Simple Summary
Peripheral blood mononuclear cells (PBMCs) are crucial for the host’s immune system. This study analyzed the PBMCs transcriptome data from Dapulian (Chinese local breed) and Landrace (Commercial breed) pigs after stimulation with polyinosinic-polycytidylic acid. Differentially expressed genes were identified through comparative analysis, followed by gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analyses; pathways such as the MHC protein complex, the IL-17 signaling pathway and Toll-like receptor signaling pathway were enriched, alongside key immune response-related genes in Dapulian pig, such as CXCL14, IL1R1, IL33, and SLA-DMA. Furthermore, an miRNA–mRNA regulatory network was constructed to elucidate the immune response in Dapulian pig. These findings highlight the transcriptomic differences between pig breeds and offer insights into the immune response regulation of antiviral responses in indigenous and modern commercial pigs.
Abstract
There is a consensus that indigenous pigs in China are more resistant than modern commercial pigs in terms of disease resistance. Generally, the immune response is an important part of anti-disease capability; however, the related mechanism in pigs is largely puzzling. Here, the public transcriptome data of peripheral blood mononuclear cells (PBMCs) from Dapulian (Chinese local breed) and Landrace (Commercial breed) pigs after stimulation with polyinosinic-polycytidylic acid (poly I:C, a conventional reagent used for simulation of the viral infection) were reanalyzed, and the immune response mechanism in different pig breeds was investigated from a transcriptomic perspective. Of note, through comparative analyses of Dapulian and Landrace pigs, the candidate genes involved in swine broad-spectrum resistance were identified, such as TIMD4, RNF128 and VCAM1. In addition, after differential gene expression, target gene identification and functional enrichment analyses, a potential regulatory network of miRNA genes associated with immune response was obtained in Dapulian pigs, including five miRNAs and 12 genes (such as ssc-miR-181a, ssc-miR-486, IL1R1 and NFKB2). This work provides new insights into the immune response regulation of antiviral responses in indigenous and modern commercial pigs.
1. Introduction
Pigs are one of the most prevalent domestic animals around the world. Domestic pig (Sus scrofa domesticus) was domesticated by humans from wild boar in Europe and Asia around 10,000 years ago [1]. Over the past few decades, with the development of intensive breeding technology, large-scale group breeding has become a model for the development of modern animal husbandry, which has resulted in a series of immune problems [2,3]. In the pig industry, diseases caused by viral or bacterial pathogens exert a limitation on growth efficiency, and infectious diseases usually cause huge economic losses, and seriously restrict the development of the industry, such as African swine fever (ASF) [4,5]. As is well known, the indigenous pig breeds in China have better disease resistance than commercial pig breeds [6]. Thus, a deeper understanding of the comprehensive regulatory network that controls disease resistance in pigs has become an urgent research topic in farm animal husbandry.
In pigs, disease resistance is usually composed of two aspects, the general and the specific anti-disease ability. The former refers to the pathogen resistance of pigs, that is, the collective comprehensive defense ability against various pathogens, and it mainly involves immune factors and immune cells in the body. The latter refers to the specific resistance of pigs to specific diseases or pathogens, and it depends on the body’s immune response. Peripheral blood mononuclear cells (PBMCs), belonging to the first defense line of the immune system, contain a wide range of immune cells, and play an integral part in the rapid response of the immune system during bacterial or viral infections [7,8]. Numerous studies have shown that PBMCs are suitable for modeling the disease pathogenesis and basis genetics underlying infectious or irritant diseases [9,10,11]. Studying the transcriptional differences in PBMCs of pigs is beneficial to understanding the genetic basis of resistance to disease.
Polyinosinic-polycytidylic acid (poly I:C) is a synthetic viral double-stranded RNA (dsRNA), which can be recognized by Toll-like receptor 3, induces IFN production, and can be used to simulate viral infection in experiments [12,13]. Poly I:C can also activate IPS-1- and TRIF-dependent pathways, leading to the activation of NK cells, which are natural immune system effector cells and mainly responsible for immune surveillance, and play an important part in preventing viral infections [14]. Poly I:C has widespread applications in experimental research of viral infection. Christopher K. Tuggle utilized poly I:C and lipopolysaccharide (LPS) to stimulate porcine alveolar macrophages (PAMs), revealing that non-stimulated and stimulated macrophages differ in terms of gene expression influenced by changes in chromatin accessibility at active regulatory regions, primarily mediated by H3K27ac modifications [15]. Mei conducted miRNA sequencing with poly I:C-stimulated and porcine reproductive and respiratory syndrome virus (PRRSV)-infected PAMs, suggesting that, compared with the poly I:C group, PRRSV slightly altered the miRNA expression profile, and this study confirmed that PRRSV is able to silence the host innate immune response at the early phase of infection [16].
RNA-seq has evolved into a major tool for studying molecular biology since its inception. With the advancement of analysis protocols, it has become possible to study genetic changes between and within species [17,18]. In pig, it has revealed that the PBMC genes’ expression profiles were altered by Poly I:C in different pig breeds, and some genes that changed in the two breeds were involved in immune-related functions, which also play an important role in tissue development [18]. Here, we focus on the immune response of pigs from the transcriptomic aspect. The porcine immune response-related genes were obtained through differential gene expression comparative analysis; after the target genes’ identification and functional enrichment, the potential miRNA–mRNA regulatory network of immune response in Dapulian pig was constructed.
2. Materials and Methods
2.1. Data Collection
In the study, the transcriptome data of PBMCs from Dapulian and Landrace pigs were collected (control group of Dapulian (DC), control group of Landrace (LC), treatment group of Dapulian (DT), treatment group of Landrace (LT)) from public reports [18,19]. Six 5-week-old piglets were selected for blood sampling; all pigs were raised in the same facility and each group had three replicates. PBMCs treated with and without poly I:C infection were cultured for 24 h and collected for sequencing. All raw transcriptome data have been downloaded from the NCBI public database, and the transcriptome data accession numbers are as follows: mRNA—PRJNA301538 and miRNA—PRJNA308253.
