Characterization of Lophiotoma leucotropis Mitochondrial Genome of Family Turridae and Phylogenetic Considerations within the Neogastropoda
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
2.2. DNA Sequencing and Assembly
2.3. Genome Annotation and Bioinformatics Analysis
2.4. Phylogenetic Analysis
3. Results
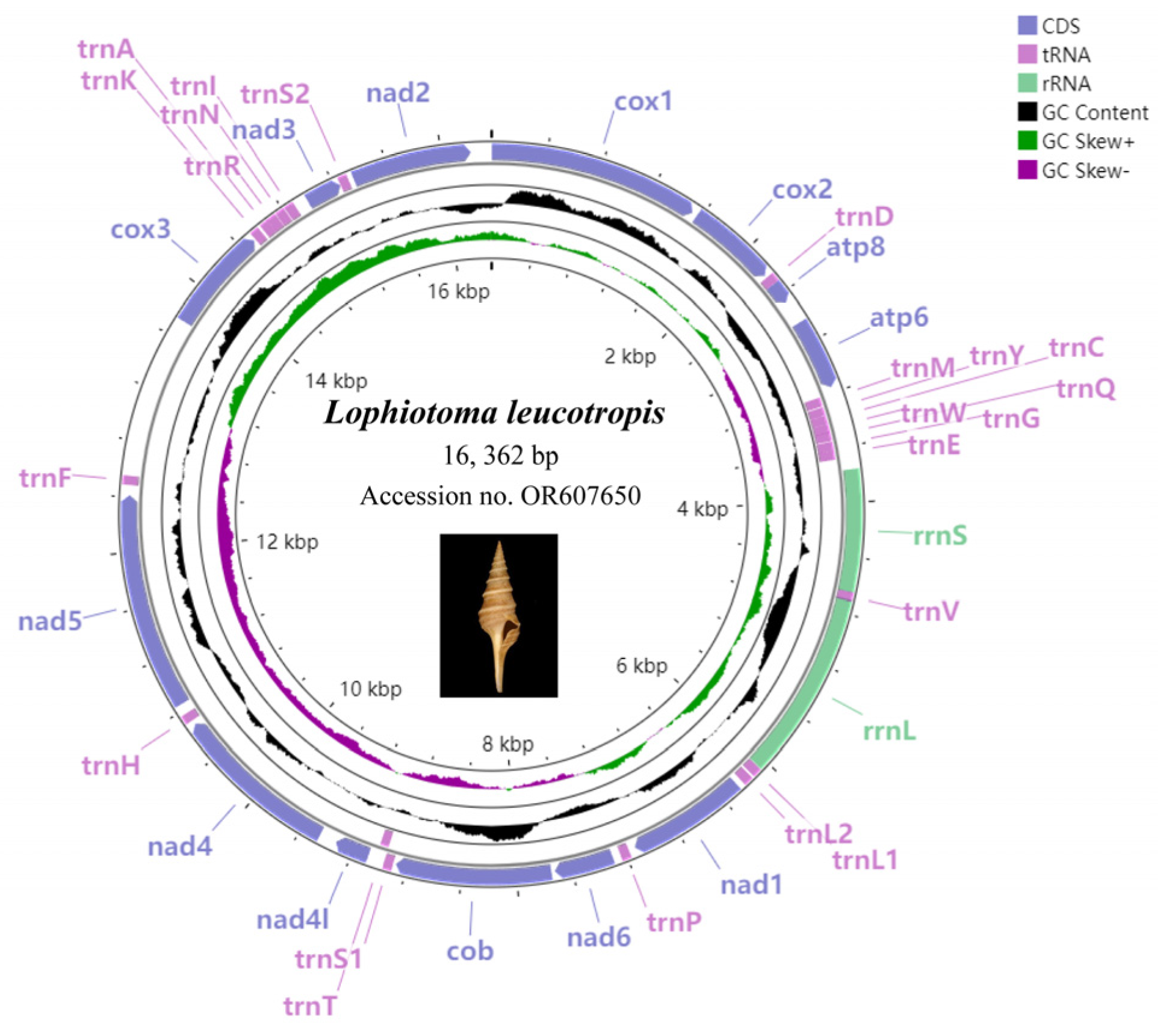
3.1. Characterization of the Mitogenome Structure
3.2. Base Content and Base Offset
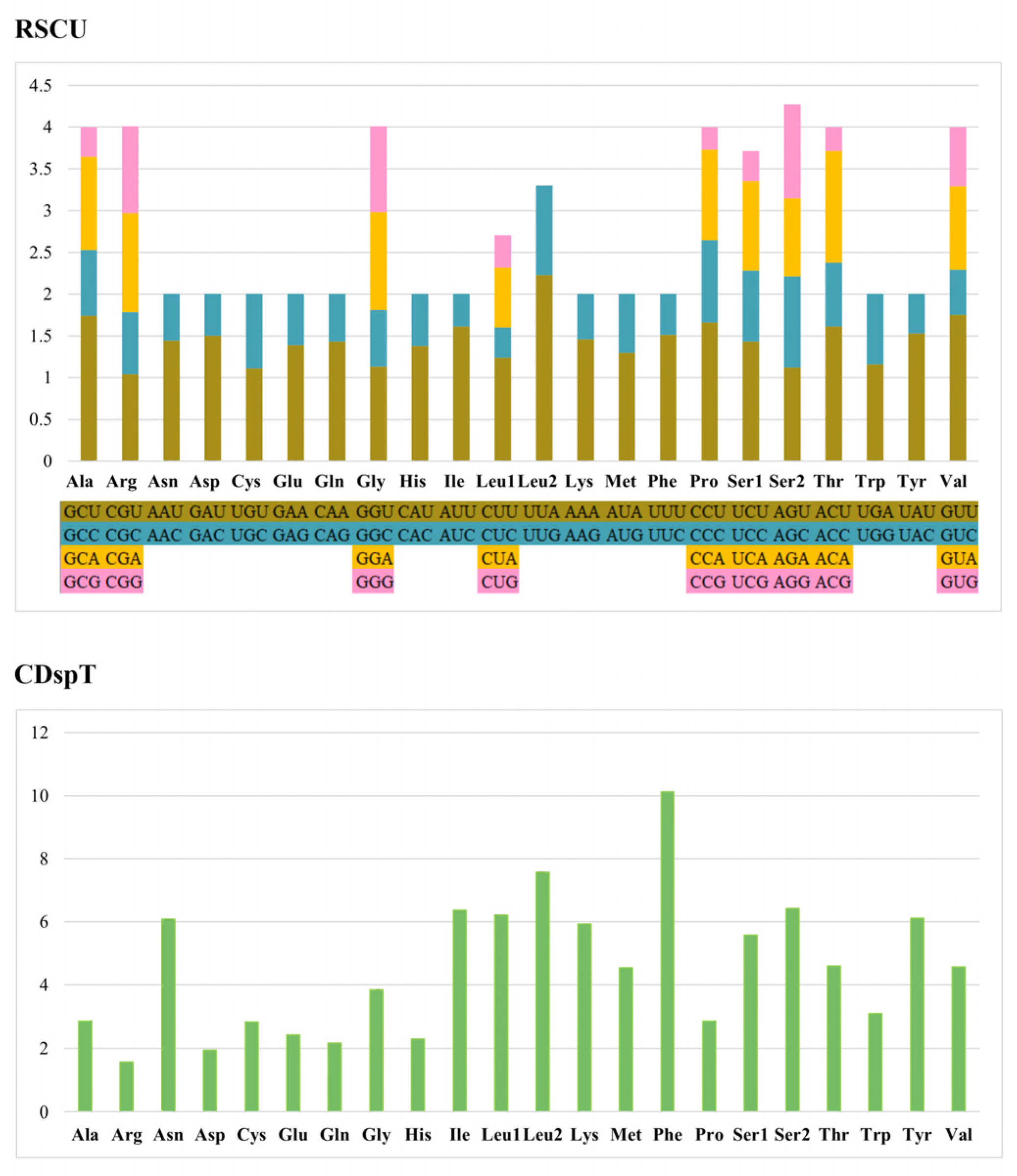
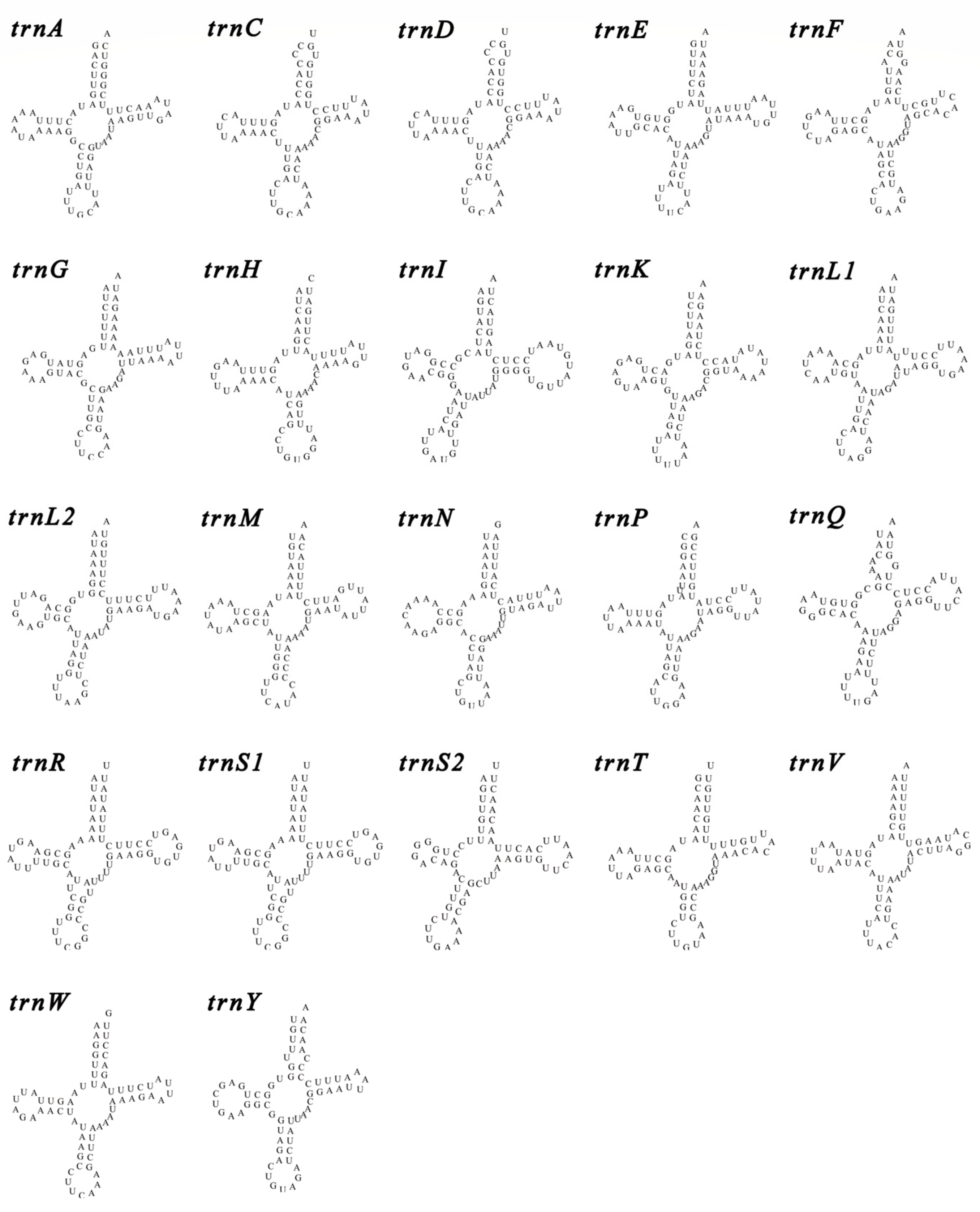
3.3. Amino Acid Content, RSCU, and tRNA
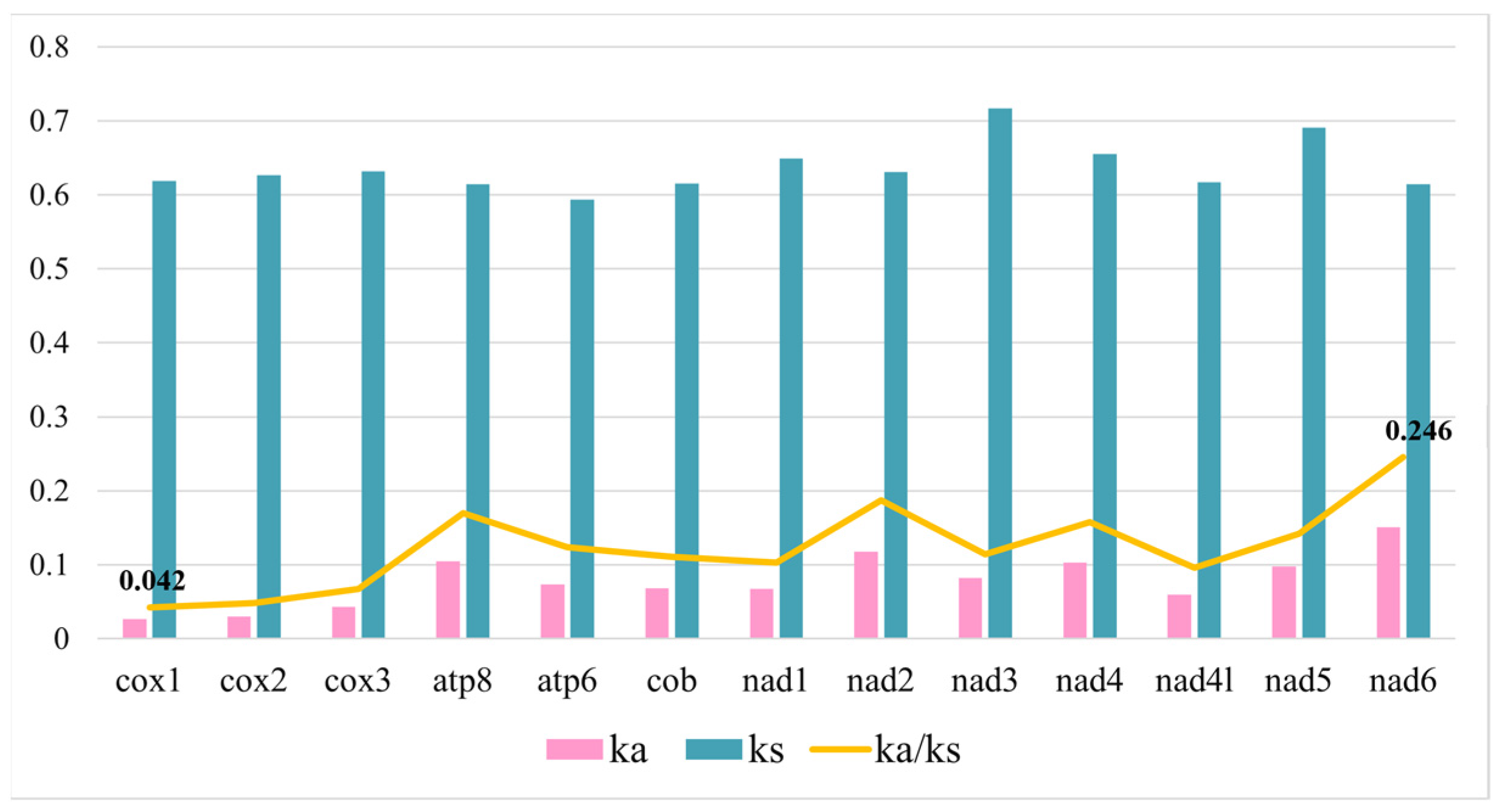
3.4. Selective Pressure Analysis
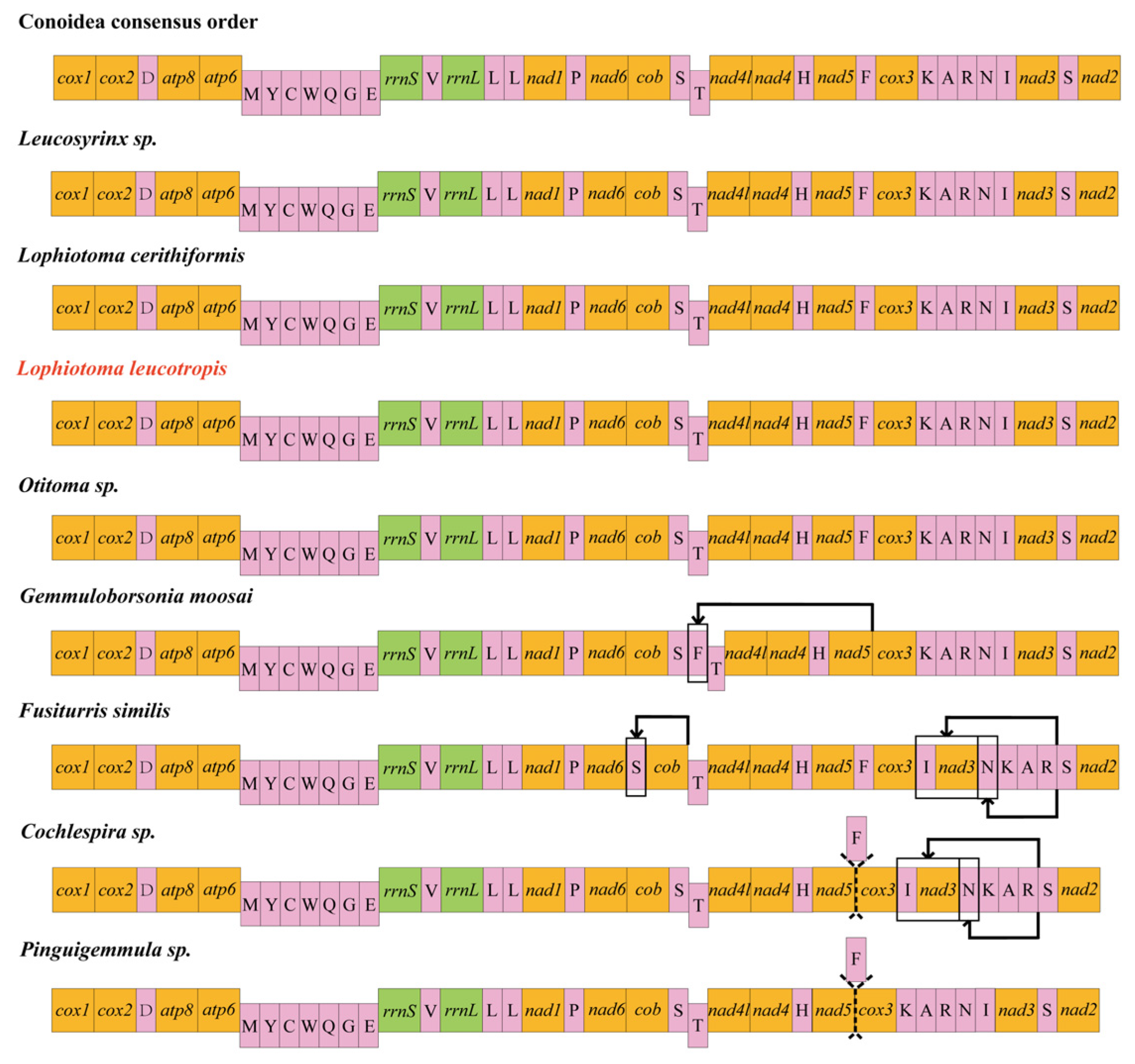
3.5. Gene Rearrangement
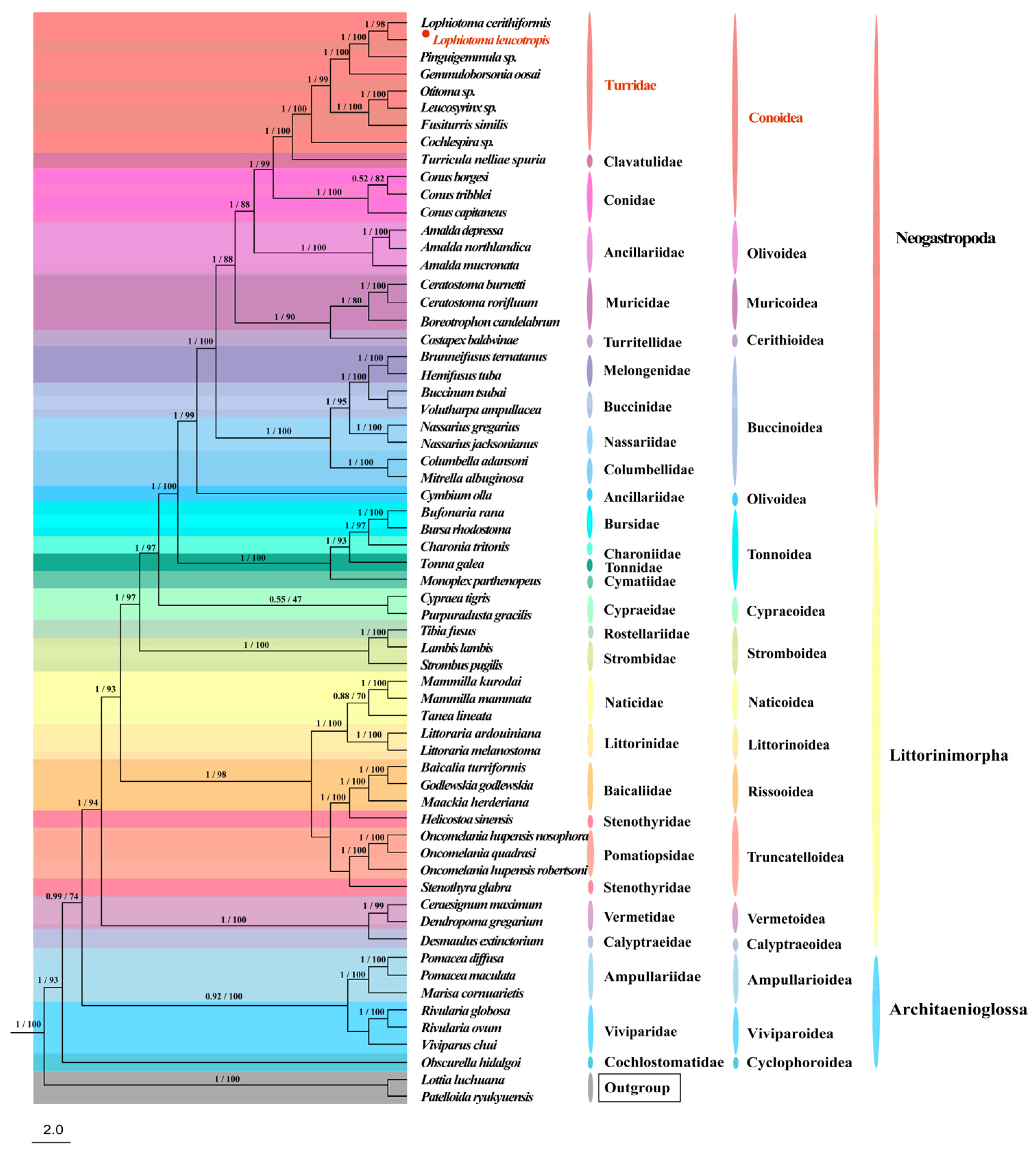
3.6. Phylogenetic Relationship
4. Discussion
4.1. Basic Features of the Mitogenome of L. leucotropis
4.2. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Adams, A. The Zoology of the Voyage of HMS Samarang; under the Command of Captain Sir Edward Belcher, during the Years 1843–1846; Reeve and Benham: London, UK, 1850. [Google Scholar]
- Wenz, W. Teil 1: Allgemeiner Teil und Prosobranchia; Handbuch der Paläozoologie, Bd. 6; Borntraeger: Berlin, Germany, 1938; pp. 1–1639. [Google Scholar]
