Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Samples
2.2. PCR Amplification Conditions and Sequencing
2.3. Genotyping of Dromedaries by TaqI PCR-RFLP
2.4. Bioinformatics
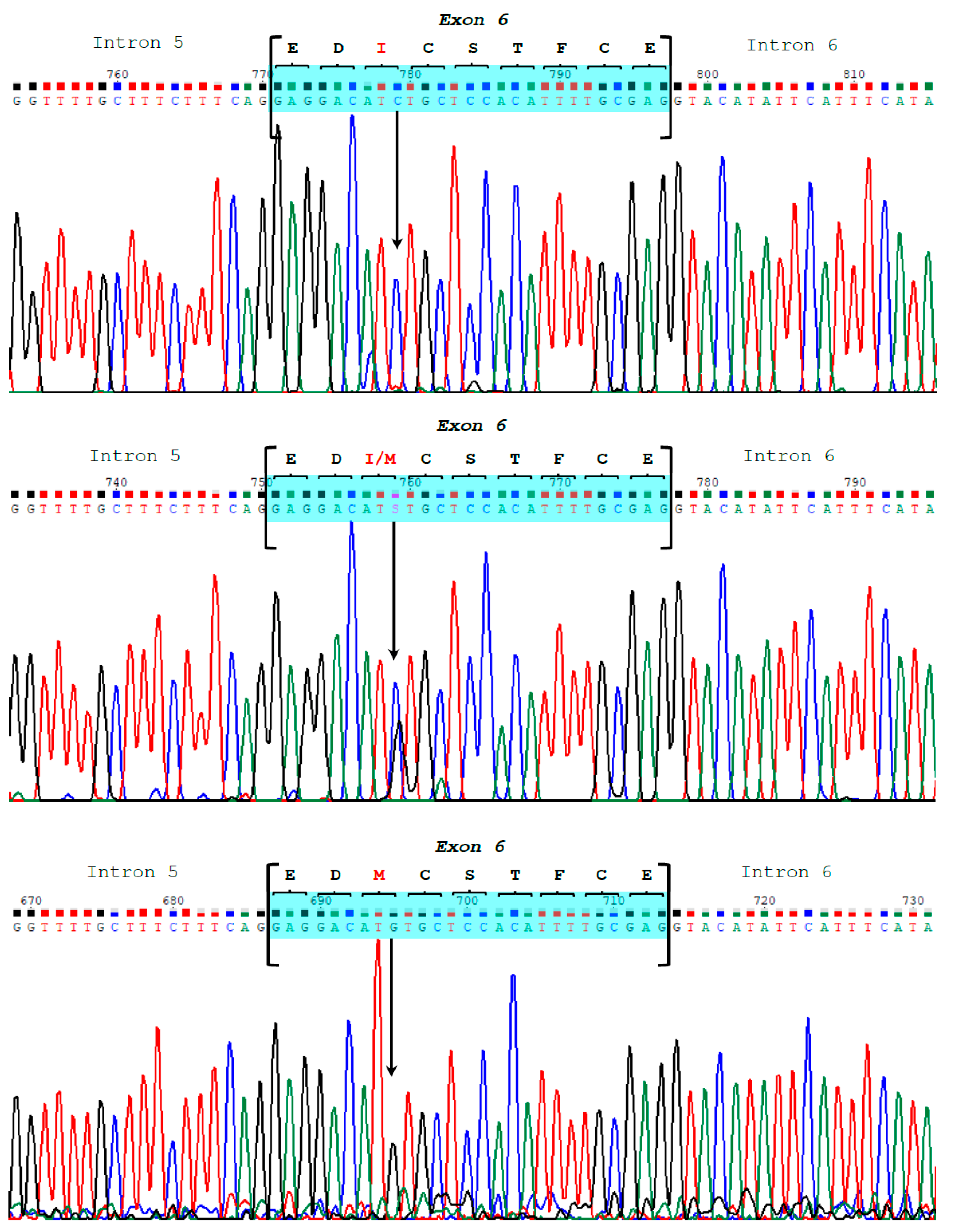
3. Results
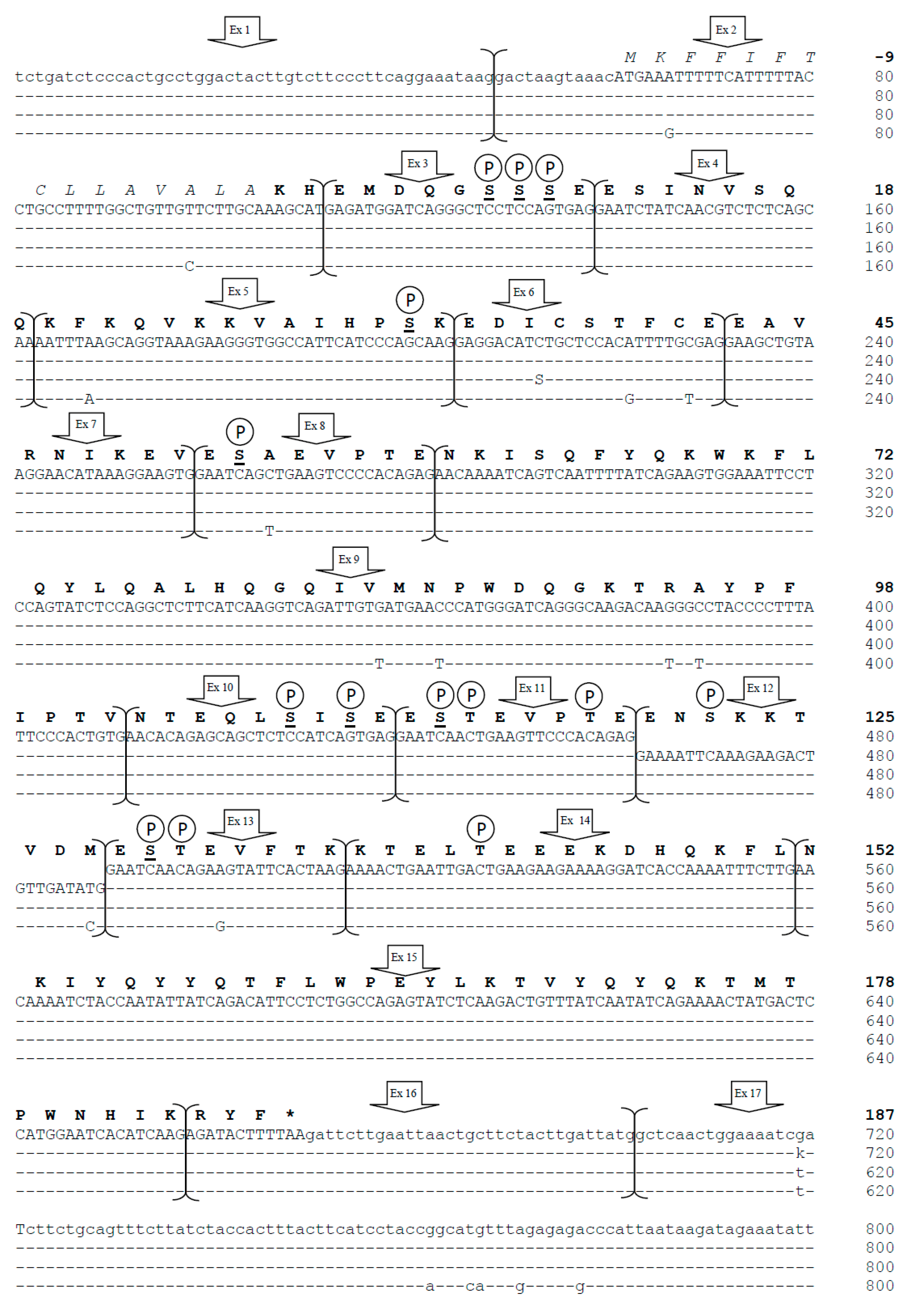
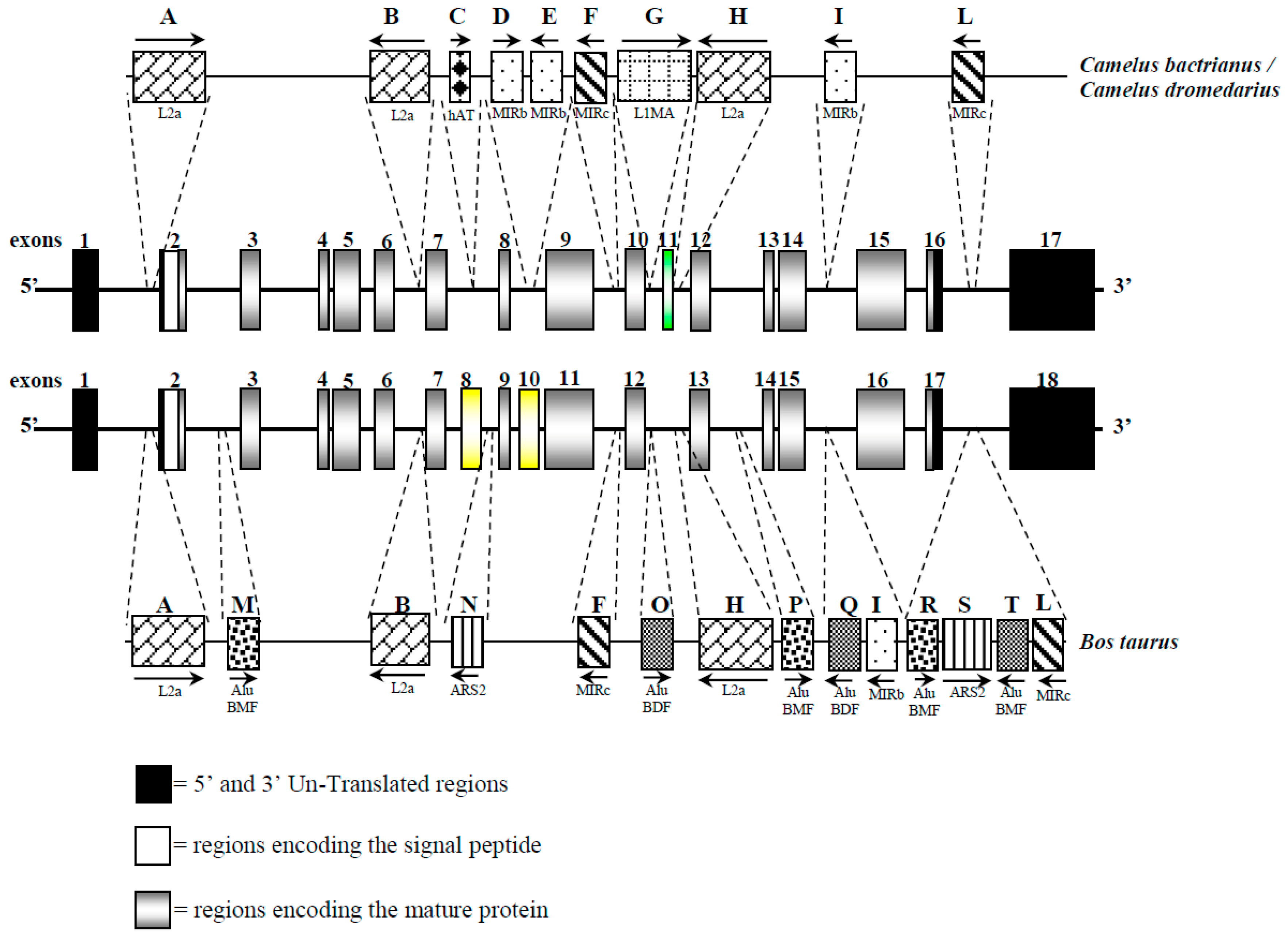
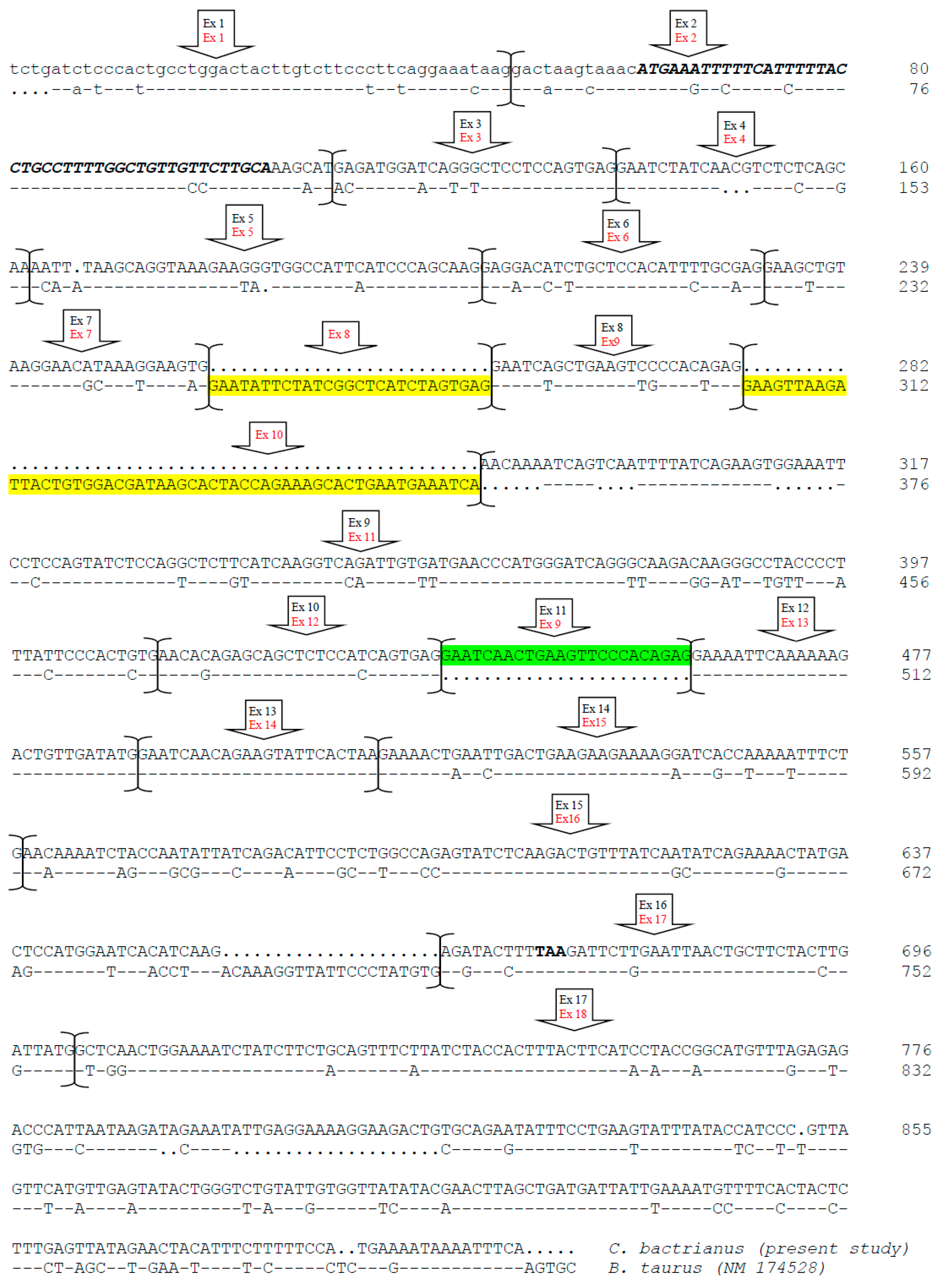
3.1. CSN1S2 Gene Structure in Camels
3.2. Genetic Variability in Bactrian and Dromedary Camels
3.3. CSN1S2 Gene Promoter
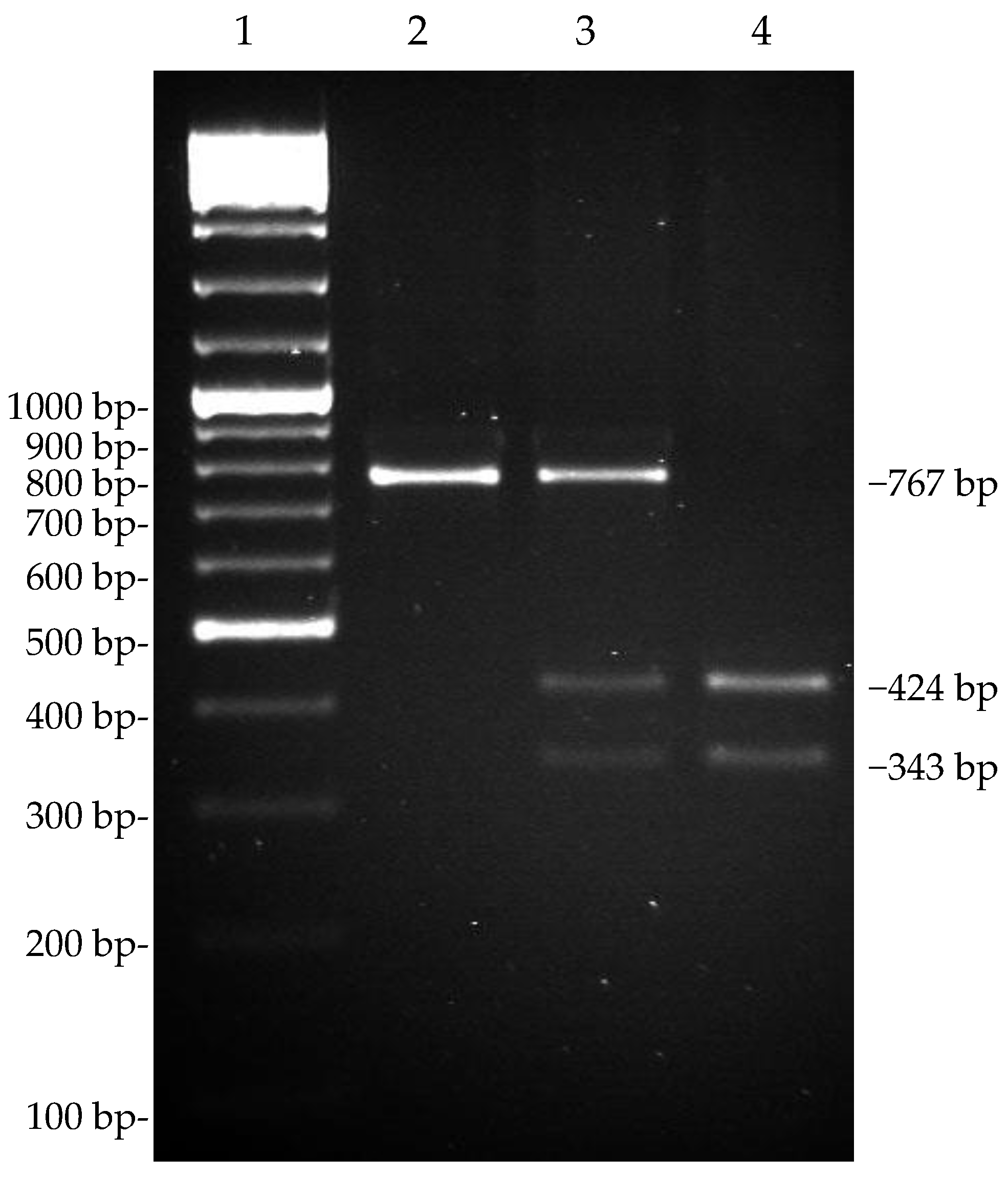
3.4. Genotyping of the SNP g.15110G>T by TaqI PCR-RFLP
3.5. Interspersed Elements and microRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feligini, M.; Bonizzi, I.; Buffoni, J.N.; Cosenza, G.; Ramunno, L. Identification and quantification of αS1, αS2, β, and κ-caseins in water buffalo milk by reverse phase-high performance liquid chromatography and mass spectrometry. J. Agric. Food Chem. 2009, 57, 2988–2992. [Google Scholar] [CrossRef]
- Rijnkels, M. Multispecies comparison of the casein gene loci and evolution of casein gene family. J. Mammary Gland Biol. Neoplasia 2002, 7, 327–345. [Google Scholar] [CrossRef]
- Pauciullo, A.; Shuiep, E.T.; Ogah, M.D.; Cosenza, G.; Di Stasio, L.; Erhardt, G. Casein Gene Cluster in Camelids: Comparative Genome Analysis and New Findings on Haplotype Variability and Physical Mapping. Front. Genet. 2019, 10, 748. [Google Scholar] [CrossRef]
- Kim, J.J.; Yu, J.; Bag, J.; Bakovic, M.; Cant, J.P. Translation attenuation via 3′ terminal codon usage in bovine csn1s2 is responsible for the difference in αs2-and β-casein profile in milk. RNA Biol. 2015, 12, 354–367. [Google Scholar] [CrossRef]
- Kappeler, S.; Farah, Z.; Puhan, Z. Sequence analysis of Camelus dromedarius milk caseins. J. Dairy Res. 1998, 65, 209–222. [Google Scholar] [CrossRef]
- Eigel, W.; Butler, J.; Ernstrom, C.; Farrell, H., Jr.; Harwalkar, V.; Jenness, R.; Whitney, R.M. Nomenclature of proteins of cow’s milk: Fifth revision. J. Dairy Sci. 1984, 67, 1599–1631. [Google Scholar] [CrossRef]
- Giambra, I.J.; Erhardt, G. Molecular genetic characterization of ovine CSN1S2 variants C and D reveal further important variability within CSN1S2. Anim. Genet. 2012, 43, 642–645. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Pappalardo, M.; Longobardi, E.; Gallo, D.; Pastore, N.; Di Gregorio, P.; Rando, A. Characterization of two new alleles at the goat CSN1S2 locus. Anim. Genet. 2001, 32, 264–268. [Google Scholar] [CrossRef]
- Ramunno, L.; Longobardi, E.; Pappalardo, M.; Rando, A.; Di Gregorio, P.; Cosenza, G.; Mariani, P.; Pastore, N.; Masina, P. An allele associated with a non-detectable amount of αs2 casein in goat milk. Anim. Genet. 2001, 32, 19–26. [Google Scholar] [CrossRef]
