Low Occurrence of Salmonella spp. in Wild Animals in Bahia, Brazil—Population Assessment and Characterization in the Caatinga and Atlantic Forest Biomes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
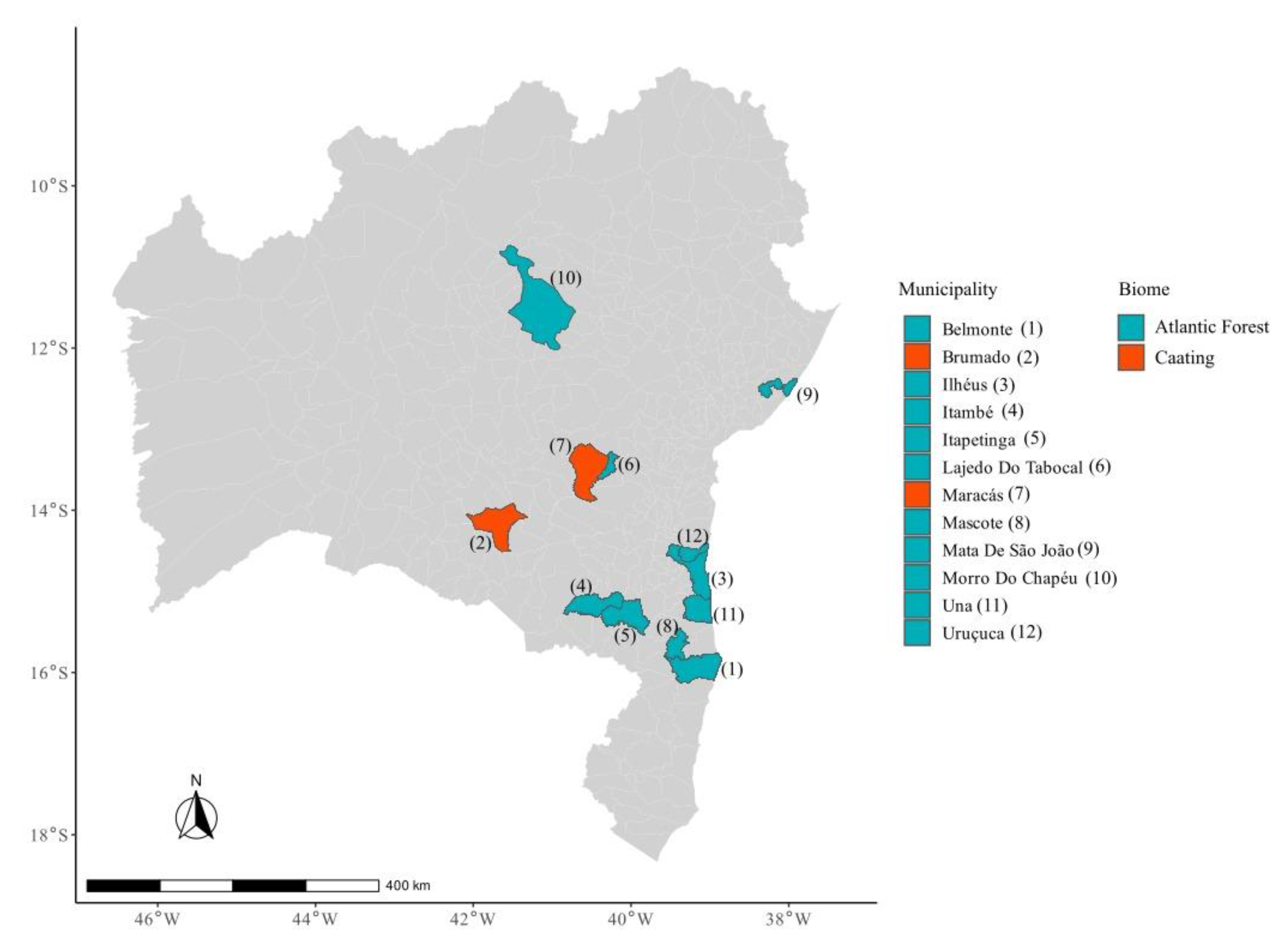
2.2. Study Area
2.3. Specimen Collection
2.4. Salmonella Isolation and Identification
2.5. Antibiotic Susceptibility Test
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rock, M.; Buntain, B.J.; Hatfield, J.M.; Hallgrímsson, B. Animal–Human Connections, “One Health”, and the Syndemic Approach to Prevention. Soc. Sci. Med. 2009, 68, 991–995. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Iovine, R.O.; Dejuste, C.; Miranda, F.; Filoni, C.; Bueno, M.G.; de Carvalho, V.M. Isolation of Escherichia coli and Salmonella spp. from Free-Ranging Wild Animals. Braz. J. Microbiol. 2015, 46, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, D.M.; Dunn, A.M.; Smith, M.J.; Telfer, S. Wildlife Diseases: From Individuals to Ecosystems. J. Anim. Ecol. 2011, 80, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Siembieda, J.L.; Kock, R.A.; McCracken, T.A.; Newman, S.H. The Role of Wildlife in Transboundary Animal Diseases. Anim. Health Res. Rev. 2011, 12, 95–111. [Google Scholar] [CrossRef]
- Hilbert, F.; Smulders, F.J.M.; Chopra-Dewasthaly, R.; Paulsen, P. Salmonella in the Wildlife-Human Interface. Food Res. Int. 2012, 45, 603–608. [Google Scholar] [CrossRef]
- López-Islas, J.J.; Méndez-Olvera, E.T.; Martínez-Gómez, D.; López-Pérez, A.M.; Orozco, L.; Suzan, G.; Eslava, C. Characterization of Salmonella spp. and E. coli Strains Isolated from Wild Carnivores in Janos Biosphere Reserve, Mexico. Animals 2022, 12, 1064. [Google Scholar] [CrossRef]
- Rubini, S.; Ravaioli, C.; Previato, S.; D’Incau, M.; Tassinari, M.; Guidi, E.; Lupi, S.; Merialdi, G.; Bergamini, M. Prevalence of Salmonella Strains in Wild Animals from a Highly Populated Area of North-Eastern Italy. Ann. Ist. Super. Sanità. 2016, 52, 277–280. [Google Scholar] [CrossRef]
- Millan, J.; Aduriz, G.; Moreno, B.; Juste, R.A.; Barral, M. Salmonella Isolates from Wild Birds and Mammals in the Basque Country (Spain). Rev. Sci. Tech. OIE 2004, 23, 905–911. [Google Scholar] [CrossRef]
- Carraro, P.E.; Barbosa, F.D.O.; Benevides, V.P.; Casas, M.R.T.; Berchieri Junior, A.; Bürger, K.P. Prevalence and Antimicrobial Resistance of Salmonella spp. Isolated from Free-Ranging Wild Boars in the State of São Paulo, Brazil. Cienc. Rural. 2022, 52, e20210263. [Google Scholar] [CrossRef]
