of Zhikong Scallop (Chlamys farreri) to Heat Stress by Improving Energy Metabolism and Antioxidant Capacity. Antioxidants 2022, 11, 1084. [Google Scholar] [CrossRef]
- Sun, J.; Liu, Z.; Quan, J.; Li, L.; Zhao, G.; Lu, J. RNA-Seq Analysis Reveals Alternative Splicing Under Heat Stress in Rainbow Trout (Oncorhynchus mykiss). Mar. Biotechnol. 2022, 24, 5–17. [Google Scholar] [CrossRef]
- Shi, K.-P.; Dong, S.-L.; Zhou, Y.-G.; Li, Y.; Gao, Q.-F.; Sun, D.-J. RNA-Seq Reveals Temporal Differences in the Transcriptome Response to Acute Heat Stress in the Atlantic Salmon (Salmo salar). Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 169–178. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Liu, Z.; Zhou, Y.; Xia, B.; Wang, Y.; Kang, Y.; Wang, J. Transcriptome Analysis Provides Insights into Hepatic Responses to Moderate Heat Stress in the Rainbow Trout (Oncorhynchus mykiss). Gene 2017, 619, 1–9. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Y.; Liang, S.; Lin, G.; Chen, S.; Yang, G. Tissue-Overlapping Response of Half-Smooth Tongue Sole (Cynoglossus semilaevis) to Thermostressing Based on Transcriptome Profiles. Gene 2016, 586, 97–104. [Google Scholar] [CrossRef]
- Barat, A.; Sahoo, P.K.; Kumar, R.; Goel, C.; Singh, A.K. Transcriptional Response to Heat Shock in Liver of Snow Trout (Schizothorax richardsonii)—A Vulnerable Himalayan Cyprinid Fish. Funct. Integr. Genom. 2016, 16, 203–213. [Google Scholar] [CrossRef]
- Bromke, M.A. Amino Acid Biosynthesis Pathways in Diatoms. Metabolites 2013, 3, 294–311. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, P.; Zhai, B.; Zhang, M.; Xiang, Y.; Fang, J.; Xu, S.; Gao, Y.; Chen, X.; Sui, X.; et al. The Emerging Role of Ferroptosis in Inflammation. Biomed. Pharmacother. 2020, 127, 110108. [Google Scholar] [CrossRef]
- Lu, X.; Alvarez, L.; Huang, Z.; Chen, L.; An, W.; Corrales, F.J.; Avila, M.A.; Kanel, G.; Mato, J.M. Methionine Adenosyltransferase 1A Knockout Mice Are Predisposed to Liver Injury and Exhibit Increased Expression of Genes Involved in Proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 5560–5565. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, J.; Yu, Z.; Li, H.; Liu, Z.; Deng, D.; Liu, J.; Chen, C.; Zhang, C.; Ou, Z.; et al. BCAT2 Shapes a Noninflamed Tumor Microenvironment and Induces Resistance to Anti-PD-1/PD-L1 Immunotherapy by Negatively Regulating Proinflammatory Chemokines and Anticancer Immunity. Adv. Sci. 2023, 10, 2207155. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Z.; Tsai, H.-I.; Liu, Y.; Gao, J.; Wang, M.; Song, L.; Cao, X.; Xu, Z.; Chen, H.; et al. Branched-Chain Amino Acid Aminotransferase 2 Regulates Ferroptotic Cell Death in Cancer Cells. Cell Death Differ. 2021, 28, 1222–1236. [Google Scholar] [CrossRef]
- Cui, H.; Guo, D.; Zhang, X.; Zhu, Y.; Wang, Z.; Jin, Y.; Guo, W.; Zhang, S. ENO3 Inhibits Growth and Metastasis of Hepatocellular Carcinoma via Wnt/β-Catenin Signaling Pathway. Front. Cell Dev. Biol. 2021, 9, 797102. [Google Scholar] [CrossRef]
- Rosemary Siafakas, A.; Richardson, D.R. Growth Arrest and DNA Damage-45 Alpha (GADD45α). Int. J. Biochem. Cell Biol. 2009, 41, 986–989. [Google Scholar] [CrossRef]
- Lv, H.; Liu, Y.; Li, H.; Yin, X.; Wang, P.; Qu, X.; Gao, Y.; Li, W.; Chu, Z. Modulation of Antioxidant Enzymes, Heat Shock Protein, and Intestinal Microbiota of Large Yellow Croaker (Larimichthys crocea) Under Acute Cold Stress. Front. Mar. Sci. 2021, 8, 725899. [Google Scholar] [CrossRef]