- Erhardt, G.; Jäger, S.; Budelli, E.; Caroli, A. Genetic polymorphism of goat αS2-casein (CSN1S2) and evidence for a further allele. Milchwissenschaft 2002, 57, 137–140. [Google Scholar]
- Lagonigro, R.; Pietrola, E.; D’Andrea, M.; Veltri, C.; Pilla, F. Molecular genetic characterization of the goat s2-casein E allele. Anim. Genet. 2001, 32, 391–393. [Google Scholar] [CrossRef]
- Farrell, H., Jr.; Jimenez-Flores, R.; Bleck, G.; Brown, E.; Butler, J.; Creamer, L.; Hicks, C.; Hollar, C.; Ng-Kwai-Hang, K.; Swaisgood, H. Nomenclature of the proteins of cows’ milk—Sixth revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, G.; Gallo, D.; Auzino, B.; Gaspa, G.; Pauciullo, A. Complete CSN1S2 Characterization, Novel Allele Identification and Association With Milk Fatty Acid Composition in River Buffalo. Front. Genet. 2021, 11, 622494. [Google Scholar] [CrossRef]
- Pauciullo, A.; Erhardt, G. Molecular Characterization of the Llamas (Lama glama) Casein Cluster Genes Transcripts (CSN1S1, CSN2, CSN1S2, CSN3) and Regulatory Regions. PLoS ONE 2015, 10, e0124963. [Google Scholar] [CrossRef]
- Ryskaliyeva, A.; Henry, C.; Miranda, G.; Faye, B.; Konuspayeva, G.; Martin, P. Alternative splicing events expand molecular diversity of camel CSN1S2 increasing its ability to generate potentially bioactive peptides. Sci. Rep. 2019, 9, 5243. [Google Scholar] [CrossRef] [PubMed]
- Mutery, A.A.; Rais, N.; Mohamed, W.K.; Abdelaziz, T. Genetic diversity in casein gene cluster in a dromedary camel (C. dromedarius) Population from the United Arab Emirates. Genes 2021, 12, 1417. [Google Scholar] [CrossRef]
- Al Haj, O.A.; Al Kanhal, H.A. Compositional, technological and nutritional aspects of dromedary camel milk. Int. Dairy J. 2010, 20, 811–821. [Google Scholar] [CrossRef]
- Letaief, N.; Bedhiaf-Romdhani, S.; Ben Salem, W.; Mohammed, A.; Gaspa, G.; Pauciullo, A. Tunisian camel casein gene characterization reveals similarities and differences with Sudanese and Nigerian populations. J. Dairy Sci. 2022, 105, 6783–6794. [Google Scholar] [CrossRef]
- Pauciullo, A.; Giambra, I.; Iannuzzi, L.; Erhardt, G. The β-casein in camels: Molecular characterization of the CSN2 gene, promoter analysis and genetic variability. Gene 2014, 547, 159–168. [Google Scholar] [CrossRef]
- Pauciullo, A.; Ogah, D.M.; Iannaccone, M.; Erhardt, G.; Di Stasio, L.; Cosenza, G. Genetic characterization of the oxytocin-neurophysin I gene (OXT) and its regulatory regions analysis in domestic Old and New World camelids. PLoS ONE 2018, 13, e0195407. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Erhardt, G.; Shuiep, E.T.S.; Lisson, M.; Weimann, C.; Wang, Z.; Zubeir, I.E.Y.M.E.; Pauciullo, A. Alpha S1-casein polymorphisms in camel (Camelus dromedarius) and descriptions of biological active peptides and allergenic epitopes. Trop. Anim. Health Prod. 2016, 48, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Pauciullo, A.; Shuiep, E.; Cosenza, G.; Ramunno, L.; Erhardt, G. Molecular characterization and genetic variability at κ-casein gene (CSN3) in camels. Gene 2013, 513, 22–30. [Google Scholar] [CrossRef]