2.2. Quality Control of Raw Data
To ensure the consistency of the processing results, the standard procedures of data preprocessing were adopted [20,21]. Firstly, raw RNA-seq data quality were assessed using FastQC (v0.11.9) [22]. The unqualified data, low-quality data and sequencing adapters were removed using the default parameters of FASTP (v0.22.0), and the obtained clean data were secondly checked by FastQC, then the high-quality data were adapted in subsequent data analyses [23].
2.3. Mapping to the Reference Genome
The porcine reference genome (GCF_000003025.6) was downloaded from the NCBI database and the high-quality data were aligned to the reference genome using STAR (v2.6.1b) software; the uniquely mapped reads were assembled into genes by utilizing the Ensembl Susscrofa annotation file (Sus_scrofa.Sscrofa11.1.111) [24]. The gene expression matrix was then constructed using featureCounts (v1.6.3) software [25]. For miRNA data, the Subjunc (v1.6.3) software was used for reads alignment, and the transcript expression matrix was generated and quantified using the featureCounts (v1.6.3) software [26]. In addition, the exonic regions were assigned for read counts, with the gene_id attribute serving as the unique identifier to aggregate exons belonging to the same genes. Fragments per kilobase of exon model per million mapped fragments (FPKM) were then calculated using a custom R script, which was applied for gene expression assessment.
2.4. Principal Components Analysis (PCA) and Sample Correlation Analysis
The scale function of R was used for normalization, and the prcomp function computes the principal components of standardized gene count by constructing a correlation matrix. For numerical stability, a singular value decomposition (SVD) is employed rather than the Eigen decomposition approach, and ggplot2 (R package, v3.4.3) is used for visualization. The cor function in the R environment was used to calculate the correlation values between samples, and pheatmap (R package, v1.0.12) was used for visualization.
2.5. Identification of Differentially Expressed Genes
To obtain differentially expressed gene and miRNA information, DESeq2 (R package, v1.38.3) was applied, a typical differential analysis tool that employs the shrinkage estimation of dispersion and fold changes to enhance the stability and interpretability of results. The analysis begins with a count matrix K, where each row i corresponds to a gene, each column j represents a sample, and each entry Kij denotes the number of sequencing reads unambiguously mapped to gene i in sample j [27]. In the results, the fold change of each gene was calculated, as well as the false discovery rate-corrected p-value, and the genes with |Log2(fold change)| > 1 and −log10(p-value) > 1.3 were regarded as differentially expressed genes (DEGs), while |Log2(fold change)| > 0 and −log10(p-value) > 1.3 are considered to be the threshold of differentially expressed miRNAs (DEMs); the results are displayed in volcano plots.
2.6. Gene Functional Enrichment
With gene symbol as the input, the functional annotation of the target gene set was performed using clusterProfiler (R package, v4.6.2) [28], and the terms were considered significant with a p-value < 0.05, plotted using ggplot2 (R package, v3.4.3) and enrichplot (R package, v1.18.4).
2.7. Network Analysis of Protein-Protein Interaction (PPI)
The PPI network of Landrace immune response-related genes in Supplementary Figure S1 was performed using the string (v12.0) online URL https://string-db.org/; the minimum required interaction score is 0.900, and the meaning of network edges represents confidence, with line thickness indicating the strength of data support.
2.8. Target Genes of miRNA Prediction
To analyze the targeting relationship between miRNAs and genes, the miRanda database was used for target gene predictions of miRNA. Firstly, the sequence alignment was performed based on sequence complementarity, followed by calculating the minimum free energy (MFE) score for each selected miRNA–mRNA pair, and the pairs with MFE scores exceeding the predefined threshold are finally selected as potential targets [29]. Subsequently, the g:Profiler tool (https://biit.cs.ut.ee/gprofiler/gost, accessed on 22 July 2024) was used to convert the gene names.
3. Results
3.1. The Different Transcriptome Modes of PBMCs with and Without Poly I:C Treatment in Pig
To explore the molecular mechanisms of immune response in native and modern commercial pigs, we collected the public transcriptome data of PBMCs from Dapulian and Landrace pigs, divided into four groups according to the poly I:C infection (treatment group of Dapulian (DT), treatment of Landrace (LT)) and not (control of Dapulian (DC), control of Landrace (LC)), and the underlying transcriptome dynamics were investigated (Figure 1A). Firstly, we performed principal component analysis and correlation analysis with all the investigated samples, and found that the PBMC samples from different pig species could be clearly separated (Figure 1B), while the control group was different from that of the poly I:C-infected group (Figure 1C and Figure S1A,B).
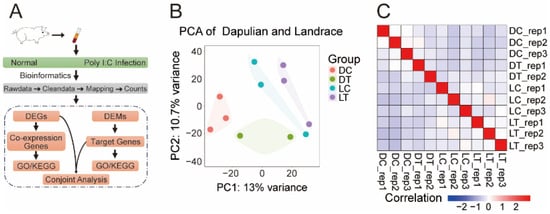
Figure 1.
Transcriptome analysis of PBMCs from Dapulian and Landrace pigs in normal pigs and with poly I:C infection. (A) The technical route of data analysis in this study; (B) PCA of transcriptome data in four groups of Dapulian and Landrace; (C) correlation analysis of the PBMCs samples between Dapulian and Landrace from normal and poly I:C-infected groups.
3.2. The DEGs Identification of PBMCs from Different Groups
To obtain insights into the effects of poly I:C on the transcriptome characteristics of PBMCs in pig, a differential analysis was performed. A total of 757 DEGs were generated in Dapulian pigs, including 607 upregulated and 150 downregulated genes (Figure 2A and Figure S1C and Supplementary Table S1). A total of 128 DEGs were obtained in Landrace; 55 genes were increased and 73 declined (Figure 2B and Figure S1D and Table S1).