- Merkevičienė, L.; Butrimaitė-Ambrozevičienė, Č.; Paškevičius, G.; Pikūnienė, A.; Virgailis, M.; Dailidavičienė, J.; Daukšienė, A.; Šiugždinienė, R.; Ruzauskas, M. Serological Variety and Antimicrobial Resistance in Salmonella Isolated from Reptiles. Biology 2022, 11, 836. [Google Scholar] [CrossRef]
- Fluit, A.C.; Schmitz, F.-J. Resistance Integrons and Super-Integrons. Clin. Microbiol. Infect. 2004, 10, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Perveen, N.; Muzaffar, S.B.; Al-Deeb, M.A. Exploring Human-Animal Host Interactions and Emergence of COVID-19: Evolutionary and Ecological Dynamics. Saudi J. Biol. Sci. 2021, 28, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Uelze, L.; Bloch, A.; Borowiak, M.; Grobbel, M.; Deneke, C.; Fischer, M.; Malorny, B.; Pietsch, M.; Simon, S.; Szabó, I.; et al. What WGS Reveals about Salmonella enterica subsp. enterica in Wildlife in Germany. Microorganisms 2021, 9, 1911. [Google Scholar] [CrossRef]
- IBGE, Instituto Brasileiro de Geografia e Estatística. Biomas e Sistema Costeiro-Marinho Do Brasil: Compatível Com a Escala 1:250,000; Coordenação de Recursos Naturais e Estudos Ambientais: Rio de Janeiro, Brazil, 2019. [Google Scholar]
- Dos Santos, E.J.E.; Azevedo, R.P.; Lopes, A.T.S.; Rocha, J.M.; Albuquerque, G.R.; Wenceslau, A.A.; Miranda, F.R.; Rodrigues, D.d.P.; Maciel, B.M. Salmonella spp. in Wild Free-Living Birds from Atlantic Forest Fragments in Southern Bahia, Brazil. BioMed Res. Int. 2020, 2020, 7594136. [Google Scholar] [CrossRef] [PubMed]
- Aabo, S.; Rasmussen, O.F.; Roseen, L.; Sørensen, P.D.; Olsen, J.E. Salmonella Identification by the Polymerase Chain Reaction. Mol. Cel. Probes 1993, 7, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Maciel, B.M.; Argôlo Filho, R.C.; Nogueira, S.S.C.; Dias, J.C.T.; Rezende, R.P. High Prevalence of Salmonella in Tegu Lizards (Tupinambis Merianae), and Susceptibility of the Serotypes to Antibiotics. Zoonoses Public Health 2010, 57, e26–e32. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- CLISI, Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement; CLSI Document M100-S24; CLSI: Wayne, NJ, USA, 2014; Volume 34. [Google Scholar]
- Dohoo, I.; Martin, W.; Stryhn, H.E. Veterinary Epidemiologic Research, 2nd ed.; University of Prince Edward Island: Charlottetown, Canada, 2010; ISBN 978-0-919013-60-5. [Google Scholar]
- Skov, M.N.; Madsen, J.J.; Rahbek, C.; Lodal, J.; Jespersen, J.B.; Jørgensen, J.C.; Dietz, H.H.; Chriél, M.; Baggesen, D.L. Transmission of Salmonella between Wildlife and Meat-Production Animals in Denmark. J. Appl. Microbiol. 2008, 105, 1558–1568. [Google Scholar] [CrossRef]
