Determining the Presence and Size of Shoulder Lesions in Sows Using Computer Vision
Abstract
:Simple Summary
Abstract
1. Introduction
- Compare the performance of deep learning models, including two versions of YOLO (5s, 5m and 8s, 8m) and two versions of FRCNN (R50 Backbone and X-101 Backbone) in the localization of shoulder lesions in various stages of development.
- Compare two traditional imaging segmentation methods (Adapting thresholding and Otsu’s method thresholding) and two deep learning-based U-Net architectures (Vanilla and Attention U-net) to segment lesion pixels and estimate size.
2. Materials and Methods
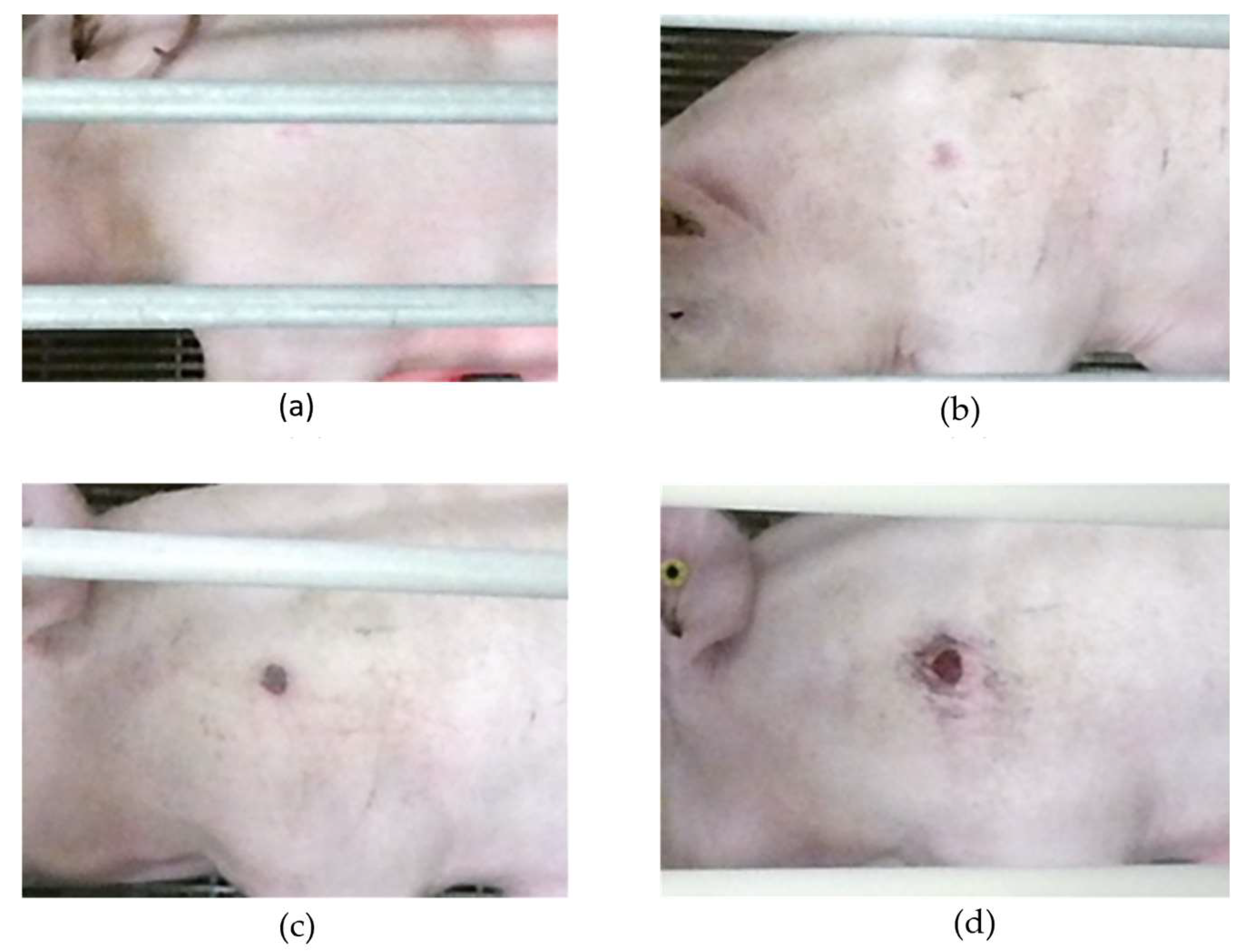
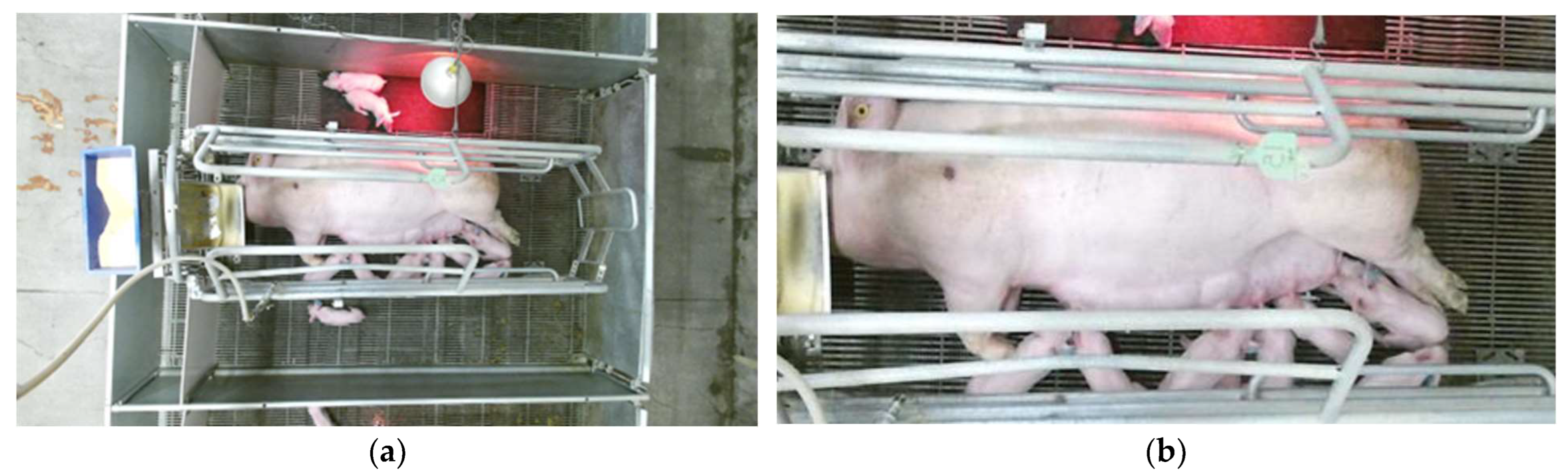
2.1. Data Collection
2.2. Lesion Localization
- YOLO (You Only Look Once) approaches object detection as a regression problem. These models are renowned for their exceptional balance between speed and accuracy, making them ideal for real-time applications. This is crucial in a production setting where rapid and reliable detection of shoulder lesions is essential. The availability of various model sizes (small and medium) allowed for the tailoring of the model to the computational resources available and the optimization of either speed or accuracy. It processes entire images in one step, providing direct predictions for bounding boxes and class probabilities. This study explored two different versions of YOLO developed by Ultralytics [20] as seen in Figure 4, namely v5 and v8, with varying architecture sizes (small “s” and medium “m”).
- Faster-RCNN is a two-stage detector, which uses a Region Proposal Network (RPN) instead of using a selective search algorithm to output object proposals. Region-of-Interest (ROI) pooling is applied to make all proposals the same size. Then, processed proposals are passed to a fully connected layer that classifies the objects in the bounding boxes. Two different backbone models, ResNet-50 and ResNet-101, pre-trained on ImageNet classification tasks were implemented to leverage their deep residual learning framework. This is particularly advantageous for capturing complex features of shoulder lesions, which might be missed by shallower networks like YOLO.

2.3. Lesion Pixel Segmentation
- TP represents the number of true positive pixels, i.e., pixels that are correctly classified as lesions in both the ground truth and predicted masks.
- FP represents the number of false positive pixels, i.e., pixels that are classified as lesions in the predicted mask but not in the ground truth.
- FN represents the number of false negative pixels, i.e., pixels that are lesions in the ground truth but not in the predicted mask.