- Ponder, W.F.; Colgan, D.J.; Healy, J.; Nützel, A.; Simone, L.R.; Strong, E.E. Caenogastropod phylogeny. In Molluscan Phylogeny; U. California Press: Berkeley, CA, USA, 2008. [Google Scholar]
- Taylor, J.D. Diets of sublittoral predatory gastropods of Hong Kong. In The Marine Flora and Fauna of Hong Kong and Southern China; Hong Kong University Press: Hong Kong, China, 1980; pp. 907–920. [Google Scholar]
- Taylor, J.D. Diets and habitats of shallow water predatory gastropods around Tolo Channel, Hong Kong. In The Malacofauna of Hong Kong Southern China; Hong Kong University Press: Hong Kong, China, 1980; pp. 163–180. [Google Scholar]
- Abbott, R.T.; Dance, S.P. Compendium of Seashells; American Malacologists, Inc.: Melbourne, FL, USA, 1986; p. 411. [Google Scholar]
- Kilburn, R.N. Turridae (Mollusca: Gastropoda) of southern Africa and Mozambique. Part 4. Subfamilies Drilliinae, Crassispirinae and Strictispirinae. Ann. Natal Mus. 1988, 29, 167–320. [Google Scholar]
- Taylor, J.D.; Morris, N.J.; Taylor, C.N. Food specialization and the evolution of predatory prosobranch gastropods. Palaeontology 1980, 23 Pt 2, 375–409. [Google Scholar]