- Bingham, E.W.; Farell, H.M., Jr. Phosphorylation of Casein by the Lactating Mammary Gland: A Review. J. Dairy Sci. 1977, 60, 1199–1207. [Google Scholar] [CrossRef]
- Mercier, J.-C. Phosphorylation of caseins, present evidence for an amino acid triplet code posttranslationally recognized by specific kinases. Biochimie 1981, 63, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.-H.; Visker, M.; Miranda, G.; Delacroix-Buchet, A.; Bovenhuis, H.; Martin, P. The relationships among bovine αS-casein phosphorylation isoforms suggest different phosphorylation pathways. J. Dairy Sci. 2016, 99, 8168–8177. [Google Scholar] [CrossRef]
- Baum, F.; Ebner, J.; Pischetsrieder, M. Identification of multiphosphorylated peptides in milk. J. Agric. Food Chem. 2013, 61, 9110–9117. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, S.Y.; Kochin, V.; Ferraris, S.E.; de Thonel, A.; Pallari, H.-M.; Corthals, G.L.; Eriksson, J.E. Reference-facilitated phosphoproteomics: Fast and reliable phosphopeptide validation by μLC-ESI-Q-TOF MS/MS. Mol. Cell. Proteom. 2007, 6, 1380–1391. [Google Scholar] [CrossRef]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kvist, A.; Rego, N.; Staaf, J.; Vallon-Christersson, J.; Luts, L.; Loman, N.; Jonsson, G.; Naya, H.; Hoglund, M. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011, 71, 78–86. [Google Scholar] [CrossRef]
- Anbanandam, A.; Albarado, D.C.; Nguyen, C.T.; Halder, G.; Gao, X.; Veeraraghavan, S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc. Natl. Acad. Sci. USA 2006, 103, 17225–17230. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.M.; Wyszomierski, S.L.; Hadsell, D. Regulation of milk protein gene expression. Annu. Rev. Nutr. 1999, 19, 407–436. [Google Scholar] [CrossRef]
- Wyszomierski, S.L.; Rosen, J.M. Cooperative Effects of STAT5 (Signal Transducer and Activator of Transcription 5) and C/EBP β (CCAAT/Enhancer-Binding Protein-β) onβ -Casein Gene Transcription Are Mediated by the Glucocorticoid Receptor. Mol. Endocrinol. 2001, 15, 228–240. [Google Scholar] [CrossRef]
- Zhao, F.-Q.; Adachi, K.; Oka, T. Involvement of Oct-1 in transcriptional regulation of β-casein gene expression in mouse mammary gland. Biochim. et Biophys. Acta (BBA)—Gene Struct. Expr. 2002, 1577, 27–37. [Google Scholar] [CrossRef]
- Zwilling, S.; Annweiler, A.; Wirth, T. The POU domains of the Oct1 and Oct2 transcription factors mediate specific interaction with TBP. Nucleic Acids Res. 1994, 22, 1655–1662. [Google Scholar] [CrossRef]