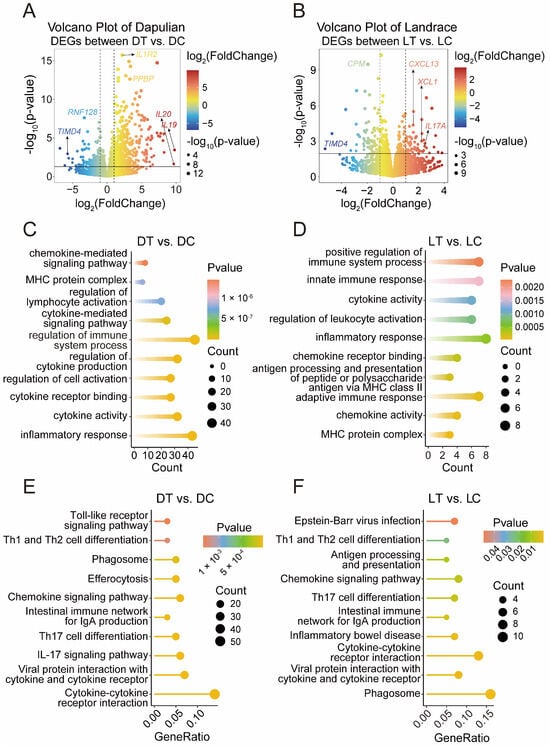
Figure 2.
DEGs analysis of PBMCs from control and treatment groups. (A) Volcano plot of DEGs between DT and DC, where red represents upregulated genes, blue represents downregulated genes, and marker genes are related to immune function. (B) Volcano plot of DEGs between LT and LC, where red represents upregulated genes, blue represents downregulated genes, and marker genes are related to immune function. (C) GO enrichment terms of DEGs in Dapulian. (D) GO enrichment pathways of DEGs in Landrace. (E) KEGG enrichment terms of DEGs in Dapulian. (F) KEGG enrichment pathways of DEGs in Landrace.
Functional enrichment analysis was then performed using gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG). For the DEGs in Dapulian pigs, the GO enrichment analysis showed they were enriched in immunological functions, such as the processes of inflammatory response, cytokine activity, the regulation of immune system processes, the regulation of lymphocyte activation, and MHC protein complex (Figure 2C and Table S2), and in the result of KEGG enrichment analysis, these DEGs were also found to be related with immunological pathways, including viral protein interactions with cytokine and cytokine receptor, the IL-17 signaling pathway, the intestinal immune network for IgA production, the chemokine signaling pathway, and the Toll-like receptor signaling pathway (Figure 2E and Table S2). Regarding the DEGs in Landrace, they were involved in immune-related functions, such as the MHC protein complex, adaptive immune response, inflammatory response, innate immune response, and the positive regulation of immune system processes (Figure 2D and Table S2), and KEGG enrichment analysis showed that they were also enriched in immune system-related pathways, such as Viral protein interaction with the cytokine and cytokine receptor, cytokine–cytokine receptor interaction, the intestinal immune network for IgA production, the chemokine signaling pathway, and antigen processing and presentation (Figure 2F and Table S2).
3.3. Key Regulatory Genes of Immune Response in Pigs
In order to identify the key regulatory genes of the immune response when pigs are infected by poly I:C, we found a total of 40 DEGs that were differentially expressed both in Dapulian and Landrace pigs (Figure 3A and Table S3), such as VCAM1, CLDN4 and C1S.
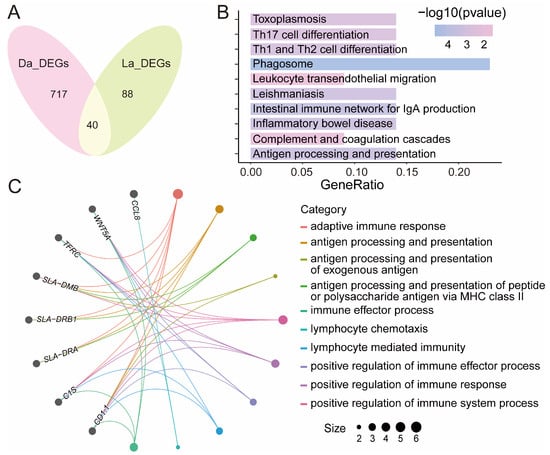
Figure 3.
Identification of key regulatory genes of immune response in pigs. (A) Venn diagram of overlapping DEGs in Dapulian and Landrace. (B) GO enrichment analysis of co-expressed DEGs between Dapulian and Landrace. (C) KEGG enrichment of co-expressed DEGs between Dapulian and Landrace.
Then, GO enrichment analysis found that these genes were also enriched in immune response-related pathways, such as adaptive immune response, antigen processing and presentation, antigen processing and presentation of the peptide or polysaccharide antigen via MHC class II, antigen processing and the presentation of exogenous antigen, and the immune effector process (Figure 3B and Table S3). In addition, these DEGs were also subjected to KEGG enrichment analysis, and it revealed that VCAM1 and CLDN4 were involved in the pathway of leukocyte transendothelial migration, C1S and C1R were related with complement and coagulation cascades, TFRC plays a role in phagosome process, and SLA-DMB, SLA-DRB1 and SLA-DRA were enriched in terms of the intestinal immune network for IgA production, antigen processing and presentation, Th1 and Th2 cell differentiation, and Th17 cell differentiation, etc. (Figure 3C and Table S3).
3.4. Functional Enrichment of Genes Specifically Expressed in Dapulian and Landrace
To explore the functions of DEGs specifically expressed in Dapulian and Landrace, we performed functional enrichment analyses on 717 and 88 DEGs unique to each species, respectively (Table S4).
Through the GO enrichment analysis of 717 DEGs specifically in Dapulian, we found that the inflammatory response, cytokine activity, cytokine receptor binding, regulation of leukocyte activation, and regulation of immune system process were enriched (Figure 4A). KEGG analysis enriched cytokine–cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, the IL-17 signaling pathway, inflammatory bowel disease, and Th17 cell differentiation pathways (Figure 4B).
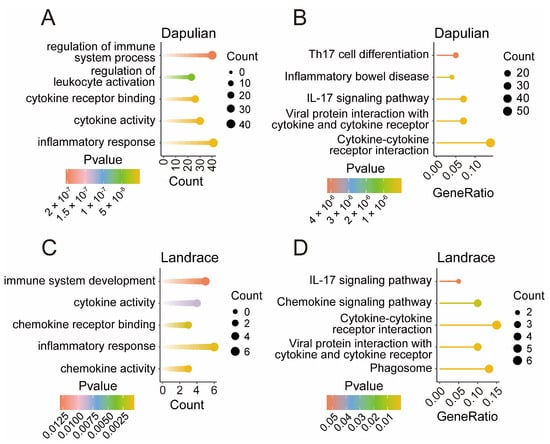
Figure 4.