- Li, B.Q. Taxonomic and Faunal Study of Turridae (Gastropoda: Neogastropoda) of the China Seas; Institute of Oceanology, Chinese Academy of Sciences: Qingdao, China, 2007. [Google Scholar]
- Casey, T.L. Notes on the Pleurotomidae with Description of Some New Genera and Species; Biodiversity Heritage Library: Washington, DC, USA, 1904. [Google Scholar]
- Powell, A.W.B. The Molluscan Families Speightiidae and Turridae: An Evaluation of the Valid Taxa, Both Recent and Fossil, with Lists of Characteristic Species. Unity Press. 1966. Available online: https://xueshu.baidu.com/usercenter/paper/show?paperid=b8b8351a6ffa301e1b929bf274f2f123&site=xueshu_se (accessed on 30 October 2023).
- McLean, J.H. A Revised Classification of the Family Turridae, with the Proposal of New Subfamilies, Genera and Subgenera from the Eastern Pacific; California Malacozoological Society: Berkeley, CA, USA, 1971. [Google Scholar]
- Taylor, J.D. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda). Bull. Nat. Hist. Mus. 1993, 59, 125–170. [Google Scholar]
- Bouchet, P. Turrid genera and mode of development: The use and abuse of protoconch morphology. Malacologia 1990, 32, 69–77. [Google Scholar]
