Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Samples
2.2. PCR Amplification Conditions and Sequencing
2.3. Genotyping of Dromedaries by TaqI PCR-RFLP
2.4. Bioinformatics
3. Results
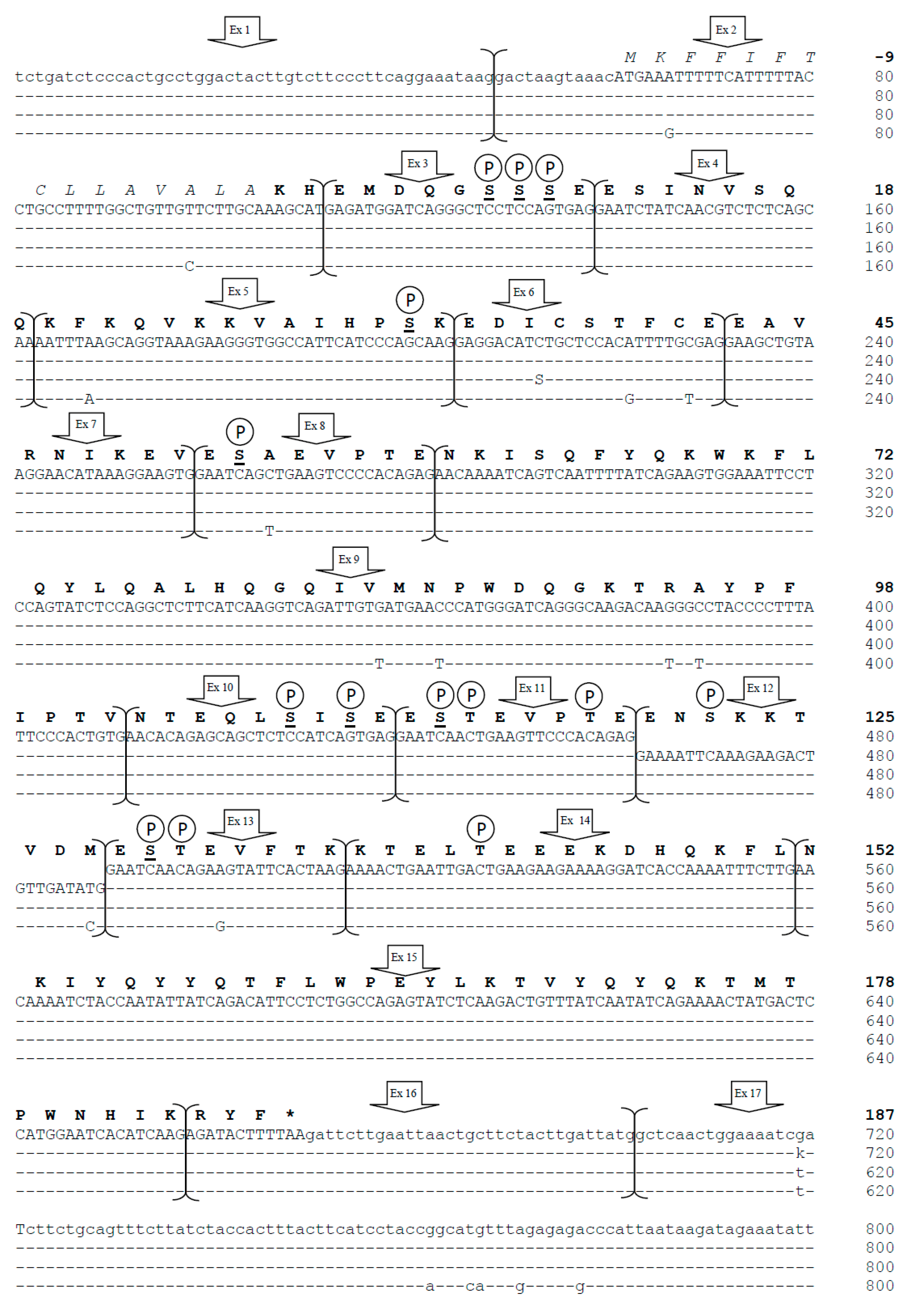
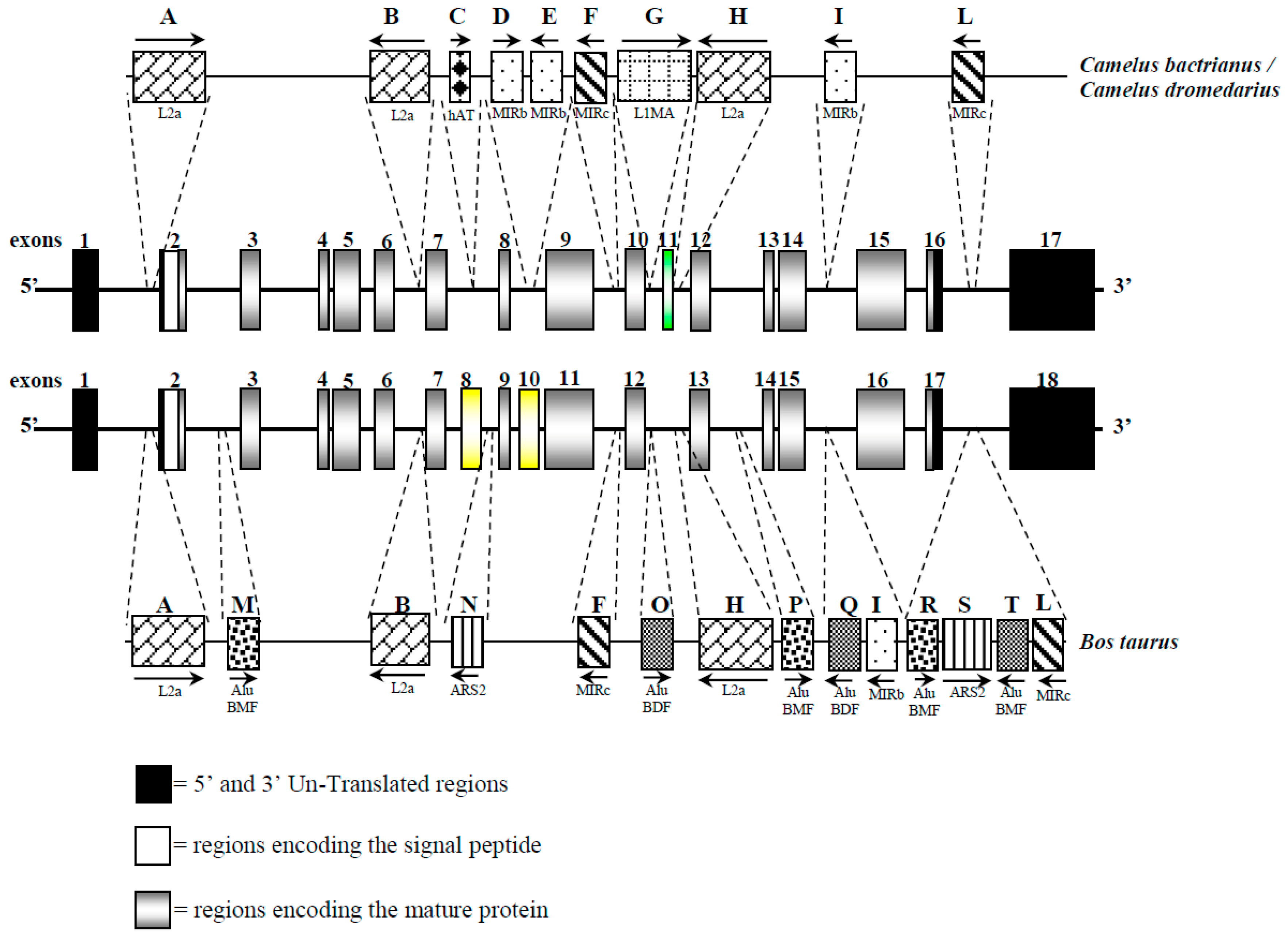
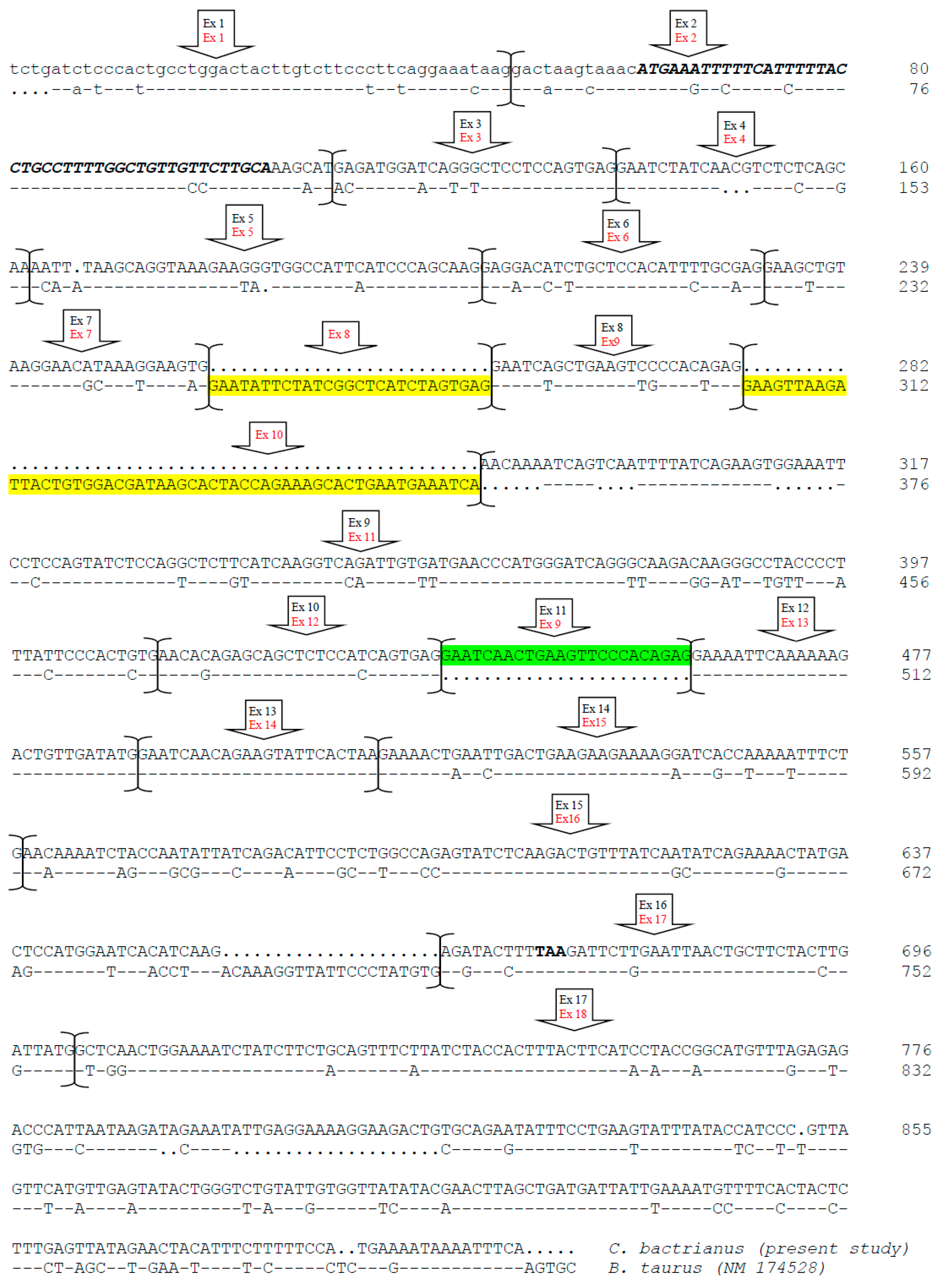
3.1. CSN1S2 Gene Structure in Camels
3.2. Genetic Variability in Bactrian and Dromedary Camels
3.3. CSN1S2 Gene Promoter
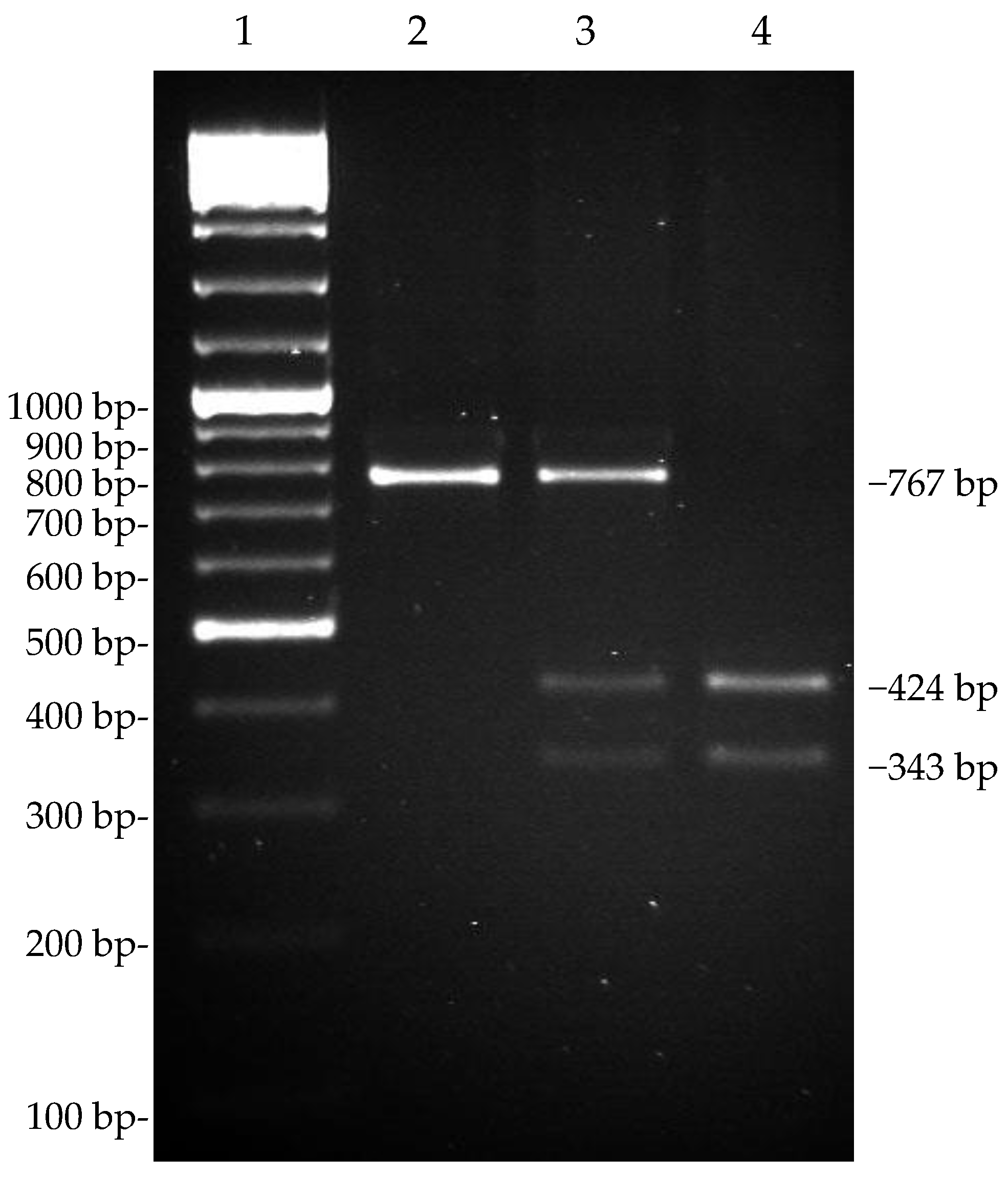
3.4. Genotyping of the SNP g.15110G>T by TaqI PCR-RFLP
3.5. Interspersed Elements and microRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feligini, M.; Bonizzi, I.; Buffoni, J.N.; Cosenza, G.; Ramunno, L. Identification and quantification of αS1, αS2, β, and κ-caseins in water buffalo milk by reverse phase-high performance liquid chromatography and mass spectrometry. J. Agric. Food Chem. 2009, 57, 2988–2992. [Google Scholar] [CrossRef]
- Rijnkels, M. Multispecies comparison of the casein gene loci and evolution of casein gene family. J. Mammary Gland Biol. Neoplasia 2002, 7, 327–345. [Google Scholar] [CrossRef]
- Pauciullo, A.; Shuiep, E.T.; Ogah, M.D.; Cosenza, G.; Di Stasio, L.; Erhardt, G. Casein Gene Cluster in Camelids: Comparative Genome Analysis and New Findings on Haplotype Variability and Physical Mapping. Front. Genet. 2019, 10, 748. [Google Scholar] [CrossRef]
- Kim, J.J.; Yu, J.; Bag, J.; Bakovic, M.; Cant, J.P. Translation attenuation via 3′ terminal codon usage in bovine csn1s2 is responsible for the difference in αs2-and β-casein profile in milk. RNA Biol. 2015, 12, 354–367. [Google Scholar] [CrossRef][Green Version]
- Kappeler, S.; Farah, Z.; Puhan, Z. Sequence analysis of Camelus dromedarius milk caseins. J. Dairy Res. 1998, 65, 209–222. [Google Scholar] [CrossRef]