- Nelson, C.; Albert, V.R.; Elsholtz, H.P.; Lu, L.I.-W.; Rosenfeld, M.G. Activation of Cell-Specific Expression of Rat Growth Hormone and Prolactin Genes by a Common Transcription Factor. Science 1988, 239, 1400–1405. [Google Scholar] [CrossRef]
- Gil-Puig, C.; Seoane, S.; Blanco, M.; Macia, M.; Garcia-Caballero, T.; Segura, C.; Perez-Fernandez, R. Pit-1 is expressed in normal and tumorous human breast and regulates GH secretion and cell proliferation. Eur. J. Endocrinol. 2005, 153, 335–344. [Google Scholar] [CrossRef]
- Renaville, R.; Gengler, N.; Vrech, E.; Prandi, A.; Massart, S.; Corradini, C.; Bertozzi, C.; Mortiaux, F.; Burny, A.; Portetelle, D. Pit-1 Gene Polymorphism, Milk Yield, and Conformation Traits for Italian Holstein-Friesian Bulls. J. Dairy Sci. 1997, 80, 3431–3438. [Google Scholar] [CrossRef]
- Kuss, A.; Gogol, J.; Bartenschlager, H.; Geldermann, H. Polymorphic AP-1 Binding Site in Bovine CSN1S1 Shows Quantitative Differences in Protein Binding Associated with Milk Protein Expression. J. Dairy Sci. 2005, 88, 2246–2252. [Google Scholar] [CrossRef]
- Karin, M.; Chang, L. Eurosterone meeting AP-1–glucocorticoid receptor crosstalk taken to a higher level. J. Endocrinol. 2001, 169, 447–451. [Google Scholar] [CrossRef]
- Olazabal, I.; Muñoz, J.; Ogueta, S.; Obregón, E.; García-Ruiz, J.P. Prolactin (PRL)-PRL Receptor System Increases Cell Proliferation Involving JNK (c-Jun Amino Terminal Kinase) and AP-1 Activation: Inhibition by Glucocorticoids. Mol. Endocrinol. 2000, 14, 564–575. [Google Scholar] [CrossRef]
- Duan, Z.-J.; Fang, X.; Rohde, A.; Han, H.; Stamatoyannopoulos, G.; Li, Q. Developmental specificity of recruitment of TBP to the TATA box of the human γ-globin gene. Proc. Natl. Acad. Sci. USA 2002, 99, 5509–5514. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.L.; Chandrasekharan, M.B.; Hall, T.C.; Crowe, A.J. Sequence and Spacing of TATA Box Elements Are Critical for Accurate Initiation from the β-Phaseolin Promoter. Perspect. Surg. 2004, 279, 8102–8110. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Ozawa, A.; Ishii, S.; Shibusawa, N.; Hashida, T.; Ishizuka, T.; Hosoya, T.; Monden, T.; Satoh, T.; Mori, M. Isolation and Characterization of the Rat Prolactin-Releasing Peptide Gene: Multiple TATA Boxes in the Promoter Region. Biochem. Biophys. Res. Commun. 2001, 281, 53–56. [Google Scholar] [CrossRef]
- Lechner, J.; Welte, T.; Tomasi, J.K.; Bruno, P.; Cairns, C.; Gustafsson, J.; Doppler, W. Promoter-dependent Synergy between Glucocorticoid Receptor and Stat5 in the Activation of β-Casein Gene Transcription. J. Biol. Chem. 1997, 272, 20954–20960. [Google Scholar] [CrossRef]
- Raught, B.; Khursheed, B.; Kazansky, A.; Rosen, J. YY1 Represses β-Casein Gene Expression by Preventing the Formation of a Lactation-Associated Complex. Mol. Cell. Biol. 1994, 14, 1752–1763. [Google Scholar] [CrossRef]