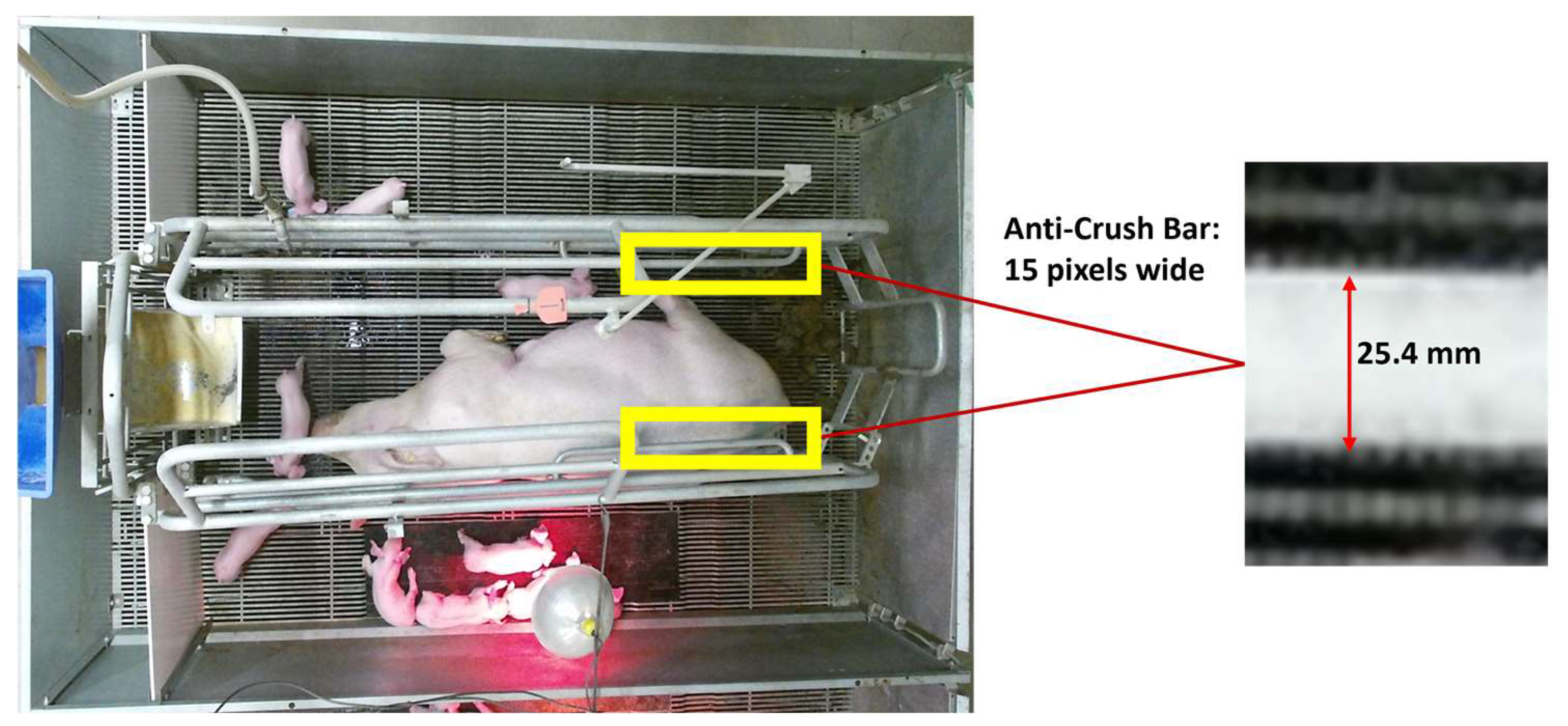
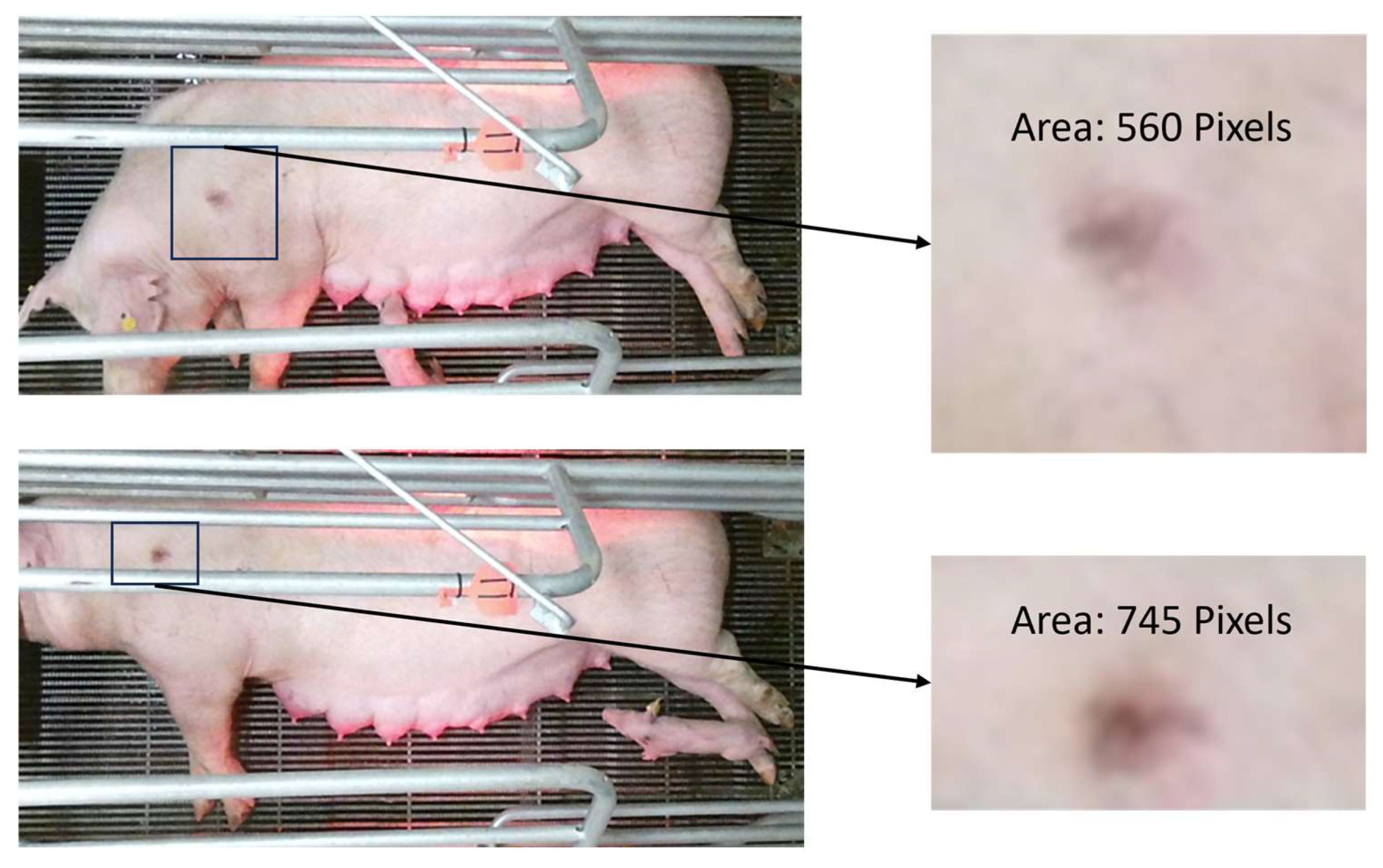
2.4. Size Referencing
3. Results
3.1. Lesion Localization
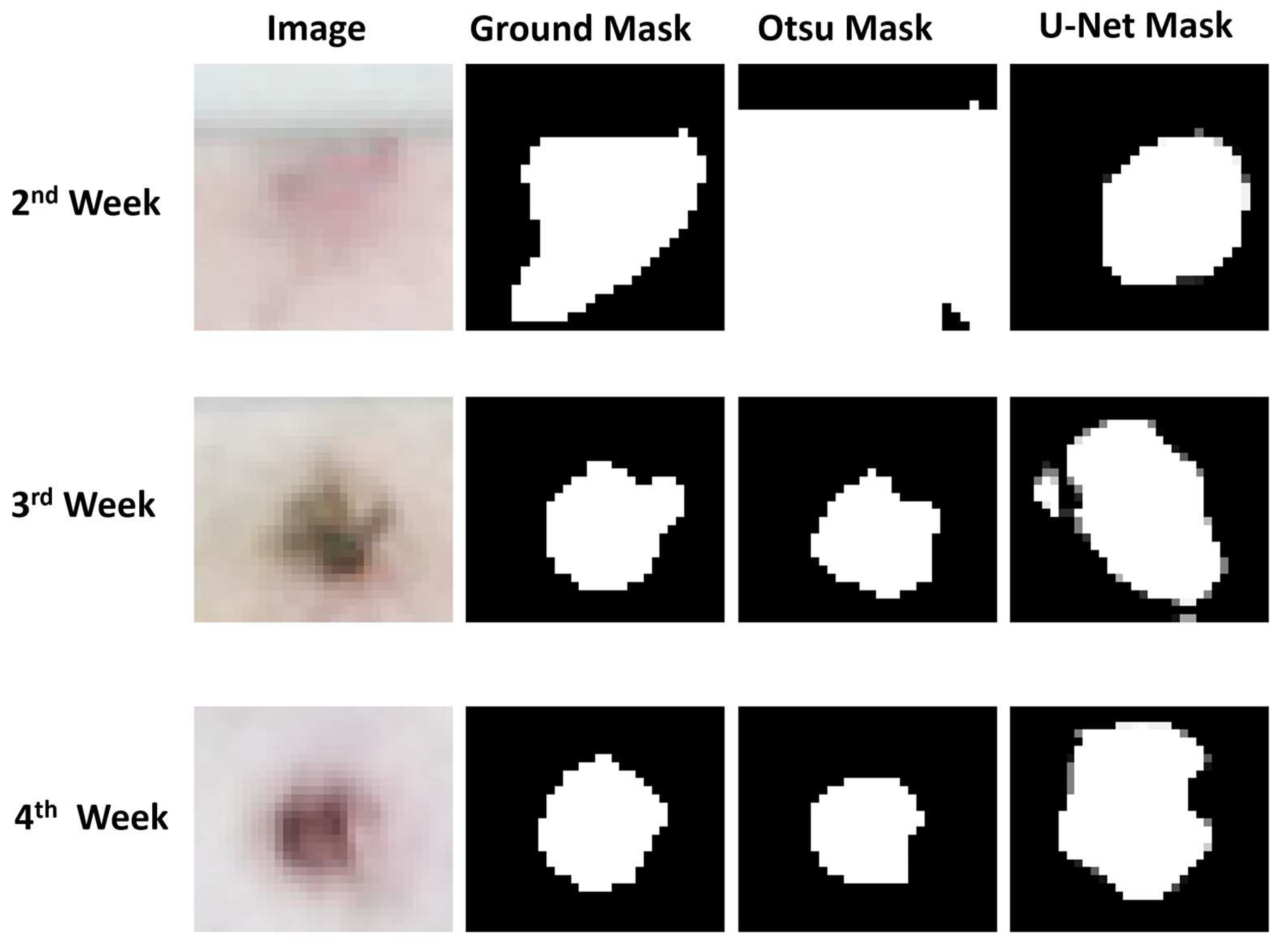
3.2. Lesion Segmentation
3.3. Challenges
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herskin, M.S.; Bonde, M.K.; Jørgensen, E.; Jensen, K.H. Decubital Shoulder Ulcers in Sows: A Review of Classification, Pain and Welfare Consequences. Animal 2011, 5, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Nola, G.T.; Vistnes, L.M. Differential Response of Skin and Muscle in the Experimental Production of Pressure Sores. Plast. Reconstr. Surg. 1980, 66, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Nordbø, Ø.; Gangsei, L.E.; Aasmundstad, T.; Grindflek, E.; Kongsro, J. The Genetic Correlation between Scapula Shape and Shoulder Lesions in Sows. J. Anim. Sci. 2018, 96, 1237–1245. [Google Scholar] [CrossRef]
- The Pig Site Nebraska Swine Report 2005: Shoulder Ulcers in Sows and Their Prevention. Available online: https://www.thepigsite.com/articles/nebraska-swine-report-2005-shoulder-ulcers-in-sows-and-their-prevention (accessed on 13 October 2022).