Functional enrichment of genes specifically expressed in Dapulian and Landrace. (A) GO enrichment analysis of specific DEGs in Dapulian. (B) KEGG enrichment analysis of specific DEGs in Dapulian. (C) GO enrichment analysis of specific DEGs in Landrace. (D) KEGG enrichment analysis of specific DEGs in Landrace.
Similarly, the GO enrichment analysis of 88 DEGs specifically in Landrace revealed chemokine activity, inflammatory response, chemokine receptor binding, cytokine activity, and immune system development (Figure 4C). KEGG analysis enriched phagosome, viral protein interaction with cytokine and cytokine receptors, cytokine–cytokine receptor interaction, the chemokine signaling pathway, and the IL-17 signaling pathways (Figure 4D).
3.5. Differential Expression Analysis of miRNA in Different Groups
In addition, to characterize the changes of miRNA in control and treatment groups, a differential expression analysis of miRNA was conducted between DT and DC, as well as LT and LC groups (Table S5). A total of 10 DEMs in Dapulian were obtained (Figure 5A)—8 DEMs (such as miR855-5p, miR1-3p and miR208b) were upregulated and 2 were downregulated (i.e., miR486 and miR181a). In Landrace, only miR1249 was increased and miR192 was decreased (Figure 5B).
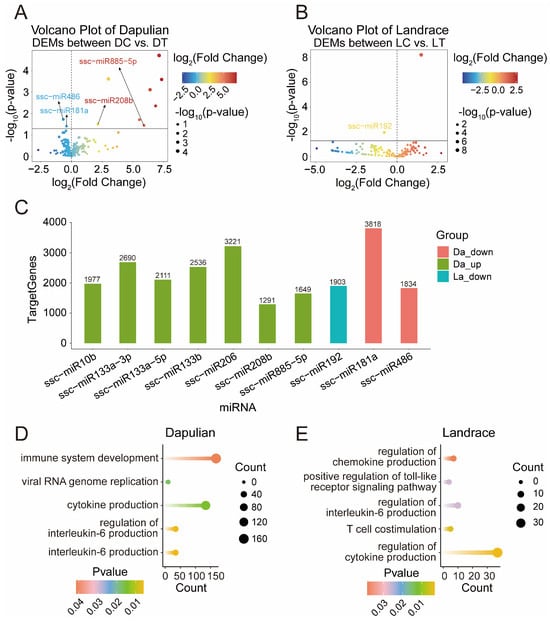
Figure 5.
Analysis of DEMs in the PBMCs control group and treatment group. (A) Volcano plot of DEMs between DT and DC; the labeled miRNAs are the miRNAs included in the potential regulatory network of miRNA–mRNA below. (B) Volcano plot of DEMs between LT and LC. (C) Number of predicted target genes of DEMs in Dapulian and Landrace; the horizontal axis is the miRNA and the vertical axis is the number of target genes predicted by DEMs. (D) GO enrichment analysis of DEM target genes in Dapulian. (E) GO enrichment analysis of DEM target genes in Landrace.
To identify miRNA target genes and functions, we used the miRanda database to predict the target genes of DEMs (Figure 5C and Table S5). The GO enrichment showed that the target genes regulated by DEMs in Dapulian were enriched in terms of interleukin-6 production, cytokine production, viral RNA genome replication and immune system development (Figure 5D and Table S5). In addition, the target genes regulated by DEMs in Landrace were enriched in the process of regulation of cytokine production, T cell costimulation, the regulation of interleukin-6 production, the positive regulation of the toll-like receptor signaling pathway and the regulation of chemokine production pathways (Figure 5E and Table S5).
3.6. Construction of Potential Regulatory Network of miRNA-mRNA Regard to Immune Response in Dapulian Pig
Given the interaction of genes and miRNA, the potential regulatory network of miRNA and genes was constructed in relation to the immune responses of pigs, which were based on the DEGs and DEMs in Dapulian. the expression levels of DEGs and DEMs were opposite to one another, because they are theoretically negatively regulated. We derived 73 miRNA–gene pairs (Figure 6A and Table S6).
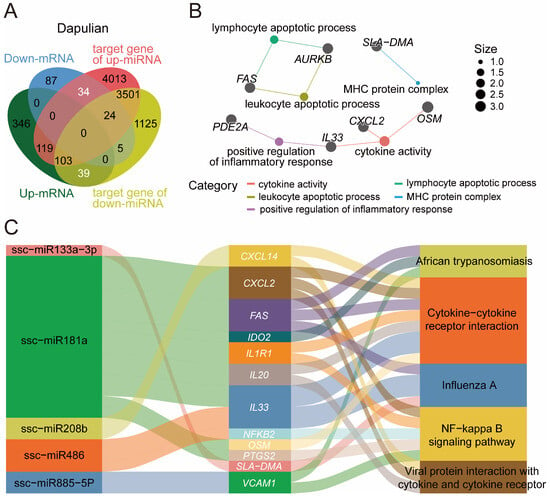
Figure 6.
Construction of a potential regulatory network of miRNA–mRNA in the immune response of Dapulian pigs. (A) Venn diagram of DEGs and DEMs in Dapulian. (B) GO enrichment analysis of downregulated DEGs, upregulated DEM target genes, upregulated DEGs, and downregulate DEM target genes in Dapulian. (C) Targeting relationship between miRNA and genes in Dapulian.