- Moral, R.; Wang, R.; Russo, I.H.; A Mailo, D.; A Lamartiniere, C.; Russo, J. The plasticizer butyl benzyl phthalate induces genomic changes in rat mammary gland after neonatal/prepubertal exposure. BMC Genom. 2007, 8, 453. [Google Scholar] [CrossRef]
- Kawasaki, K.; Lafont, A.-G.; Sire, J.-Y. The Evolution of Milk Casein Genes from Tooth Genes before the Origin of Mammals. Mol. Biol. Evol. 2011, 28, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and diversity of trasposable elements in vertebrate genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef]
- Goodier, J.L.; Kazazian, H.H., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef]
- Okada, N. Evolution of tRNA-derived SINEs. In The Impact of Short Interspersed Elements (SINEs) on the Host Genome; Springer: New York, NY, USA, 1995. [Google Scholar]
- Lenstra, A.J.; Boxtel, J.A.F.V.; A Zwaagstra, K.; Schwerin, M. Short interspersed nuclear element (SINE) sequences of the Bovidae. Anim. Genet. 1993, 24, 33–39. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Rando, A.; Illario, R.; Gallo, D.; Di Berardino, D.; Masina, P. The goat αs1-casein gene: Gene structure and promoter analysis. Gene 2004, 334, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Program, N.C.S.; Zhao, S.; Bailey, J.A.; Sahinalp, S.C.; Alkan, C.; Tuzun, E.; Green, E.D.; Eichler, E.E. Analysis of Primate Genomic Variation Reveals a Repeat-Driven Expansion of the Human Genome. Genome Res. 2003, 13, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Honey, J.G. Family Camelidae. In The Evolution of Artiodactyls; Johns Hopkins University Press: Baltimore, MD, USA, 2007; pp. 177–188. [Google Scholar]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; AbuTarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef] [PubMed]
- Bibi, F.; Bukhsianidze, M.; Gentry, A.W.; Geraads, D.; Kostopoulos, D.S.; Vrba, E.S. The fossil record and evolution of Bovidae: State of the field. Palaeontol. Electron. 2009, 12, 10A. [Google Scholar]
- Vrba, E.S. Phylogenetic analysis and classification of fossil and recent Alcelaphini Mammalia: Bovidae. Biol. J. Linn. Soc. 1979, 11, 207–228. [Google Scholar] [CrossRef]
- Groenen, M.; Dijkhof, R.; Verstege, A.; van der Poel, J. The complete sequence of the gene encoding bovine α2-casein. Gene 1993, 123, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Parveen, S.; Zhu, P.; Shafique, L.; Lan, H.; Xu, D.; Ashraf, S.; Ashraf, S.; Sherazi, M.; Liu, Q. Molecular Characterization and Phylogenetic Analysis of Casein Gene Family in Camelus ferus. Genes 2023, 14, 256. [Google Scholar] [CrossRef]
- Pauciullo, A.; Gauly, M.; Cosenza, G.; Wagner, H.; Erhardt, G. Lama glama αS1-casein: Identification of new polymorphisms in the CSN1S1 gene. J. Dairy Sci. 2017, 100, 1282–1289. [Google Scholar] [CrossRef]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef]