- Ponder, W.F. Classification of the Caenogastropoda and Heterostropha-a list of the family-group names and higher taxa. Prosobranch Phylogeny Malacol. Rev. 1988, 4, 288–326. [Google Scholar]
- Ponder, W.F. The origin and evolution of the Neogastropoda. Malacologia 1973, 12, 295–338. [Google Scholar]
- Bouvier, E.L. Système Nerveux, Morphologie Générale et Classification des Gastéropodes Prosobranches; G. Masson: Paris, France, 1887. [Google Scholar]
- Ponder, W.F.; Lindberg, D.R. Gastropod phylogeny—Challenges for the 90s. In Origin evolutionary radiation of the Mollusca; Oxford Academic: Oxford, UK, 1996; pp. 135–154. [Google Scholar]
- Ponder, W.F.; Lindberg, D.R. Towards a phylogeny of gastropod molluscs: An analysis using morphological characters. Zool. J. Linn. Soc. 1997, 119, 83–265. [Google Scholar]
- Colgan, D.J.; Ponder, W.F.; Eggler, P.E. Gastropod evolutionary rates and phylogenetic relationships assessed using partial 28S rDNA and histone H3 sequences. Zool. Scr. 2000, 29, 29–63. [Google Scholar]
- Cunha, R.L.; Grande, C.; Zardoya, R. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol. Biol. 2009, 9, 210. [Google Scholar]