- Rioja-Lang, F.C.; Seddon, Y.M.; Brown, J.A. Shoulder Lesions in Sows: A Review of Their Causes, Prevention, and Treatment. J. Swine Health Prod. 2018, 26, 101–107. [Google Scholar] [CrossRef]
- Gaab, T.; Nogay, E.; Pierdon, M. Development and Progression of Shoulder Lesions and Their Influence on Sow Behavior. Animals 2022, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Paudel, S.; Brown-Brandl, T.; Rohrer, G.; Sharma, S.R. Individual Pigs’ Identification Using Deep Learning. In Proceedings of the 2023 ASABE Annual International Meeting, Omaha, NE, USA, 9–12 July 2023; p. 1. [Google Scholar]
- Staveley, L.M.; Zemitis, J.E.; Plush, K.J.; D’Souza, D.N. Infrared Thermography for Early Identification and Treatment of Shoulder Sores to Improve Sow and Piglet Welfare. Animals 2022, 12, 3136. [Google Scholar] [CrossRef]
- Shao, H.; Pu, J.; Mu, J. Pig-Posture Recognition Based on Computer Vision: Dataset and Exploration. Animals 2021, 11, 1295. [Google Scholar] [CrossRef]
- Xu, Z.; Tian, F.; Zhou, J.; Zhou, J.; Bromfield, C.; Lim, T.T.; Safranski, T.J.; Yan, Z.; Calyam, P. Posture Identification for Stall-Housed Sows around Estrus Using a Robotic Imaging System. Comput. Electron. Agric. 2023, 211, 107971. [Google Scholar] [CrossRef]
- Wang, R.; Gao, Z.; Li, Q.; Zhao, C.; Gao, R.; Zhang, H.; Li, S.; Feng, L. Detection Method of Cow Estrus Behavior in Natural Scenes Based on Improved YOLOv5. Agriculture 2022, 12, 1339. [Google Scholar] [CrossRef]
- Yap, M.H.; Hachiuma, R.; Alavi, A.; Brüngel, R.; Cassidy, B.; Goyal, M.; Zhu, H.; Rückert, J.; Olshansky, M.; Huang, X.; et al. Deep Learning in Diabetic Foot Ulcers Detection: A Comprehensive Evaluation. Comput. Biol. Med. 2021, 135, 104596. [Google Scholar] [CrossRef]
- Wu, J.; Chen, E.Z.; Rong, R.; Li, X.; Xu, D.; Jiang, H. Skin Lesion Segmentation with C-UNet. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2785–2788. [Google Scholar] [CrossRef]
- Kumar, B.N.; Mahesh, T.R.; Geetha, G.; Guluwadi, S. Redefining Retinal Lesion Segmentation: A Quantum Leap With DL-UNet Enhanced Auto Encoder-Decoder for Fundus Image Analysis. IEEE Access 2023, 11, 70853–70864. [Google Scholar] [CrossRef]
- Mosquera-Berrazueta, L.; Perez, N.; Benitez, D.; Grijalva, F.; Camacho, O.; Herrera, M.; Marrero-Ponce, Y. Red-Unet: An Enhanced U-Net Architecture to Segment Tuberculosis Lesions on X-Ray Images. In Proceedings of the 2023 IEEE 13th International Conference on Pattern Recognition Systems (ICPRS), Guayaquil, Ecuador, 4–7 July 2023. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, Z.; Tu, W.; Zhu, Y. Computed Tomography Image Segmentation of Irregular Cerebral Hemorrhage Lesions Based on Improved U-Net. J. Radiat. Res. Appl. Sci. 2023, 16, 100638. [Google Scholar] [CrossRef]
- Bagheri, F.; Tarokh, M.J.; Ziaratban, M. Semantic Segmentation of Lesions from Dermoscopic Images Using Yolo-Deeplab Networks. Int. J. Eng. Trans. B Appl. 2021, 34, 458–469. [Google Scholar] [CrossRef]
- Jensen, D.B.; Pedersen, L.J. Automatic Counting and Positioning of Slaughter Pigs within the Pen Using a Convolutional Neural Network and Video Images. Comput. Electron. Agric. 2021, 188, 106296. [Google Scholar] [CrossRef]
- Leonard, S.M.; Xin, H.; Brown-Brandl, T.M.; Ramirez, B.C.; Johnson, A.K.; Dutta, S.; Rohrer, G.A. Effects of Farrowing Stall Layout and Number of Heat Lamps on Sow and Piglet Behavior. Appl. Anim. Behav. Sci. 2021, 239, 105334. [Google Scholar] [CrossRef]
- Ren, H.; Wang, Y.; Dong, X. Binarization Algorithm Based on Side Window Multidimensional Convolution Classification. Sensors 2022, 22, 5640. [Google Scholar] [CrossRef] [PubMed]