- Eigel, W.; Butler, J.; Ernstrom, C.; Farrell, H., Jr.; Harwalkar, V.; Jenness, R.; Whitney, R.M. Nomenclature of proteins of cow’s milk: Fifth revision. J. Dairy Sci. 1984, 67, 1599–1631. [Google Scholar] [CrossRef]
- Giambra, I.J.; Erhardt, G. Molecular genetic characterization of ovine CSN1S2 variants C and D reveal further important variability within CSN1S2. Anim. Genet. 2012, 43, 642–645. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Pappalardo, M.; Longobardi, E.; Gallo, D.; Pastore, N.; Di Gregorio, P.; Rando, A. Characterization of two new alleles at the goat CSN1S2 locus. Anim. Genet. 2001, 32, 264–268. [Google Scholar] [CrossRef]
- Ramunno, L.; Longobardi, E.; Pappalardo, M.; Rando, A.; Di Gregorio, P.; Cosenza, G.; Mariani, P.; Pastore, N.; Masina, P. An allele associated with a non-detectable amount of αs2 casein in goat milk. Anim. Genet. 2001, 32, 19–26. [Google Scholar] [CrossRef]
- Erhardt, G.; Jäger, S.; Budelli, E.; Caroli, A. Genetic polymorphism of goat αS2-casein (CSN1S2) and evidence for a further allele. Milchwissenschaft 2002, 57, 137–140. [Google Scholar]
- Lagonigro, R.; Pietrola, E.; D’Andrea, M.; Veltri, C.; Pilla, F. Molecular genetic characterization of the goat s2-casein E allele. Anim. Genet. 2001, 32, 391–393. [Google Scholar] [CrossRef]
- Farrell, H., Jr.; Jimenez-Flores, R.; Bleck, G.; Brown, E.; Butler, J.; Creamer, L.; Hicks, C.; Hollar, C.; Ng-Kwai-Hang, K.; Swaisgood, H. Nomenclature of the proteins of cows’ milk—Sixth revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, G.; Gallo, D.; Auzino, B.; Gaspa, G.; Pauciullo, A. Complete CSN1S2 Characterization, Novel Allele Identification and Association With Milk Fatty Acid Composition in River Buffalo. Front. Genet. 2021, 11, 622494. [Google Scholar] [CrossRef]