- Cosenza, G.; Pauciullo, A.; Macciotta, N.P.P.; Apicella, E.; Steri, R.; La Battaglia, A.; Jemma, L.; Coletta, A.; Di Berardino, D.; Ramunno, L. Mediterranean river buffalo CSN1S1 gene: Search for polymorphisms and association studies. Anim. Prod. Sci. 2015, 55, 654–660. [Google Scholar] [CrossRef]
- Pauciullo, A.; Martorello, S.; Carku, K.; Versace, C.; Coletta, A.; Cosenza, G. A novel duplex ACRS-PCR for composite CSN1S1–CSN3 genotype discrimination in domestic buffalo. Ital. J. Anim. Sci. 2021, 20, 1264–1269. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Rando, A.; Pauciullo, A.; Illario, R.; Gallo, D.; Di Berardino, D.; Masina, P. Comparative analysis of gene sequence of goat CSN1S1 F and N alleles and characterization of CSN1S1 transcript variants in mammary gland. Gene 2005, 345, 289–299. [Google Scholar] [CrossRef]
- Fitak, R.R.; Mohandesan, E.; Corander, J.; Burger, P.A. The de novo genome assembly and annotation of a female domestic dromedary of North African origin. Mol. Ecol. Resour. 2016, 16, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Fitak, R.R.; Mohandesan, E.; Corander, J.; Yadamsuren, A.; Chuluunbat, B.; Abdelhadi, O.; Raziq, A.; Nagy, P.; Walzer, C.; Faye, B.; et al. Genomic signatures of domestication in Old World camels. Commun. Biol. 2020, 3, 316. [Google Scholar] [CrossRef] [PubMed]

| Position | Nucleotide | Bactrian Present Study (OQ730238) | Bactrian Genome (NW_011517196) | Nucleotide | Dromedary Present Study (OQ730239) | Dromedary Genome (NW_011591251) |
|---|---|---|---|---|---|---|
| Promoter | 311 | R | G | 311 | G | G |
| 674 | R | G | 674 | G | G | |
| Intron 1 | 845 | Y | T | 845 | T | T |
| 865 | W | T | 865 | T | T | |
| 905 | Y | C | 905 | C | C | |
| 1086 | R | A | 1086 | A | A | |
| 1197 | R | G | 1197 | G | G | |
| 1294 | R | G | 1294 | G | G | |
| 1399 | R | G | 1399 | G | G | |
| 1568 | T | T | 1568 | K | T | |
| Intron 3 | 2848 | R | A | 2857 | A | A |
| 2854 | G | G | 2863 | R | G | |
| Intron 5 | 3530 | Y | T | 3539 | T | T |
| 3587 | W | A | 3596 | A | A | |
| Exon 6 | 3639 | S | C | 3648 | C | C |
| Intron 6 | 3757 | G | G | 3766 | R | G |
| 4429 | C | C | 4438 | Y | C | |
| Intron 7 | 5412 | R | G | 5421 | G | G |
| 5798 | G | G | 5807 | K | G | |
| Intron 8 | 6028 | A | A | 6037 | R | A |
| 6281 | G | G | 6290 | R | G | |
| 7198 | A | A | 7236 | M | A | |
| Intron 9 | 7942 | G | G | 7980 | K | G |
| 8139 | S | C | 8177 | C | C | |
| 8169 | W | T | 8207 | T | T | |
| Intron 10 | 8623 | C | T | 8659 | Y | T |
| 8767 | A | A | 8793 | R | A | |
| Intron 11 | 9662 | T | T | 9698 | Y | T |
| Intron 12 | 9883 | T | T | 9919 | W | T |