- Jocher, G.; Chaurasia, A.; Stoken, A.; Borovec, J.; NanoCode012; Kwon, Y.; Michael, K.; Tao, X.; Fang, J.; Imyhxy; et al. Ultralytics/Yolov5: V7.0—YOLOv5 SOTA Realtime Instance Segmentation. Zenodo 2022. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation BT—Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Brüngel, R.; Friedrich, C.M. DETR and YOLOv5: Exploring Performance and Self-Training for Diabetic Foot Ulcer Detection. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 148–153. [Google Scholar]
- Teixeira, D.L.; Boyle, L.A.; Enríquez-Hidalgo, D. Skin Temperature of Slaughter Pigs with Tail Lesions. Front. Vet. Sci. 2020, 7, 198. [Google Scholar] [CrossRef]
- Li, R.; Ji, Z.; Hu, S.; Huang, X.; Yang, J.; Li, W. Tomato Maturity Recognition Model Based on Improved YOLOv5 in Greenhouse. Agronomy 2023, 13, 603. [Google Scholar] [CrossRef]
- Dai, M.; Dorjoy, M.M.H.; Miao, H.; Zhang, S. A New Pest Detection Method Based on Improved YOLOv5m. Insects 2023, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Li, S.; Bai, Q.; Yang, J.; Zhang, X.; Li, Z.; Duan, Z. Object Detection Method for Grasping Robot Based on Improved Yolov5. Micromachines 2021, 12, 1273. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Mou, L.; Zhu, X.X.; Mandal, M. Skin Lesion Segmentation Based on Improved U-Net. In Proceedings of the 2019 IEEE Canadian Conference of Electrical and Computer Engineering (CCECE), Edmonton, AB, Canada, 5–8 May 2019; pp. 1–4. [Google Scholar]
- Tong, X.; Wei, J.; Sun, B.; Su, S.; Zuo, Z.; Wu, P. ASCU-Net: Attention Gate, Spatial and Channel Attention U-Net for Skin Lesion Segmentation. Diagnostics 2021, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, X.; Wu, H.; Zhou, Y.; Sun, X.; Yu, S.; Yu, S.; Liu, H. A Fast 2-D Otsu Lung Tissue Image Segmentation Algorithm Based on Improved PSO. Microprocess. Microsyst. 2021, 80, 103527. [Google Scholar] [CrossRef]
- Yusoff, A.K.M.; Raof, R.A.A.; Mahrom, N.; Noor, S.S.M.; Zakaria, F.F.; Len, P. Enhancement and Segmentation of Ziehl Neelson Sputum Slide Images Using Contrast Enhancement and Otsu Threshold Technique. J. Adv. Res. Appl. Sci. Eng. Technol. 2023, 30, 282–289. [Google Scholar] [CrossRef]



| Week | Number of Images |
|---|---|
| 1 | 93 |
| 2 | 293 |
| 3 | 269 |
| 4 | 169 |
| Architecture | mAP@0.5 | mAP@0.5:0.95::0.05 |
|---|---|---|
| YOLOv5s | 0.91 | 0.46 |
| YOLOv5m | 0.92 | 0.48 |
| YOLOv8s | 0.84 | 0.31 |
| YOLOv8m | 0.81 | 0.35 |
| FRCNN–R50 Backbone | 0.26 | 0.12 |
| FRCNN–X-101 Backbone | 0.56 | 0.14 |
| Segmentation Method | Dice Coefficient | ||
|---|---|---|---|
| 2nd Week | 3rd Week | 4th Week | |
| Adaptive Thresholding | 0.38 | 0.20 | 0.16 |
| Otsu’s Method | 0.66 | 0.83 | 0.82 |
| Vanilla U-Net | 0.72 | 0.63 | 0.61 |
| Attention U-Net | 0.68 | 0.63 | 0.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bery, S.; Brown-Brandl, T.M.; Jones, B.T.; Rohrer, G.A.; Sharma, S.R. Determining the Presence and Size of Shoulder Lesions in Sows Using Computer Vision. Animals 2024, 14, 131. https://doi.org/10.3390/ani14010131
Bery S, Brown-Brandl TM, Jones BT, Rohrer GA, Sharma SR. Determining the Presence and Size of Shoulder Lesions in Sows Using Computer Vision. Animals. 2024; 14(1):131. https://doi.org/10.3390/ani14010131
Chicago/Turabian StyleBery, Shubham, Tami M. Brown-Brandl, Bradley T. Jones, Gary A. Rohrer, and Sudhendu Raj Sharma. 2024. "Determining the Presence and Size of Shoulder Lesions in Sows Using Computer Vision" Animals 14, no. 1: 131. https://doi.org/10.3390/ani14010131
APA StyleBery, S., Brown-Brandl, T. M., Jones, B. T., Rohrer, G. A., & Sharma, S. R. (2024). Determining the Presence and Size of Shoulder Lesions in Sows Using Computer Vision. Animals, 14(1), 131. https://doi.org/10.3390/ani14010131