- Pauciullo, A.; Erhardt, G. Molecular Characterization of the Llamas (Lama glama) Casein Cluster Genes Transcripts (CSN1S1, CSN2, CSN1S2, CSN3) and Regulatory Regions. PLoS ONE 2015, 10, e0124963. [Google Scholar] [CrossRef][Green Version]
- Ryskaliyeva, A.; Henry, C.; Miranda, G.; Faye, B.; Konuspayeva, G.; Martin, P. Alternative splicing events expand molecular diversity of camel CSN1S2 increasing its ability to generate potentially bioactive peptides. Sci. Rep. 2019, 9, 5243. [Google Scholar] [CrossRef] [PubMed]
- Mutery, A.A.; Rais, N.; Mohamed, W.K.; Abdelaziz, T. Genetic diversity in casein gene cluster in a dromedary camel (C. dromedarius) Population from the United Arab Emirates. Genes 2021, 12, 1417. [Google Scholar] [CrossRef]
- Al Haj, O.A.; Al Kanhal, H.A. Compositional, technological and nutritional aspects of dromedary camel milk. Int. Dairy J. 2010, 20, 811–821. [Google Scholar] [CrossRef]
- Letaief, N.; Bedhiaf-Romdhani, S.; Ben Salem, W.; Mohammed, A.; Gaspa, G.; Pauciullo, A. Tunisian camel casein gene characterization reveals similarities and differences with Sudanese and Nigerian populations. J. Dairy Sci. 2022, 105, 6783–6794. [Google Scholar] [CrossRef]
- Pauciullo, A.; Giambra, I.; Iannuzzi, L.; Erhardt, G. The β-casein in camels: Molecular characterization of the CSN2 gene, promoter analysis and genetic variability. Gene 2014, 547, 159–168. [Google Scholar] [CrossRef]
- Pauciullo, A.; Ogah, D.M.; Iannaccone, M.; Erhardt, G.; Di Stasio, L.; Cosenza, G. Genetic characterization of the oxytocin-neurophysin I gene (OXT) and its regulatory regions analysis in domestic Old and New World camelids. PLoS ONE 2018, 13, e0195407. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Erhardt, G.; Shuiep, E.T.S.; Lisson, M.; Weimann, C.; Wang, Z.; Zubeir, I.E.Y.M.E.; Pauciullo, A. Alpha S1-casein polymorphisms in camel (Camelus dromedarius) and descriptions of biological active peptides and allergenic epitopes. Trop. Anim. Health Prod. 2016, 48, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Pauciullo, A.; Shuiep, E.; Cosenza, G.; Ramunno, L.; Erhardt, G. Molecular characterization and genetic variability at κ-casein gene (CSN3) in camels. Gene 2013, 513, 22–30. [Google Scholar] [CrossRef]
- Bingham, E.W.; Farell, H.M., Jr. Phosphorylation of Casein by the Lactating Mammary Gland: A Review. J. Dairy Sci. 1977, 60, 1199–1207. [Google Scholar] [CrossRef]
- Mercier, J.-C. Phosphorylation of caseins, present evidence for an amino acid triplet code posttranslationally recognized by specific kinases. Biochimie 1981, 63, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.-H.; Visker, M.; Miranda, G.; Delacroix-Buchet, A.; Bovenhuis, H.; Martin, P. The relationships among bovine αS-casein phosphorylation isoforms suggest different phosphorylation pathways. J. Dairy Sci. 2016, 99, 8168–8177. [Google Scholar] [CrossRef]
- Baum, F.; Ebner, J.; Pischetsrieder, M. Identification of multiphosphorylated peptides in milk. J. Agric. Food Chem. 2013, 61, 9110–9117. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, S.Y.; Kochin, V.; Ferraris, S.E.; de Thonel, A.; Pallari, H.-M.; Corthals, G.L.; Eriksson, J.E. Reference-facilitated phosphoproteomics: Fast and reliable phosphopeptide validation by μLC-ESI-Q-TOF MS/MS. Mol. Cell. Proteom. 2007, 6, 1380–1391. [Google Scholar] [CrossRef]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kvist, A.; Rego, N.; Staaf, J.; Vallon-Christersson, J.; Luts, L.; Loman, N.; Jonsson, G.; Naya, H.; Hoglund, M. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011, 71, 78–86. [Google Scholar] [CrossRef]