- Vogler, B.R.; Zurfluh, K.; Mattmann, P.; Schmitt, K.; Albini, S. Low Occurrence of Salmonella spp. in Wild Birds from a Swiss Rehabilitation Centre. Vet. Rec. Open 2021, 8, e17. [Google Scholar] [CrossRef]
- Botti, V.; Navillod, F.V.; Domenis, L.; Orusa, R.; Pepe, E.; Robetto, S.; Guidetti, C. Salmonella spp. and Antibiotic-Resistant Strains in Wild Mammals and Birds in North-Western Italy from 2002 to 2010. Vet. Ital. 2013, 49, 195–202. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Zając, M.; Kamińska, E.; Bomba, A.; Żmudzki, J.; Jabłoński, A.; Wasyl, D. Salmonella and Antimicrobial Resistance in Wild Rodents—True or False Threat? Pathogens 2020, 9, 771. [Google Scholar] [CrossRef]
- Dos Santos, E.J.E.; Lopes, A.T.S.; Maciel, B.M. Salmonella in Wild Animals: A Public Health Concern. In Enterobacteria; Bhonchal Bhardwaj, S., Ed.; IntechOpen: London, UK, 2022; Volume 136, ISBN 978-1-80355-310-8. [Google Scholar]
- Ribas, A.; Saijuntha, W.; Agatsuma, T.; Prantlová, V.; Poonlaphdecha, S. Rodents as a Source of Salmonella Contamination in Wet Markets in Thailand. Vector Borne Zoonotic Dis. 2016, 16, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.H.; Che, X.; Paulick, A.; Guo, C.; Lee, B.; Muller, D.; Uhlemann, A.-C.; Lowy, F.D.; Corrigan, R.M.; Lipkin, W.I. New York City House Mice (Mus Musculus) as Potential Reservoirs for Pathogenic Bacteria and Antimicrobial Resistance Determinants. mBio 2018, 9, e00624-18. [Google Scholar] [CrossRef] [PubMed]
- Mares, M. Current Topics in Salmonella and Salmonellosis; IntechOpen: London, UK, 2017; Volume 284, ISBN 978-953-51-3066-6. [Google Scholar]
- Backhans, A.; Jacobson, M.; Hansson, I.; Lebbad, M.; Lambertz, S.T.; Gammelgård, E.; Saager, M.; Akande, O.; Fellström, C. Occurrence of Pathogens in Wild Rodents Caught on Swedish Pig and Chicken Farms. Epidemiol. Infect. 2013, 141, 1885–1891. [Google Scholar] [CrossRef]
- De Barros, B.D.C.V.; Chagas, E.N.; Bezerra, L.W.; Ribeiro, L.G.; Duarte Júnior, J.W.B.; Pereira, D.; da Penha Junior, E.T.; Silva, J.R.; Bezerra, D.A.M.; Bandeira, R.S.; et al. Rotavirus A in Wild and Domestic Animals from Areas with Environmental Degradation in the Brazilian Amazon. PLoS ONE 2018, 13, e0209005. [Google Scholar] [CrossRef]
- Carusi, L.C.P.; Farace, M.I.; Ribicich, M.M.; Villafañe, I.E.G. Reproduction and Parasitology of Didelphis albiventris (Didelphimorphia) in an Agroecosystem Landscape in Central Argentina. Mammalia 2009, 73, 89–97. [Google Scholar] [CrossRef]
- Casagrande, R.A.; Lopes, L.F.L.; Reis, E.M.d.; Rodrigues, D.d.P.; Matushima, E.R. Isolation of Salmonella enterica in opossum (Didelphis aurita and Didelphis albiventris) of the São Paulo State, Brazil. Cienc. Rural. 2011, 41, 492–496. [Google Scholar] [CrossRef]