- Singh, S.P.; Ahmad, T.; Sharma, J.; Chakrabarti, R. Effect of Temperature on Food Consumption, Immune System, Antioxidant Enzymes, and Heat Shock Protein 70 of Channa Punctata (Bloch, 1793). Fish Physiol. Biochem. 2021, 47, 79–91. [Google Scholar] [CrossRef]
- Zunino, B.; Rubio-Patiño, C.; Villa, E.; Meynet, O.; Proics, E.; Cornille, A.; Pommier, S.; Mondragón, L.; Chiche, J.; Bereder, J.-M.; et al. Hyperthermic Intraperitoneal Chemotherapy Leads to an Anticancer Immune Response via Exposure of Cell Surface Heat Shock Protein 90. Oncogene 2016, 35, 261–268. [Google Scholar] [CrossRef]
- Qiu, X.-B.; Shao, Y.-M.; Miao, S.; Wang, L. The Diversity of the DnaJ/Hsp40 Family, the Crucial Partners for Hsp70 Chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Mahanty, A.; Mohanty, S.; Mohanty, B.P. Dietary Supplementation of Curcumin Augments Heat Stress Tolerance through Upregulation of Nrf-2-Mediated Antioxidative Enzymes and Hsps in Puntius Sophore. Fish Physiol. Biochem. 2017, 43, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liang, X.; Zhang, Y.; Li, Y.; Cao, X.; Gao, J. Cloning of Three Heat Shock Protein Genes (HSP70, HSP90α and HSP90β) and Their Expressions in Response to Thermal Stress in Loach (Misgurnus anguillicaudatus) Fed with Different Levels of Vitamin C. Fish Shellfish Immunol. 2017, 66, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Shi, Y.; Shan, Z.; Yang, L.; Wang, X.; Shi, L. Bioaccumulation, Oxidative Stress and HSP70 Expression in Cyprinus Carpio L. Exposed to Microcystin-LR under Laboratory Conditions. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2012, 155, 483–490. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequences (5′-3′) |
|---|---|
| Hsp90aa1 | F: GGACCATCGCCAAAT |
| R: GGCAGAGTAGAAACCCAC | |
| Odc1 | F: GGATCATTTATGCCAACC |
| R: TCCGAGCAACCTTCATT | |
| Acod1 | F: CGACCTTACGACGTGAACAT |
| R: TTTAGCCAGACCCAACACAC | |
| Cyp51a1 | F: ACGACGATGAGATTTCCG |
| R: CTTGCTTCTGTTCGCTGTA | |
| Hspa5 | F: ACGGGCAACAAGAACAAGAT |
| R: GGTTCTTGAGTGAGTAGGCG | |
| Il1rl2 | F: GGGTGTCTAAGGGTTGG |
| R: GCACTTCCTCTGTGGGTT | |
| Myd88 | F: GAGAGAATCCCACGCAGAAG |
| R: TTATGCTTCGATCCTGGTGC | |
| Msmo | F: AGGCTTCGTGTTCCAGTT |
| R: CCAGTCGTAAGGGATGCTA | |
| Dhcr24 | F: AGTGGTGTTTGATGCCTAC |
| R: TTCCTCCCTCCTTCTTC | |
| Oplah | F: TTCACCGCACGCTACAT |
| R: GGCCAAAGTCCCGATT | |
| β-actin | F: AGGCTGGATTCGCTGGAGATGAT |
| R: TGGTGACAATACCGTGCTCAATGG |
| Gene ID | Gene Name | Gene Annotation | log2 (FC) |
|---|---|---|---|
| gene-Cfus20961 | gprin3 | G protein-regulated inducer of neurite outgrowth 3 | 8.3369 |
| gene-Cfus12438 | chs2 | chitin synthase2 | 8.1574 |
| gene-Cfus21522 | pprc1 | Peroxisome proliferator-activated receptor gamma coactivator-related protein 1 | 8.0864 |
| gene-Cfus01065 | robo2 | roundabout homolog 2-like | 7.7606 |
| gene-Cfus18848 | entpd2 | ectonucleoside triphosphate diphosphohydrolase 2 | 7.2640 |
| gene-Cfus18283 | ptx | pentraxin fusion protein-like | 6.8802 |
| gene-Cfus01755 | porcn | protein-serine O-palmitoleoyltransferase porcupine-like | 6.8777 |
| gene-Cfus03155 | tuba1c | tubulin alpha-1C chain | 6.7665 |
| gene-Cfus03755 | arpp21 | cAMP regulated phosphoprotein 21 | 6.5925 |
| gene-Cfus06190 | trim25 | E3 ubiquitin/ISG15 ligase TRIM25-like | 6.4865 |