- Anbanandam, A.; Albarado, D.C.; Nguyen, C.T.; Halder, G.; Gao, X.; Veeraraghavan, S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc. Natl. Acad. Sci. USA 2006, 103, 17225–17230. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.M.; Wyszomierski, S.L.; Hadsell, D. Regulation of milk protein gene expression. Annu. Rev. Nutr. 1999, 19, 407–436. [Google Scholar] [CrossRef]
- Wyszomierski, S.L.; Rosen, J.M. Cooperative Effects of STAT5 (Signal Transducer and Activator of Transcription 5) and C/EBP β (CCAAT/Enhancer-Binding Protein-β) onβ -Casein Gene Transcription Are Mediated by the Glucocorticoid Receptor. Mol. Endocrinol. 2001, 15, 228–240. [Google Scholar] [CrossRef][Green Version]
- Zhao, F.-Q.; Adachi, K.; Oka, T. Involvement of Oct-1 in transcriptional regulation of β-casein gene expression in mouse mammary gland. Biochim. et Biophys. Acta (BBA)—Gene Struct. Expr. 2002, 1577, 27–37. [Google Scholar] [CrossRef]
- Zwilling, S.; Annweiler, A.; Wirth, T. The POU domains of the Oct1 and Oct2 transcription factors mediate specific interaction with TBP. Nucleic Acids Res. 1994, 22, 1655–1662. [Google Scholar] [CrossRef]
- Nelson, C.; Albert, V.R.; Elsholtz, H.P.; Lu, L.I.-W.; Rosenfeld, M.G. Activation of Cell-Specific Expression of Rat Growth Hormone and Prolactin Genes by a Common Transcription Factor. Science 1988, 239, 1400–1405. [Google Scholar] [CrossRef]
- Gil-Puig, C.; Seoane, S.; Blanco, M.; Macia, M.; Garcia-Caballero, T.; Segura, C.; Perez-Fernandez, R. Pit-1 is expressed in normal and tumorous human breast and regulates GH secretion and cell proliferation. Eur. J. Endocrinol. 2005, 153, 335–344. [Google Scholar] [CrossRef]