- Miao, J.; Feng, J.T.; Liu, X.J.; Yan, C.R.; Ye, Y.Y.; Li, J.J.; Xu, K.D.; Guo, B.Y.; Lü, Z.M. Sequence comparison of the mitochondrial genomes of five brackish water species of the family Neritidae: Phylogenetic implications and divergence time estimation. Ecol. Evol. 2022, 12, e8984. [Google Scholar] [CrossRef]
- Feng, J.T.; Xia, L.P.; Yan, C.R.; Miao, J.; Ye, Y.Y.; Li, J.J.; Guo, B.Y.; Lü, Z.M. Characterization of four mitochondrial genomes of family Neritidae (Gastropoda: Neritimorpha) and insight into its phylogenetic relationships. Sci. Rep. 2021, 11, 11748. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L.; Macey, J.R.; Medina, M. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 2005, 395, 311–348. [Google Scholar] [PubMed]
- Gu, M.B. Tlas of Famous Shell in the World for Identification and Appreciation; Henan Scientific and Technological Publishing House: Zhengzhou, China, 2005. [Google Scholar]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Swofford, D. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0 b10; ScienceOpen, Inc.: Boston, MA, USA, 2002. [Google Scholar]
- Posada, D. Modeltest: A Tool to Select the Best-Fit Model of Nucleotide Substitution. Version 3.7. 2005. Available online: https://dposada.webs.uvigo.es/ (accessed on 30 October 2023).