| gene-Cfus08092 | LOC106488566 | RNA-directed DNA polymerase from mobile element jockey-like | −6.7983 |
| gene-Cfus13395 | cacna1h | voltage-dependent T-type calcium channel subunit alpha-1H-like | −6.6817 |
| gene-Cfus06402 | nceh1 | neutral cholesterol ester hydrolase 1-like | −6.4654 |
| gene-Cfus20062 | cps1 | carbamoyl-phosphate synthase 1 | −6.2394 |
| gene-Cfus02255 | ankrd13d | ankyrin repeat domain 13D | −5.9840 |
| gene-Cfus15168 | chrna2 | neuronal acetylcholine receptor subunit alpha-2-like | −5.4795 |
| gene-Cfus19983 | lrrc58 | leucine rich repeat containing 58 | −5.4091 |
| gene-Cfus13163 | vmo1 | vitelline membrane outer layer protein 1 homolog | −5.2657 |
| gene-Cfus16454 | mast2 | microtubule associated serine/threonine kinase 2 | −5.0676 |
| gene-Cfus17407 | sbk1 | serine/threonine-protein kinase SBK1-like | −4.8348 |
| Pathway/ Gene Name | Gene Annotation | log2 (FC) | Up/ Down |
|---|---|---|---|
| Biosynthesis of amino acids | |||
| mat2a | Methionine adenosyltransferase 2A | 3.0678 | Up |
| mat1a | S-adenosylmethionine synthase isoform type-1 | 2.2197 | Up |
| bcat2 | Branched-chain-amino-acid aminotransferase, mitochondrial | 2.4590 | Up |
| eno3 | Enolase 3 | 2.1771 | Up |
| shmt1 | Serine hydroxymethyltransferase, cytosolic-like | 2.0500 | Up |
| srr | L-threonine ammonia-lyase-like | 2.8882 | Up |
| arg2 | Arginase-2, mitochondrial | 2.2139 | Up |
| psat1 | Phosphoserine aminotransferase 1 | −2.1117 | Down |
| phgdh | Phosphoglycerate dehydrogenase | −2.1841 | Down |
| tkt | Transketolase | −2.6636 | Down |
| cps1 | Carbamoyl-phosphate synthase 1 | −6.2394 | Down |
| Protein processing in endoplasmic reticulum | |||
| hsp90a.1 | Heat shock protein HSP 90-alpha | 2.7554 | Up |
| hspa8 | Heat shock cognate 71 kDa protein | 2.0996 | Up |
| hyou1 | Hypoxia upregulated 1 | 2.2399 | Up |
| ube2g1 | Ubiquitin-conjugating enzyme E2 G1-like | 2.1523 | Up |
| cryaa | Crystallin alpha A | 2.7842 | Up |
| ern2 | Endoplasmic reticulum to nucleus signaling 2 | 3.5579 | Up |
| dnajai | DnaJ homolog subfamily A member 1-like | 2.8502 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Tian, C.; Yang, Z.; Huang, C.; Jiao, K.; Yang, L.; Duan, C.; Zhang, Z.; Li, G. Effects of Chronic Heat Stress on Growth, Apoptosis, Antioxidant Enzymes, Transcriptomic Profiles, and Immune-Related Genes of Hong Kong Catfish (Clarias fuscus). Animals 2024, 14, 1006. https://doi.org/10.3390/ani14071006
Liu Y, Tian C, Yang Z, Huang C, Jiao K, Yang L, Duan C, Zhang Z, Li G. Effects of Chronic Heat Stress on Growth, Apoptosis, Antioxidant Enzymes, Transcriptomic Profiles, and Immune-Related Genes of Hong Kong Catfish (Clarias fuscus). Animals. 2024; 14(7):1006. https://doi.org/10.3390/ani14071006
Chicago/Turabian StyleLiu, Yong, Changxu Tian, Zhihua Yang, Cailin Huang, Kaizhi Jiao, Lei Yang, Cunyu Duan, Zhixin Zhang, and Guangli Li. 2024. "Effects of Chronic Heat Stress on Growth, Apoptosis, Antioxidant Enzymes, Transcriptomic Profiles, and Immune-Related Genes of Hong Kong Catfish (Clarias fuscus)" Animals 14, no. 7: 1006. https://doi.org/10.3390/ani14071006
APA StyleLiu, Y., Tian, C., Yang, Z., Huang, C., Jiao, K., Yang, L., Duan, C., Zhang, Z., & Li, G. (2024). Effects of Chronic Heat Stress on Growth, Apoptosis, Antioxidant Enzymes, Transcriptomic Profiles, and Immune-Related Genes of Hong Kong Catfish (Clarias fuscus). Animals, 14(7), 1006. https://doi.org/10.3390/ani14071006