- Reinthaler, F.F.; Posch, J.; Feierl, G.; Wüst, G.; Haas, D.; Ruckenbauer, G.; Mascher, F.; Marth, E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Z.B.; Zeng, Z.L.; Yang, X.W.; Huang, Y.; Liu, J.H. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017, 38, 55–80. [Google Scholar] [CrossRef]
- Jechalke, S.; Heuer, H.; Siemens, J.; Amelung, W.; Smalla, K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014, 22, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Parmar, T.K.; Rawtani, D.; Agrawal, Y.K. Bioindicators: The Natural Indicator of Environmental Pollution. Front. Life Sci. 2016, 9, 110–118. [Google Scholar] [CrossRef]
| Biome | Age | Sex | Municipality | Season | Sampling Date | Anthropization (%) | Serovars | Species | Class |
|---|---|---|---|---|---|---|---|---|---|
| Atlantic Forest | Adult | F | Ilhéus | Rainy | 27 July 2017 | 51.8 | Agona | Ceratopipra rubrocapilla | Avian |
| Atlantic Forest | Adult | F | Itapetinga | Dry | 24 August 2018 | 47.0 | O:16 | Didelphis albiventris | Mammalia |
| Atlantic Forest | Young | F | Itapetinga | Rainy | 5 February 2019 | 79.2 | O:16 | Didelphis albiventris | Mammalia |
| Atlantic Forest | Adult | M | Itambé | Rainy | 7 February 2019 | 70.3 | Muenchen | Cerradomys vivoi | Mammalia |
| Municipality | Family (n) | Biome | Anthropization (%) by Family | Anthropization (%) by Municipality |
|---|---|---|---|---|
| Belmonte | Cricetidae (33) | Atlantic Forest | 39.8 ± 1.51 | 39.9 ± 1.54 |
| Didelphidae (12) | 40.6 ± 1.62 | |||
| Sciuridae (2) | 38.8 ± 0.00 | |||
| Brumado | Sphaerodactylidae (11) | Caatinga | 17.2 ± 0.00 | 17.2 ± 0.00 |
| Ilhéus | Callithrichidae (12) | Atlantic Forest | 21.7 ± 0.00 | 46.2 ± 12.1 |
| Columbidae (5) | 52.1 ± 0.62 | |||
| Dendrocolaptidae (8) | 52.5 ± 0.72 | |||
| Fringillidae (1) | 51.85 | |||
| Onychorhynchidae (1) | 51.85 | |||
| Passerellidae (1) | 51.85 | |||
| Pipridae (10) | 51.8 ± 0.00 | |||
| Thraupidae (5) | 51.8 ± 0.00 | |||
| Trochilidae (2) | 51.8 ± 0.00 | |||
| Turdidae (15) | 52.3 ± 0.70 | |||
| Typhlopidae (1) | 45.4 | |||
| Tyraniidae (1) | 51.8 | |||
| Itambé | Cricetidae (18) | Atlantic Forest | 57.4 ± 12.4 | 56.4 ± 11.8 |
| Didelphidae (36) | 56.2 ± 11.6 | |||
| Itapetinga | Cricetidae (38) | Atlantic Forest | 72.1 ± 11.2 | 68.9 ± 13.9 |
| Didelphidae (84) | 67.4 ± 14.8 | |||
| Lajedo do Tabocal | Cricetidae (56) | Atlantic Forest | 28.7 ± 7.89 | 30.6 ± 8.54 |
| Didelphidae (10) | 36.4 ± 9.49 | |||
| Echimidae (14) | 34.2 ± 7.91 | |||
| Maracás | Cricetidae (46) | Atlantic Forest | 58.6 ± 11.1 | 64.7 ± 21.4 |