- Jaa, N. MrModeltest 2.3. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rambaut, A. Figtree Version 1.4. 0. Revisado. 2015. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 30 October 2023).
- Bandyopadhyay, P.K.; Stevenson, B.J.; Cady, M.T.; Olivera, B.M.; Wolstenholme, D.R. Complete mitochondrial DNA sequence of a Conoidean gastropod, Lophiotoma (Xenuroturris) cerithiformis: Gene order and gastropod phylogeny. Toxicon 2006, 48, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Qi, L.; Kong, L.F.; Li, Q. Mitogenomics reveals phylogenetic relationships of Patellogastropoda (Mollusca, Gastropoda) and dynamic gene rearrangements. Zool. Scr. 2022, 51, 147–160. [Google Scholar] [CrossRef]
- Feng, J.; Guo, Y.; Yan, C.; Ye, Y.; Li, J.; Guo, B.; Lü, Z. Comparative analysis of the complete mitochondrial genomes in two limpets from Lottiidae (Gastropoda: Patellogastropoda): Rare irregular gene rearrangement within Gastropoda. Sci. Rep. 2020, 10, 19277. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.F.; Li, Y.N.; Kocot, K.M.; Yang, Y.; Qi, L.; Li, Q.; Halanych, K.M. Mitogenomics reveals phylogenetic relationships of Arcoida (Mollusca, Bivalvia) and multiple independent expansions and contractions in mitochondrial genome size. Mol. Phylogenet. Evol. 2020, 150, 106857. [Google Scholar] [CrossRef]
- Li, Q.; Kong, L.F.; Yu, H. Evolution of mitochondrial gene arrangements in Arcidae (Bivalvia: Arcida) and their phylogenetic implications. Mol. Phylogenet. Evol. 2020, 150, 106879. [Google Scholar]
- Heralde, F.M., III; Kantor, Y.I.; Astilla, M.A.Q.; Lluisma, A.O.; Geronimo, R.; Aliño, P.M.; Watkins, M.; Corneli, P.S.; Olivera, B.M.; Santos, A.D. The Indo-Pacific Gemmula species in the subfamily Turrinae: Aspects of field distribution, molecular phylogeny, radular anatomy and feeding ecology. Philipp. Sci. Lett. 2010, 3, 20105. [Google Scholar] [CrossRef]
- Li, B.; Li, X. Report on the turrid genera Gemmula, Lophiotoma and Ptychosyrinx (Gastropoda: Turridae: Turrinae) from the China seas. Zootaxa 2008, 1778, 1–25. [Google Scholar] [CrossRef]
- Solanki, D.A.; Kanejiya, J.R.; Gohil, B.M. Turricula nelliae spuria, Hedley, 1992 (Mollusca: Gastropod: Clavatulidae): Range extension and new country record. Cibtech J. Zool. 2017, 6, 14–19. [Google Scholar]
- Riedel, F. Ursprung und Evolution de “höheren” Casenogastropoda: Eine paläobiologische Konzeption; Fachbereich Geowissenschaften, FU Berlin: Berlin, Germany, 2000. [Google Scholar]
- Colgan, D.J.; Ponder, W.F.; Beacham, E.; Macaranas, J.M. Molecular phylogenetic studies of Gastropoda based on six gene segments representing coding or non-coding and mitochondrial or nuclear DNA. Molluscan Res. 2003, 23, 123–148. [Google Scholar] [CrossRef]
- Colgan, D.J.; Ponder, W.F.; Beacham, E.; Macaranas, J.M. Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca). Mol. Phylogenet. Evol. 2007, 42, 717–737. [Google Scholar] [CrossRef] [PubMed]
- Osca, D.; Templado, J.; Zardoya, R. Caenogastropod mitogenomics. Mol. Phylogenet. Evol. 2015, 93, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Kantor, Y.I.; Sysoev, A.; Couloux, A.; Meyer, C.; Rawlings, T.; Todd, J.; Bouchet, P. The dragon tamed? A molecular phylogeny of the Conoidea (Gastropoda). J. Mollus. Stud. 2011, 77, 259–272. [Google Scholar] [CrossRef]
- Puillandre, N.; Samadi, S.; Boisselier, M.C.; Sysoev, A.; Kantor, Y.I.; Cruaud, C.; Couloux, A.; Bouchet, P. Starting to unravel the toxoglossan knot: Molecular phylogeny of the “turrids” (Neogastropoda: Conoidea). Mol. Phylogenet. Evol. 2008, 47, 1122–1134. [Google Scholar] [CrossRef]
- Uribe, J.E.; Zardoya, R.; Puillandre, N. Phylogenetic relationships of the conoidean snails (Gastropoda: Caenogastropoda) based on mitochondrial genomes. Mol. Phylogenet. Evol. 2018, 127, 898–906. [Google Scholar] [CrossRef]