- Renaville, R.; Gengler, N.; Vrech, E.; Prandi, A.; Massart, S.; Corradini, C.; Bertozzi, C.; Mortiaux, F.; Burny, A.; Portetelle, D. Pit-1 Gene Polymorphism, Milk Yield, and Conformation Traits for Italian Holstein-Friesian Bulls. J. Dairy Sci. 1997, 80, 3431–3438. [Google Scholar] [CrossRef]
- Kuss, A.; Gogol, J.; Bartenschlager, H.; Geldermann, H. Polymorphic AP-1 Binding Site in Bovine CSN1S1 Shows Quantitative Differences in Protein Binding Associated with Milk Protein Expression. J. Dairy Sci. 2005, 88, 2246–2252. [Google Scholar] [CrossRef]
- Karin, M.; Chang, L. Eurosterone meeting AP-1–glucocorticoid receptor crosstalk taken to a higher level. J. Endocrinol. 2001, 169, 447–451. [Google Scholar] [CrossRef]
- Olazabal, I.; Muñoz, J.; Ogueta, S.; Obregón, E.; García-Ruiz, J.P. Prolactin (PRL)-PRL Receptor System Increases Cell Proliferation Involving JNK (c-Jun Amino Terminal Kinase) and AP-1 Activation: Inhibition by Glucocorticoids. Mol. Endocrinol. 2000, 14, 564–575. [Google Scholar] [CrossRef]
- Duan, Z.-J.; Fang, X.; Rohde, A.; Han, H.; Stamatoyannopoulos, G.; Li, Q. Developmental specificity of recruitment of TBP to the TATA box of the human γ-globin gene. Proc. Natl. Acad. Sci. USA 2002, 99, 5509–5514. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.L.; Chandrasekharan, M.B.; Hall, T.C.; Crowe, A.J. Sequence and Spacing of TATA Box Elements Are Critical for Accurate Initiation from the β-Phaseolin Promoter. Perspect. Surg. 2004, 279, 8102–8110. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Ozawa, A.; Ishii, S.; Shibusawa, N.; Hashida, T.; Ishizuka, T.; Hosoya, T.; Monden, T.; Satoh, T.; Mori, M. Isolation and Characterization of the Rat Prolactin-Releasing Peptide Gene: Multiple TATA Boxes in the Promoter Region. Biochem. Biophys. Res. Commun. 2001, 281, 53–56. [Google Scholar] [CrossRef]