| Intron 14 | 11329 | A | A | 11368 | M | A |
| 12073 | Y | C | 12112 | C | C | |
| Intron 15 | 12261 | A | A | 12300 | W | A |
| Intron 16 | 13842 | Y | T | 13881 | T | T |
| Exon 17 | 15069 | T | T | 15110 | K | T |
| Total | 18 | 16 |
| Transcription Factor | Consensus Motif | Signal Sequence | Strand | Score | C. bactrianus | C. dromedarius |
|---|---|---|---|---|---|---|
| AP-1 | NNTGACTCANN | CCTGACTCCCT | + | 0.913 | −677/−667 | −677/−667 |
| TEC1 | TNCATTCYWW | TTCATTCCAT | + | 0.985 | −620/−629 | - |
| AP-4 | NNCAGCTGNN | CACAGCTGGT | + | 0.989 | −591/−582 | −591/−582 |
| Oct-1 | CWNAWTKWSATRYN | CACAATTAAATATG | + | 0.946 | −573/−560 | −573/−560 |
| Pit-1a | NNGAATATKCANNNN | AATATGAATATTATT | − | 0.944 | −565/−551 | −565/−551 |
| C/EBP-β | RNRTKNNGMAAKNN | AAGTTAAGAAAGTA | + | 0.908 | −527/−514 | −527/−514 |
| AP-4 | NNCAGCTGNN | GAGAGCTGAG | − | 0.934 | −482/−473 | −482/−473 |
| C/EBP-β * | RNRTKNNGMAAKNN | GACTTGCATAAGACT | − | 0.909 | −453/−439 | - |
| YY1 | CCATNTWNNNW | CCATATTTTTA | + | 0.899 | −436/−426 | −436/−426 |
| Pit-1a | TGAATAWNWA | TGAATATGAA | + | 0.859 | −404/−395 | −404/−395 |
| Oct-1 | CWNAWTKWSATRYN | AATATGAAAAATGT | − | 0.847 | −402/−389 | −402/−389 |
| Oct-1 | NNNRTAATNANNN | GTATTAATGAAAT | + | 0.870 | −378/−366 | −378/−366 |
| Oct-1 | CWNAWTKWSATRYN | CACATCCAAAATAT | − | 0.890 | −356/−343 | −356/−343 |
| C/EBP-α | NNTKTGGWNANNN | TATTTGTTTAAAG | + | 0.901 | −333/−321 | −333/−321 |
| STAT5A | TTCCCRKAA | TTCTAGGAA | − | 0.956 | −290/−282 | −290/−282 |
| Hfh1 | NAWTGTTTATWT | AAAAAAAAAATC | − | 0.924 | −283/−272 | −283/−272 |
| C/EBP-α | TRRCCAATSRN | GAACCACACAG | + | 0.799 | −269/−259 | −269/−259 |
| HNF-3/FOXA1 | NNNTRTTTRYTY | CTATAAATAATT | − | 0.861 | −222/−211 | −222/−211 |
| GR | NTGCGTRGGCGK | ATTCCTACACAC | + | 0.787 | −213/−202 | −213/−202 |
| TATA box | NCTATAAAAR | ACTATAAAAT | + | 0.964 | −203/−194 | −203/−194 |
| STATx | TTCCCRKAA | TTCTTATAA | + | 0.905 | −187/−179 | −187/−179 |
| STAT5A | TTCCCRKAA | TCCTTGGAA | + | 0.813 | −147/−139 | −147/−139 |
| STAT5A | TTCCCRKAA | TTCTTAGAA | + | 0.912 | −91/−83 | −91/−83 |
| TATA box | WTATAAAW | ATTTAAAT | + | 0.856 | −24/−16 | −24/−16 |
| Total elements | 24 | 22 | ||||
| Genotype Distribution | Allelic Frequency | |||||
|---|---|---|---|---|---|---|
| TT | GT | GG | Total | T | G | |
| Observed | 83 | 61 | 13 | 157 | 0.723 | 0.277 |
| Expected | 82.05 | 62.89 | 12.05 | |||
| Camelus bactrianus CSN1S2 GenBank ID: OQ730238 | Camelus dromedarius CSN1S2 GenBank ID: OQ730239 | Bos taurus CSN1S2 GenBank ID: M94327.1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Name | Strand | Position | Name | Strand | Position | Name | Strand | ||||
| Intron | Nucleotide | Intron | Nucleotide | Intron | Nucleotide | |||||||
| LINEs | ||||||||||||
| LINE 1 | ||||||||||||