- Zou, S.M. DNA Barcoding and Phylogeny of Neogadtropoda; Ocean University of China: Qingdao, China, 2014. [Google Scholar]
| Order | Superfamily | Family | Species | Accession No. |
|---|---|---|---|---|
| Littorinimorpha | Calyptraeoidea | Calyptraeidae | Desmaulus extinctorium | NC_079658 |
| Cypraeoidea | Cypraeidae | Cypraea tigris | MK783263 | |
| Purpuradusta gracilis | NC_072228 | |||
| Littorinoidea | Littorinidae | Littoraria ardouiniana | NC_066085 | |
| Littoraria melanostoma | NC_064398 | |||
| Naticoidea | Naticidae | Mammilla kurodai | NC_046596 | |
| Mammilla mammata | NC_046597 | |||
| Tanea lineata | NC_050662 | |||
| Rissooidea | Baicaliidae | Baicalia turriformis | NC_035869 | |
| Godlewskia godlewskia | NC_035870 | |||
| Maackia herderiana | NC_035871 | |||
| Stromboidea | Rostellariidae | Tibia fusus | NC_065371 | |
| Strombidae | Lambis lambis | NC_071230 | ||
| Strombus pugilis | NC_059922 | |||
| Tonnoidea | Tonnidae | Tonna galea | NC_082277 | |
| Cymatiidae | Monoplex parthenopeus | NC_013247 | ||
| Bursidae | Bufonaria rana | NC_054277 | ||
| Bursa rhodostoma | MW316792 | |||
| Charoniidae | Charonia tritonis | NC_082220 | ||
| Truncatelloidea | Pomatiopsidae | Oncomelania hupensis nosophora | LC276226 | |
| Oncomelania hupensis robertsoni | LC276228 | |||
| Oncomelania quadrasi | LC276227 | |||
| Stenothyridae | Stenothyra glabra | NC_080968 | ||
| Helicostoa sinensis | OP503947 | |||
| Vermetoidea | Vermetidae | Ceraesignum maximum | NC_014583 | |
| Dendropoma gregarium | NC_014580 | |||
| Neogastropoda | Buccinoidea | Columbellidae | Mitrella albuginosa | NC_066081 |
| Columbella adansoni | KP716637 | |||
| Buccinidae | Buccinum tsubai | NC_063974 | ||
| Volutharpa ampullacea | NC_067974 | |||
| Melongenidae | Brunneifusus ternatanus | NC_059758 | ||
| Hemifusus tuba | MN462591 | |||
| Nassariidae | Nassarius gregarius | NC_062791 | ||
| Nassarius jacksonianus | NC_041548 | |||
| Conoidea | Turridae | Cochlespira sp. | MH308394 | |
| Fusiturris similis | NC_013242 | |||
| Gemmuloborsonia moosai | NC_038183 | |||
| Leucosyrinx sp. | NC_038185 | |||
| Lophiotoma cerithiformis | NC_008098 | |||
| • Lophiotoma leucotropis | OR607650 | |||
| Otitoma sp. | MH308405 | |||
| Pinguigemmula sp. | MH308408 | |||
| Conidae | Conus borgesi | EU827198 | ||
| Conus capitaneus | NC_030354 | |||
| Conus tribblei | NC_027957 | |||
| Clavatulidae | Turricula nelliae spuria | MK251986 | ||
| Muricoidea | Muricidae | Boreotrophon candelabrum | NC_046505 | |
| Ceratostoma burnetti | NC_046569 | |||
| Ceratostoma rorifluum | NC_046526 | |||
| Cerithioidea | Turritellidae | Costapex baldwinae | MW044625 | |
| Olivoidea | Ancillariidae | Amalda mucronata | NC_061910 | |
| Amalda northlandica | GU196685 | |||
| Amalda depressa | MN400051 | |||
| Architaenioglossa | Ampullarioidea | Ampullariidae | Marisa cornuarietis | NC_025334 |
| Pomacea diffusa | NC_041142 | |||
| Pomacea maculata | NC_027503 | |||
| Cyclophoroidea | Cochlostomatidae | Obscurella hidalgoi | NC_028004 | |
| Viviparoidea | Viviparidae | Rivularia globosa | NC_066479 | |
| Rivularia ovum | NC_066478 | |||
| Viviparus chui | NC_035733 |
| Gene | Position | Strand | Size | Codon | Anticodon | Intergenic Nucleotides | |||
|---|---|---|---|---|---|---|---|---|---|
| From | To | Length | Amino Acid | Start | Stop | ||||
| cox1 | 1 | 1545 | + | 1545 | 515 | ATG | TAA | 11 | |
| cox2 | 1557 | 2243 | + | 687 | 229 | ATG | TAA | −2 | |
| trnD | 2242 | 2309 | + | 68 | GTC | 0 | |||
| atp8 | 2310 | 2468 | + | 159 | 53 | ATG | TAA | 2 | |
| atp6 | 2471 | 3166 | + | 696 | 232 | ATG | TAG | 33 | |