| Cricetidae (77) | Caatinga | 63.5 ± 24.8 | ||
| Didelphidae (1) | Atlantic Forest | 40.5 | ||
| Didelphidae (34) | Caatinga | 72.0 ± 21.6 | ||
| Echimidae (16) | 76.3 ± 18.0 | |||
| Muridae (1) | Atlantic Forest | 40.5 | ||
| Mascote | Cricetidae (8) | Atlantic Forest | 85.1 ± 11.7 | 87.2 ± 9.36 |
| Didelphidae (11) | 88.7 ± 7.51 | |||
| Mata de São João | Bradypodidae (11) | Atlantic Forest | 8.52 ± 2.43 | 8.52 ± 2.42 |
| Morro do Chapéu | Phyllodactylidae (1) | Atlantic Forest | 41.1 | 41.1 ± 0.00 |
| Scincidae (1) | 41.1 | |||
| Tropiduridae (2) | 41.1 ± 0.00 | |||
| Una | Callithrichidae (9) | Atlantic Forest | 48.4 ± 0.00 | 12.0 ± 14.4 |
| Columbidae (1) | 5.61 | |||
| Cricetidae (6) | 10.3 ± 10.6 | |||
| Dendrocolaptidae (10) | 5.61 ± 0.00 | |||
| Didelphidae (9) | 11.1 ± 6.84 | |||
| Furnariidae (1) | 5.61 | |||
| Grallariidae (1) | 5.61 | |||
| Picidae (1) | 5.61 | |||
| Pipridae (23) | 5.61 ± 0.00 | |||
| Rhynchocyclidae (1) | 5.61 | |||
| Turdidae (9) | 5.58 ± 0.09 | |||
| Tyraniidae (1) | 5.61 | |||
| Uruçuca | Dendrocolaptidae (6) | Atlantic Forest | 15.4 ± 0.00 | 15.4 ± 0.00 |
| Pipridae (1) | 15.4 ± 0.00 | |||
| Turdidae (8) | 15.4 ± 0.00 | |||
| Tyraniidae (2) | 15.4 ± 0.00 |
| Relative Frequency (%) | Absolute Frequency (n) | Species | Family | Class |
|---|---|---|---|---|
| 14.1 | 95 | Didelphis albiventris | Didelphidae | Mammalia |
| 10.1 | 68 | Necromys lasiurus | Cricetidae | Mammalia |
| 9.20 | 62 | Calomys expulsus | Cricetidae | Mammalia |
| 6.53 | 44 | Cerradomys vivoi | Cricetidae | Mammalia |
| 5.79 | 39 | Marmosops incanus | Didelphidae | Mammalia |
| 4.75 | 32 | Hylaeamys seuanezi | Cricetidae | Mammalia |
| 4.60 | 31 | Gracilinanus agilis | Didelphidae | Mammalia |
| 3.41 | 23 | Rhipidomys mastacalis | Cricetidae | Mammalia |
| 3.41 | 23 | Wiedomys pyrrhorhinos | Cricetidae | Mammalia |
| 3.12 | 21 | Leontopithecus chrysomelas | Callithrichidae | Mammalia |
| 2.67 | 18 | Turdus leucomelas | Turdidae | Avian |
| 2.67 | 18 | Thrichomys aff laurentius | Echimidae | Mammalia |
| 2.37 | 16 | Marmosa murina | Didelphidae | Mammalia |
| 1.93 | 13 | Turdus rufiventris | Turdidae | Avian |
| 1.78 | 12 | Ceratopipra rubrocapilla | Pipridae | Avian |
| 1.63 | 11 | Bradypus torquatus | Bradypodidae | Mammalia |
| 1.63 | 11 | Coleodactylus meridionalis | Sphaerodactylidae | Reptilia |
| 1.34 | 9 | Dixiphia pipra | Pipridae | Avian |
| 1.19 | 8 | Xiphorhynchus fuscus | Dendrocolaptidae | Avian |
| 1.19 | 8 | Oligoryzomys nigripes | Cricetidae | Mammalia |
| 1.19 | 8 | Trinomys albispinus | Echimidae | Mammalia |