Firstly, regarding these 73 genes, we found they were enriched in lymphocyte apoptotic process, leukocyte apoptotic process, positive regulation of inflammatory response, cytokine activity and MHC protein complex (Figure 6B and Table S6). In addition, based on the regulatory effects of miRNA on predicted targeting genes, we constructed the potential regulatory network of pairing miRNA–mRNA for the immune response process in Dapulian pigs. In the results, it was shown that ssc-miR133a-3p may regulate the SLA-DMA gene, and they were enriched in the Influenza A pathway. CXCL2, FAS, IDO2, IL1R1, IL20, IL33, OSM and PTGS2 genes may be regulated by ssc-miR181a, and are enriched in the NF-kappa B signaling pathway, cytokine-cytokine receptor interaction, African trypanosomiasis, viral protein interaction with cytokine and cytokine receptor, and the Influenza A pathway. Furthermore, we have discovered the targeted interactions between ssc-miR208b and CXCL2, ssc-miR486 and IL33 and NFKB2, and ssc-miR885-5p and VCAM1. These genes are implicated in pathways that play a role in immune modulation (Figure 6C and Table S6).
4. Discussion
The environmental challenges brought about by the large-scale farming technology make disease resistance more and more important in pig production practice. The Dapulian pig is a local Chinese breed and has stronger disease resistance than commercial pigs [18]. Indeed, during the period of PRRSV infection, Duroc × Landrace × Yorkshire (DLY) pigs show a number of clinical features that are typical of the disease, while Dapulian pigs only show mild disease symptoms [30]. Here, the difference in immune response was investigated in Dapulian and Landrace pig based on the transcriptomic data of PBMCs from normal and Poly I:C-treated groups, and the miRNA–gene regulatory network potentially associated with immune response in Dapulian pig was constructed and assessed.
Firstly, the immune response of viral infection in pigs was investigated from the aspect of PBMCs transcriptomic data. Through gene differential expression analysis, 757 DEGs in Dapulian (such as TIMD4, RNF128 and IL1R2) were obtained, and 128 DEGs in Landrace (such as TIMD4, CPM and CXCL13) were related to immune response. Among these genes, XCL1 and CXCL13 are chemokines that inhibit the apoptosis of immune cells during viral infection and play an important part in inflammatory response and immune regulation [31,32,33]. IL1R2 is an anti-inflammatory antagonist that can prevent immune disorders and systemic inflammation caused by IL1. It is naturally present on neutrophils, B cells, monocytes and macrophages, and is associated with various inflammatory diseases, such as arthritis and ulcerative colitis [34]. The TIMD4 gene is differentially expressed both in Dapulian and Landrace pigs. As a member of the cell immunoglobulin domain and mucin domain (TIM) gene family, it is a phosphatidylserine receptor, and plays an integral role in the immune response [35]. Gao et al. showed that TIMD4 can be inhibited by macrophages in vitro and prevent the production of cytokines through the NF-κB signaling pathway [36]. In addition, RNF128, also known as GRAIL, is an E3 ubiquitin ligase, which is rapidly activated to be expressed in anergic T cells when infections happen [37]. Gao et al. showed that virus infection induces RNF128 expression, which interacts with TBK1 via E3 ubiquitin ligase, enhancing K63-linked ubiquitination in response to RNA or DNA viruses. A deficiency in RNF128 impairs the phosphorylation of IRF3, IFN-β production, K63-linked ubiquitination, and TBK1 activation [38]. Tian et al. also found that RNF128 can inhibit the inflammatory response as a negative regulator of IL-3/STAT5 signaling. In addition, RNF128 facilitates the lysosomal degradation of IL-3Rα by catalyzing its K27-linked polyubiquitination in a ligase activity-dependent fashion. This finding also confirmed that RNF128 could inhibit macrophage activation and chemotaxis, thereby reducing excessive inflammatory response [39].
Specially, from the difference in the results regarding immune response genes, we may obtain some clues regarding the higher disease resistance of Dapulian pigs compared to western commercial pig breeds. Studies have shown that the superior immune function of Dapulian pigs compared to commercial pigs may be associated with the expressions of genes such as CXCL8; upon stimulation with oligodeoxynucleotides containing unmethylated CpG motifs (CpG ODN), the mRNA levels of the cytokine IL-8 (CXCL8) in the PBMCs of Dapulian pigs were significantly higher than those observed in Landrace pigs [40]. Hui Wang et al. also found that the expression levels of ADAM8 and CD59 genes in whole blood were significantly lower in Landrace compared to Dapulian piglets at the neonatal stage [41]. In our results, the CXCL8, ADAM8, and CD59 genes were upregulated in DT vs. DC groups, but showed no significant differences in LT vs. LC groups. This further supports the hypothesis that these three genes may be associated with the higher immunity observed in Dapulian pigs. In addition, our results reveal that ISG20 and IRF1 were upregulated in the DT vs. DC group, but their expressions showed no significant differences in the LT vs. LC group. The ISG20 gene, known as an interferon-stimulated gene (ISG), exhibits increased expression levels consistent with the findings of Bang Liu et al. Furthermore, IRF1 is a member of the interferon regulatory factors (IRFs) family; PRRSV can suppress the expression of antiviral genes by disrupting IRF1 gene transcription [42,43]. Therefore, the differential expressions of ISG20 and IRF1 genes may be one of the reasons for the varying immune responses observed between Dapulian and Landrace pigs.
The differential expressions suggest some miRNAs may be involved in the immune responses in pig, such as ssc-miR486, ssc-miR-181a, ssc-miR-885-5p, ssc-miR208b and ssc-miR192. As reported, the differential expressions of miRNA abundance in lung tissues of PCV2 (porcine circovirus type 2)-infected and PCV2-uninfected LW (Laiwu, a Chinese indigenous pig) and YL (Yorkshire × Landrace) pigs were investigated, and the qRT-PCR validation revealed that there were significant differences in the expressions of ssc-miR486 and ssc-miR-192 before and after infection [44]. Another study found that increasing miR-181a expression in mature T cells enhanced their sensitivity to peptide antigens, whereas inhibiting expression in immature T cells reduced sensitivity [45]. Also, ssc-miR-181a was differentially expressed in the duodenum of E. coli F18-sensitive and -resistant weaned piglets, which was verified by qRT-PCR [46]. Moreover, ssc-miR-885-5p was found to play an immune role in PDCoV (porcine del-tacoronavirus) infection in pigs through competitively binding mRNAs with lncRNA [47]; miR-208b is capable of inhibiting type I IFN signaling in HCV (Hepatitis C vi-rus)-infected hepatocytes, and the inhibition of miR-208b with miRNA inhibitors during HCV infection increased the expression of IFNAR1 (interferon alpha and beta receptor subunit 1). In our results, ssc-miR208b expression was significantly different in the DC and DT groups, which may influence the immune reaction in Dapulian pigs [48].