| trnM | 3200 | 3268 | - | 69 | CAT | 8 | |||
| trnY | 3277 | 3342 | - | 66 | GTA | 1 | |||
| trnC | 3344 | 3407 | - | 64 | GCA | 0 | |||
| trnW | 3408 | 3474 | - | 67 | TCA | −2 | |||
| trnQ | 3473 | 3537 | - | 65 | TTG | 5 | |||
| trnG | 3543 | 3609 | - | 67 | TCC | −1 | |||
| trnE | 3609 | 3677 | - | 69 | TTC | 1 | |||
| rrnS | 3764 | 4646 | + | 883 | −3 | ||||
| trnV | 4644 | 4710 | + | 67 | TCA | −10 | |||
| rrnL | 4701 | 6060 | + | 1360 | 3 | ||||
| trnL1 | 6064 | 6132 | + | 69 | TAG | 16 | |||
| trnL2 | 6149 | 6218 | + | 70 | TAA | 0 | |||
| nad1 | 6219 | 7160 | + | 942 | 314 | ATG | TAA | 11 | |
| trnP | 7172 | 7239 | + | 68 | TGG | 13 | |||
| nad6 | 7253 | 7741 | + | 489 | 163 | ATT | TAA | 3 | |
| cob | 7745 | 8884 | + | 1140 | 380 | ATG | TAA | 19 | |
| trnS1 | 8898 | 8963 | + | 66 | TGA | 0 | |||
| trnT | 8964 | 9029 | - | 66 | TGT | 11 | |||
| nad4l | 9041 | 9337 | + | 297 | 99 | ATG | TAG | −7 | |
| nad4 | 9331 | 10,671 | + | 1341 | 447 | ATG | TAA | 31 | |
| trnH | 10,703 | 10,768 | + | 66 | GTG | 27 | |||
| nad5 | 10,796 | 12,484 | + | 1689 | 563 | ATT | TAA | 1 | |
| trnF | 12,486 | 12,551 | + | 66 | GAA | 0 | |||
| D-loop | 12,552 | 13,723 | + | 1172 | 0 | ||||
| cox3 | 13,724 | 14,503 | + | 780 | 260 | ATG | TAA | 12 | |
| trnK | 14,516 | 14,582 | + | 67 | TTT | 19 | |||
| trnA | 14,602 | 14,668 | + | 67 | TGC | −1 | |||
| trnR | 14,668 | 14,737 | + | 70 | TCG | 3 | |||
| trnN | 14,741 | 14,809 | + | 69 | GTT | 6 | |||
| trnI | 14,816 | 14,886 | + | 71 | GAT | 2 | |||
| nad3 | 14,889 | 15,242 | + | 354 | 118 | ATG | TAG | 1 | |
| trnS2 | 15,244 | 15,311 | + | 68 | GCT | 48 | |||
| nad2 | 15,360 | 16,346 | + | 987 | 329 | ATT | TAA | 15 | |
| Gene | A% | C% | G% | T% | A + T% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|
| Mito | 30.93 | 15.07 | 15.73 | 38.27 | 69.20 | −0.11 | 0.02 |
| cox1 | 26.98 | 16.57 | 18.40 | 37.98 | 64.96 | −0.17 | 0.05 |
| cox2 | 29.66 | 15.90 | 17.74 | 36.70 | 66.36 | −0.11 | 0.05 |
| atp8 | 32.70 | 10.69 | 13.84 | 42.77 | 75.47 | −0.13 | 0.13 |
| atp6 | 27.55 | 15.22 | 15.22 | 44.51 | 72.06 | −0.24 | 0.00 |
| nad1 | 27.43 | 15.63 | 17.71 | 39.24 | 66.67 | −0.18 | 0.06 |
| nad6 | 29.37 | 13.99 | 11.89 | 44.76 | 74.13 | −0.21 | −0.08 |
| cob | 26.86 | 18.44 | 15.87 | 38.83 | 65.69 | −0.18 | −0.07 |
| nad4l | 30.52 | 14.86 | 16.06 | 38.55 | 69.07 | −0.12 | 0.04 |
| nad4 | 29.86 | 17.70 | 13.48 | 38.96 | 68.82 | −0.13 | −0.14 |
| nad5 | 30.27 | 19.57 | 12.81 | 37.35 | 67.62 | −0.10 | −0.21 |
| cox3 | 24.74 | 14.71 | 21.09 | 39.45 | 64.19 | −0.23 | 0.18 |
| nad3 | 24.03 | 12.40 | 17.83 | 45.74 | 69.77 | −0.31 | 0.18 |
| nad2 | 26.76 | 12.57 | 17.19 | 43.48 | 70.24 | −0.24 | 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Miao, J.; Li, J.; Ye, Y. Characterization of Lophiotoma leucotropis Mitochondrial Genome of Family Turridae and Phylogenetic Considerations within the Neogastropoda. Animals 2024, 14, 192. https://doi.org/10.3390/ani14020192
Jiang X, Miao J, Li J, Ye Y. Characterization of Lophiotoma leucotropis Mitochondrial Genome of Family Turridae and Phylogenetic Considerations within the Neogastropoda. Animals. 2024; 14(2):192. https://doi.org/10.3390/ani14020192
Chicago/Turabian StyleJiang, Xinqin, Jing Miao, Jiji Li, and Yingying Ye. 2024. "Characterization of Lophiotoma leucotropis Mitochondrial Genome of Family Turridae and Phylogenetic Considerations within the Neogastropoda" Animals 14, no. 2: 192. https://doi.org/10.3390/ani14020192
APA StyleJiang, X., Miao, J., Li, J., & Ye, Y. (2024). Characterization of Lophiotoma leucotropis Mitochondrial Genome of Family Turridae and Phylogenetic Considerations within the Neogastropoda. Animals, 14(2), 192. https://doi.org/10.3390/ani14020192