| 1.04 | 7 | Dendrocincla turdina | Dendrocolaptidae | Avian |
| 1.04 | 7 | Manacus manacus | Pipridae | Avian |
| 1.04 | 7 | Mus musculus | Cricetidae | Mammalia |
| 1.04 | 7 | Oligoryzomys stramineus | Cricetidae | Mammalia |
| 0.89 | 6 | Machaeropterus regulus | Pipridae | Avian |
| 0.89 | 6 | Cryptonanus agricolai | Didelphidae | Mammalia |
| 0.74 | 5 | Leptotila rufaxilla | Columbidae | Avian |
| 0.59 | 4 | Glyphorynchus spirurus | Dendrocolaptidae | Avian |
| 0.59 | 4 | Trinomys sp. | Echimidae | Mammalia |
| 0.45 | 3 | Monodelphis americana | Didelphidae | Mammalia |
| 7.01 | 48 | Others | - | Avian/Mammalia/Reptilia |
| 100 | 674 | Total | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, E.J.E.d.; Lopes, A.T.S.; Fehlberg, H.F.; Rocha, J.M.; Brito Júnior, P.d.A.; Bernardes, F.C.S.; Costa, T.d.S.O.; Guilherme, E.A.; Vleeschouwer, K.M.D.; Oliveira, L.d.C.; et al. Low Occurrence of Salmonella spp. in Wild Animals in Bahia, Brazil—Population Assessment and Characterization in the Caatinga and Atlantic Forest Biomes. Animals 2024, 14, 21. https://doi.org/10.3390/ani14010021
Santos EJEd, Lopes ATS, Fehlberg HF, Rocha JM, Brito Júnior PdA, Bernardes FCS, Costa TdSO, Guilherme EA, Vleeschouwer KMD, Oliveira LdC, et al. Low Occurrence of Salmonella spp. in Wild Animals in Bahia, Brazil—Population Assessment and Characterization in the Caatinga and Atlantic Forest Biomes. Animals. 2024; 14(1):21. https://doi.org/10.3390/ani14010021
Chicago/Turabian StyleSantos, Eliege Jullia Eudoxia dos, Amanda Teixeira Sampaio Lopes, Hllytchaikra Ferraz Fehlberg, Josiane Moreira Rocha, Pedro de Alcântara Brito Júnior, Fernanda Coelho Simas Bernardes, Thaise da Silva Oliveira Costa, Elisa Arcanjo Guilherme, Kristel Myriam De Vleeschouwer, Leonardo de Carvalho Oliveira, and et al. 2024. "Low Occurrence of Salmonella spp. in Wild Animals in Bahia, Brazil—Population Assessment and Characterization in the Caatinga and Atlantic Forest Biomes" Animals 14, no. 1: 21. https://doi.org/10.3390/ani14010021
APA StyleSantos, E. J. E. d., Lopes, A. T. S., Fehlberg, H. F., Rocha, J. M., Brito Júnior, P. d. A., Bernardes, F. C. S., Costa, T. d. S. O., Guilherme, E. A., Vleeschouwer, K. M. D., Oliveira, L. d. C., Rosa, B. F., Amorim, B. S. d., Filho, L. M. C., Rios, E. O., Ferreira, S. S., Rodrigues, D. d. P., Albuquerque, G. R., Miranda, F. R., Alvarez, M. R. D. V., ... Maciel, B. M. (2024). Low Occurrence of Salmonella spp. in Wild Animals in Bahia, Brazil—Population Assessment and Characterization in the Caatinga and Atlantic Forest Biomes. Animals, 14(1), 21. https://doi.org/10.3390/ani14010021