In addition, a potential regulatory network of miRNA-mRNA in the immune response process was constructed in Dapulian, which included five miRNAs and 12 genes. Among them, CXCL2 and CXCL14 are chemokines, whose expression levels change when they are infected with a virus [49]. Interleukin 20 (IL-20) and interleukin 33 (IL-33) are both cytokines. IL-33 is a tissue-derived nuclear cytokine that belongs to the IL-1 family of cytokines, which acts as a trigger for alarm signals during cell damage or tissue damage, and sends a warning to immune cells expressing the ST2 receptor (IL-1RL1) [50]. VCAM1 is a cell adhesion molecule, and IL1R1 is an IL-1 receptor; they play an essential role in inflammation and immunity [51,52]. The MHC gene is closely related to the immune system and called swine leukocyte antigen (SLA) in pigs [53,54]. A study conducted on two pig breeds found that the SLA gene has a certain connection with porcine reproductive and respiratory syndrome, porcine pseudorabies and mycoplasma pneumonia [55]. It is suggested that these proteins are persistently expressed on the surfaces of almost every nucleated cell, serving to present peptides to CD8+ cytotoxic T cells and to interface with natural killer (NK) cells [56]. Some studies have also found that SLA haplotypes are not only related to pre-weaning mortality caused by diarrhea, but also differ in relation to parasite resistance [57,58].
Furthermore, there were certain defects and limitations in the study. In one aspect, the transcriptome data of PBMCs were derived from six 5-week-old piglets of Landrace (two male and one female) and Dapulian (one male and two female) breeds; each group included three repeats and the number of experiment animals was relatively limited. One repeat from the DT group was eliminated because of sample discretion, thus the results were weak from the point of view of statistical significance. Besides this, the results of this study were obtained via data analysis, and lack experimental verification. Further experiments are still required to confirm and strengthen the conclusions of this study.
5. Conclusions
In this study, based on a comparative analysis of transcriptome data in Dupulian pigs with and without poly I:C stimulation, the candidate genes associated with disease resistance, such as TIMD4, RNF128, and IL1R2, were highlighted, and the miRNA–gene regulatory network related to the immune response of Dapulian pigs was built. This study not only found changes in immune-related genes in Dapulian and Landrace pigs after virus infection, but also explored the specific changes in immune-related genes in Daplian, providing a new scientific basis for the future breeding of pig disease resistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani14233546/s1, Figure S1: Sample correlation and DEGs in Dapulian and Landrace, and immune-related genes specific to Landrace. Table S1: DEGs in Dapulian and Landrace. Table S2: GO and KEGG of DEGs in Dapulian and Landrace. Table S3: Shared DEGs between Dapulian and Landrace, and enrichment analysis of these DEGs. Table S4: DEGs specifically expressed in Dapulian and Landrace, and enrichment analysis of these DEGs. Table S5: Analysis of DEMs in Dapulian and Landrace, and enrichment analysis of their target genes. Table S6: Key immune response-related genes in Dapulian and functional enrichment analysis.
Author Contributions
Conceptualization, J.W. and W.S.; data curation, J.W. and T.W.; funding acquisition, J.W. and W.S.; project administration, J.W. and W.S.; resources, S.Z., M.Z., X.Z., W.S. and J.W.; supervision, J.W. and W.S.; writing—review and editing, J.W. and W.S.; formal analysis, T.W.; investigation, T.W., S.Z., M.Z. and X.Z.; visualization, J.W., T.W. and Z.T.; writing—original draft, T.W.; methodology, Z.T. and M.Y.; software, Z.T. and M.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Natural Science Foundation of Shandong Province (ZR2021QC041), the Key Research and Development Program of Shandong Province (2022LZGC003 and 2021LZGC001), the Start-up Fund for High-level Talents of Qingdao Agricultural University (6651122011), and Taishan Scholar Construction Foundation of Shandong Province (ts20190946).
Institutional Review Board Statement
Not applicable. Because the data we used were downloaded from public databases and this study did not involve animal experiments.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Larson, G.; Dobney, K.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef] [PubMed]
- Prunier, A.; Heinonen, M.; Quesnel, H. High physiological demands in intensively raised pigs: Impact on health and welfare. Animal 2010, 4, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Cobanovic, N.; Magrin, L. Editorial: Health and welfare problems of farm animals: Prevalence, risk factors, consequences and possible prevention solutions. Front. Vet. Sci. 2023, 10, 1238852. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Siengsanan-Lamont, J.; Halasa, T.; Young, J.R.; Ward, M.P.; Douangngeun, B.; Theppangna, W.; Khounsy, S.; Toribio, J.; Bush, R.D.; et al. The impact of African swine fever virus on smallholder village pig production: An outbreak investigation in Lao PDR. Transbound. Emerg. Dis. 2021, 68, 2897–2908. [Google Scholar] [CrossRef]
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef]
- Qin, M.; Li, C.; Li, Z.; Chen, W.; Zeng, Y. Genetic diversities and differentially selected regions between Shandong indigenous pig breeds and western pig breeds. Front. Genet. 2020, 10, 1351. [Google Scholar] [CrossRef]