| 10 | 8635/8784 | G:L1MA10 | + | 10 | 8613/8834 | G:L1MA10 | + | |||||
| LINE 2 | ||||||||||||
| 1 | 1270/1320 | A: L2a | + | 1 | 1270/1319 | A: L2a | + | 1 | 3953/4000 | A:L2a | + | |
| 6 | 4198/4270 | B:L2a | − | 6 | 4207/4279 | B:L2a | − | 6 | 8069/8151 | B:L2a | − | |
| 11 | 9528/9624 | H:L2a | − | 11 | 9564/9659 | H:L2a | − | 12 | 12212/12306 | H:L2a | − | |
| DNA elements | ||||||||||||
| hAT-Charlie | 7 | 5319/5370 | C:MER5A | + | 7 | 5328/5379 | C:MER5A | + | ||||
| SINEs | ||||||||||||
| MIRs | ||||||||||||
| 8 | 6562/6785 | D:MIRb | + | 8 | 6572/6794 | D:MIRb | + | |||||
| 8 | 7330/7385 | E:MIRb | − | 8 | 7368/7423 | E:MIRb | − | |||||
| 9 | 8093/8232 | F:MIRc | − | 9 | 8131/8270 | F:MIRc | − | 11 | 10742/10888 | F:MIRc | − | |
| 14 | 11593/11775 | I:MIRb | − | 14 | 11632/11814 | I:MIRb | − | 15 | 14773/15100 | I:MIRb | − | |
| 16 | 13828/13938 | L:MIRc | − | 16 | 13867/14030 | L:MIRc | − | 17 | 19404/19481 | L:MIRc | − | |
| Alu/B1 | 2 | 4894/5086 | M:BMF | + | ||||||||
| 8 | 9534/9772 | N:ARS2 | + | |||||||||
| 12 | 11563/11967 | O:BDF | + | |||||||||
| 13 | 13489/13703 | P:BMF | + | |||||||||
| 15 | 14664/14947 | Q:BDF | + | |||||||||
| 17 | 16405/16608 | R:BMF | + | |||||||||
| 17 | 17300/17827 | S:ARS2 | − | |||||||||
| 17 | 19128/19403 | T:BMF | + | |||||||||
| miRNA Name | miRNA Sequence | Seed Location | Custom Target Sequence | Target Score | |
|---|---|---|---|---|---|
| T/T | G/G | ||||
| hsa-miR-298 | AGCAGAAGCAGGGAGGUUCUCCCA | 20 | CTTCTGCA | 93 | 87 |
| hsa-miR-4418 | CACUGCAGGACUCAGCAG | 23 | CTGCAGT | 83 | 76 |
| hsa-miR-3158-5p | CCUGCAGAGAGGAAGCCCUUC | 22 | TCTGCAG | 81 | 65 |
| hsa-miR-548av-3p | AAAACUGCAGUUACUUUUGC | 25 | GCAGTTT | 66 | - |
| hsa-miR-4662a-3p | AAAGAUAGACAAUUGGCUAAAU | 16 | CTATCTT | 52 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauciullo, A.; Versace, C.; Gaspa, G.; Letaief, N.; Bedhiaf-Romdhani, S.; Fulgione, A.; Cosenza, G. Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification. Animals 2023, 13, 2805. https://doi.org/10.3390/ani13172805
Pauciullo A, Versace C, Gaspa G, Letaief N, Bedhiaf-Romdhani S, Fulgione A, Cosenza G. Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification. Animals. 2023; 13(17):2805. https://doi.org/10.3390/ani13172805
Chicago/Turabian StylePauciullo, Alfredo, Carmine Versace, Giustino Gaspa, Neyrouz Letaief, Sonia Bedhiaf-Romdhani, Andrea Fulgione, and Gianfranco Cosenza. 2023. "Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification" Animals 13, no. 17: 2805. https://doi.org/10.3390/ani13172805
APA StylePauciullo, A., Versace, C., Gaspa, G., Letaief, N., Bedhiaf-Romdhani, S., Fulgione, A., & Cosenza, G. (2023). Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification. Animals, 13(17), 2805. https://doi.org/10.3390/ani13172805