- Kofanova, O.; Bellora, C.; Quesada, R.A.; Bulla, A.; Panadero-Fajardo, S.; Keipes, M.; Shea, K.; Stone, M.; Lescuyer, P.; Betsou, F. IL8 and EDEM3 gene expression ratio indicates peripheral blood mononuclear cell (PBMC) quality. J. Immunol. Methods 2019, 465, 13–19. [Google Scholar] [CrossRef]
- Bolen, C.R.; Uduman, M.; Kleinstein, S.H. Cell subset prediction for blood genomic studies. BMC Bioinform. 2011, 12, 258. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, J.; Liu, K.; Gao, L.; Wu, H.; Ma, L.; Zhou, J.; Liu, Z.; Han, J.-D.J. Human PBMC scRNA-seq–based aging clocks reveal ribosome to inflammation balance as a single-cell aging hallmark and super longevity. Sci. Adv. 2023, 9, eabq7599. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Zhong, Y.; Chen, X.; Li, Z.; Li, R.; Qin, P.; Wang, S.; Yin, J.; Liu, S. Pan-cancer scRNA-seq analysis reveals immunological and diagnostic significance of the peripheral blood mononuclear cells. Human Mol. Genet. 2024, 33, 342–354. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Zhang, J.; Zhang, Z.; Guo, H.; Wei, D.; Wu, C.; Hai, T.; Sun, H.-X.; Zhao, Y. Single-cell transcriptomic analysis reveals transcriptional and cell subpopulation differences between human and pig immune cells. Genes Genom. 2024, 46, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Fortier, M.E.; Kent, S.; Ashdown, H.; Poole, S.; Boksa, P.; Luheshi, G.N. The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 287, R759–R766. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Seya, T. TLR3: Interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv. Rev. 2008, 60, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Purdy, A.K.; Campbell, K.S. Natural killer cells and cancer. Regulation by the killer cell Ig-like receptors (KIR). Zhongguo Fei Ai Za Zhi 2010, 13, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Uribe, J.; Liu, H.; Byrne, K.A.; Bond, Z.F.; Loving, C.L.; Tuggle, C.K. Changes in H3K27ac at Gene Regulatory Regions in Porcine Alveolar Macrophages Following LPS or PolyIC Exposure. Front. Genet. 2020, 11, 817. [Google Scholar] [CrossRef]
- Wu, J.; Ji, Z.; Qiao, M.; Peng, X.; Wu, H.; Song, Z.; Zhao, H.; Liu, G.; Li, F.; Mei, S. MicroRNA transcriptome analysis of poly I:C-stimulated and PRRSV-infected porcine alveolar macrophages. J. Appl. Genet. 2019, 60, 375–383. [Google Scholar] [CrossRef]
- Zhang, F.-L.; Zhang, S.-E.; Sun, Y.-J.; Wang, J.-J.; Shen, W. Comparative transcriptomics uncover the uniqueness of oocyte development in the donkey. Front. Genet. 2022, 13, 839207. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Wang, H.; Wang, H.; Liu, J.-F.; Wu, Y.; Guo, J. Transcriptomic analysis identifies candidate genes and gene sets controlling the response of porcine peripheral blood mononuclear cells to poly I: C stimulation. G3 Genes Genomes Genet. 2016, 6, 1267–1275. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Wang, H.; Guo, J.; Wang, H.; Wu, Y.; Liu, J. MicroRNA Transcriptome of Poly I:C-Stimulated Peripheral Blood Mononuclear Cells Reveals Evidence for MicroRNAs in Regulating Host Response to RNA Viruses in Pigs. Int. J. Mol. Sci. 2016, 17, 1601. [Google Scholar] [CrossRef]
- Zhang, F.L.; Zhu, W.M.; He, T.R.; Zhao, Y.T.; Ge, W.; Tan, J.H.; Shen, W. Comparative transcriptomic analysis reveals that TPX2 and AURXA are involved in porcine PCV2 infection. Gene 2022, 834, 146649. [Google Scholar] [CrossRef]
- Yu, M.; Feng, Y.; Yan, J.; Zhang, X.; Tian, Z.; Wang, T.; Wang, J.; Shen, W. Transcriptomic regulatory analysis of skeletal muscle development in landrace pigs. Gene 2024, 915, 148407. [Google Scholar] [CrossRef] [PubMed]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Wang, S.; Kim, J.; Jiang, X.; Brunner, S.F.; Ohno-Machado, L. GAMUT: GPU accelerated microRNA analysis to uncover target genes through CUDA-miRanda. BMC Med. Genom. 2014, 7 (Suppl. S1), S9. [Google Scholar] [CrossRef]
- Xing, J.; Xing, F.; Zhang, C.; Zhang, Y.; Wang, N.; Li, Y.; Yang, L.; Jiang, C.; Zhang, C.; Wen, C.; et al. Genome-wide gene expression profiles in lung tissues of pig breeds differing in resistance to porcine reproductive and respiratory syndrome virus. PLoS ONE 2014, 9, e86101. [Google Scholar] [CrossRef]
- Song, G.; Zhang, Y.; Gao, H.; Fu, Y.; Chen, Y.; Yin, Y.; Xu, K. Differences in Immune Characteristics and Related Gene Expression in Spleen among Ningxiang, Berkshire Breeds and Their Hybrid Pigs. Genes 2024, 15, 205. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, Y.; Jiang, S.; Sui, M.; Wang, C.; Kang, L.; Sun, Y.; Jiang, Y. Suppression of lymphocyte apoptosis in spleen by CXCL13 after porcine circovirus type 2 infection and regulatory mechanism of CXCL13 expression in pigs. Vet. Res. 2019, 50, 17. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Takahama, Y. XCL1 and XCR1 in the immune system. Microbes Infect. 2012, 14, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Peters, V.A.; Joesting, J.J.; Freund, G.G. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav. Immun. 2013, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Tang, Z.L.; Yang, S.; Cao, J.Y.; Li, K. Molecular characteristics of the porcine TIMD4 gene and its association analysis. Biochem. Genet. 2012, 50, 538–548. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, P.; Xu, Y.; Gao, L.; Wang, H.; Jia, X.; Ma, H.; Liang, X.; Ma, C.; Gao, L. Tim-4 protects mice against lipopolysaccharide-induced endotoxic shock by suppressing the NF-kappaB signaling pathway. Lab. Investig. 2016, 96, 1189–1197. [Google Scholar] [CrossRef]
- Anandasabapathy, N.; Ford, G.S.; Bloom, D.; Holness, C.; Paragas, V.; Seroogy, C.; Skrenta, H.; Hollenhorst, M.; Fathman, C.G.; Soares, L. GRAIL: An E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 2003, 18, 535–547. [Google Scholar] [CrossRef]
- Song, G.; Liu, B.; Li, Z.; Wu, H.; Wang, P.; Zhao, K.; Jiang, G.; Zhang, L.; Gao, C. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat. Immunol. 2016, 17, 1342–1351. [Google Scholar] [CrossRef]
- Yu, J.; Li, J.; Shen, A.; Liu, Z.; He, T.S. E3 ubiquitin ligase RNF128 negatively regulates the IL-3/STAT5 signaling pathway by facilitating K27-linked polyubiquitination of IL-3Ralpha. Cell Commun. Signal 2024, 22, 254. [Google Scholar] [CrossRef]
- Hu, J.; Yang, D.; Wang, H.; Li, C.; Zeng, Y.; Chen, W. CpG Oligodeoxynucleotides Induce Differential Cytokine and Chemokine Gene Expression Profiles in Dapulian and Landrace Pigs. Front. Microbiol. 2016, 7, 1992. [Google Scholar] [CrossRef]
- Hu, J.; Yang, D.; Chen, W.; Li, C.; Wang, Y.; Zeng, Y.; Wang, H. Whole Blood Transcriptome Sequencing Reveals Gene Expression Differences between Dapulian and Landrace Piglets. BioMed Res. Int. 2016, 2016, 7907980. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.; Wang, R.; Qiao, S.; Wan, B.; Wang, Y.; Liu, M.; Shi, X.; Guo, J.; Zhang, G. Antibody-dependent enhancement of PRRSV infection down-modulates TNF-alpha and IFN-beta transcription in macrophages. Vet. Immunol. Immunopathol. 2013, 156, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Meng, X.; Zhen, Y.; Zhang, Y.; Hu, X.; Zhang, Q.; Zhou, X.; Liu, B. Integration of Transcriptome and Proteome in Lymph Nodes Reveal the Different Immune Responses to PRRSV Between PRRSV-Resistant Tongcheng Pigs and PRRSV-Susceptible Large White Pigs. Front. Genet. 2022, 13, 800178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, L.; Li, Y.; Jiang, P.; Wang, Y.; Wang, P.; Kang, L.; Wang, Y.; Sun, Y.; Jiang, Y. Identification and characterization of microRNA in the lung tissue of pigs with different susceptibilities to PCV2 infection. Vet. Res. 2018, 49, 18. [Google Scholar] [CrossRef]
- Li, Q.J.; Chau, J.; Ebert, P.J.; Sylvester, G.; Min, H.; Liu, G.; Braich, R.; Manoharan, M.; Soutschek, J.; Skare, P.; et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007, 129, 147–161. [Google Scholar] [CrossRef]
- Ye, L.; Su, X.; Wu, Z.; Zheng, X.; Wang, J.; Zi, C.; Zhu, G.; Wu, S.; Bao, W. Analysis of differential miRNA expression in the duodenum of Escherichia coli F18-sensitive and -resistant weaned piglets. PLoS ONE 2012, 7, e43741. [Google Scholar] [CrossRef]
- Tang, X.; Lan, T.; Wu, R.; Zhou, Z.; Chen, Y.; Sun, Y.; Zheng, Y.; Ma, J. Analysis of long non-coding RNAs in neonatal piglets at different stages of porcine deltacoronavirus infection. BMC Vet. Res. 2019, 15, 111. [Google Scholar] [CrossRef]
- Jarret, A.; McFarland, A.P.; Horner, S.M.; Kell, A.; Schwerk, J.; Hong, M.; Badil, S.; Joslyn, R.C.; Baker, D.P.; Carrington, M.; et al. Hepatitis-C-virus-induced microRNAs dampen interferon-mediated antiviral signaling. Nat. Med. 2016, 22, 1475–1481. [Google Scholar] [CrossRef]
- Wysocki, M.; Chen, H.; Steibel, J.P.; Kuhar, D.; Petry, D.; Bates, J.; Johnson, R.; Ernst, C.W.; Lunney, J.K. Identifying putative candidate genes and pathways involved in immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) infection. Anim. Genet. 2012, 43, 328–332. [Google Scholar] [CrossRef]
- Cayrol, C.; Girard, J.P. Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol. Rev. 2018, 281, 154–168. [Google Scholar] [CrossRef]
- Ou, R.; Zhang, M.; Huang, L.; Flavell, R.A.; Koni, P.A.; Moskophidis, D. Regulation of immune response and inflammatory reactions against viral infection by VCAM-1. J. Virol. 2008, 82, 2952–2965. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boraschi, D.; Italiani, P.; Weil, S.; Martin, M.U. The family of the interleukin-1 receptors. Immunol. Rev. 2018, 281, 197–232. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.P.; Rohrer, G.A.; Alexander, L.J.; Troyer, D.L.; Kirby-Dobbels, K.R.; Janzen, M.A.; Cornwell, D.L.; Louis, C.F.; Schook, L.B.; Beattie, C.W. Directed integration of the physical and genetic linkage maps of swine chromosome 7 reveals that the SLA spans the centromere. Genome Res. 1995, 5, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.E.; Ho, C.S.; Ando, A.; Rogel-Gaillard, C.; Charles, M.; Tector, M.; Tector, A.J.; Lunney, J.K. Importance of the Major Histocompatibility Complex (Swine Leukocyte Antigen) in Swine Health and Biomedical Research. Annu. Rev. Anim. Biosci. 2020, 8, 171–198. [Google Scholar] [CrossRef]
- Wimmers, K.; Schellander, K.; Ponsuksili, S. BF, HP, DQB and DRB are associated with haemolytic complement activity, acute phase protein reaction and antibody response in the pig. Vet. Immunol. Immunopathol. 2004, 99, 215–228. [Google Scholar] [CrossRef]
- Hewitt, E.W. The MHC class I antigen presentation pathway: Strategies for viral immune evasion. Immunology 2003, 110, 163–169. [Google Scholar] [CrossRef]
- Lunney, J.K.; Murrell, K.D. Immunogenetic analysis of Trichinella spiralis infections in swine. Vet. Parasitol. 1988, 29, 179–193. [Google Scholar] [CrossRef]
- Madden, K.B.; Moeller, R.F., Jr.; Douglass, L.W.; Goldman, T.; Lunney, J.K. Trichinella spiralis: Genetic basis and kinetics of the anti-encysted muscle larval response in miniature swine. Exp. Parasitol. 1993, 77, 23–35. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).