- Lechner, J.; Welte, T.; Tomasi, J.K.; Bruno, P.; Cairns, C.; Gustafsson, J.; Doppler, W. Promoter-dependent Synergy between Glucocorticoid Receptor and Stat5 in the Activation of β-Casein Gene Transcription. J. Biol. Chem. 1997, 272, 20954–20960. [Google Scholar] [CrossRef]
- Raught, B.; Khursheed, B.; Kazansky, A.; Rosen, J. YY1 Represses β-Casein Gene Expression by Preventing the Formation of a Lactation-Associated Complex. Mol. Cell. Biol. 1994, 14, 1752–1763. [Google Scholar] [CrossRef]
- Moral, R.; Wang, R.; Russo, I.H.; A Mailo, D.; A Lamartiniere, C.; Russo, J. The plasticizer butyl benzyl phthalate induces genomic changes in rat mammary gland after neonatal/prepubertal exposure. BMC Genom. 2007, 8, 453. [Google Scholar] [CrossRef]
- Kawasaki, K.; Lafont, A.-G.; Sire, J.-Y. The Evolution of Milk Casein Genes from Tooth Genes before the Origin of Mammals. Mol. Biol. Evol. 2011, 28, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and diversity of trasposable elements in vertebrate genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef]
- Goodier, J.L.; Kazazian, H.H., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef]
- Okada, N. Evolution of tRNA-derived SINEs. In The Impact of Short Interspersed Elements (SINEs) on the Host Genome; Springer: New York, NY, USA, 1995. [Google Scholar]
- Lenstra, A.J.; Boxtel, J.A.F.V.; A Zwaagstra, K.; Schwerin, M. Short interspersed nuclear element (SINE) sequences of the Bovidae. Anim. Genet. 1993, 24, 33–39. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Rando, A.; Illario, R.; Gallo, D.; Di Berardino, D.; Masina, P. The goat αs1-casein gene: Gene structure and promoter analysis. Gene 2004, 334, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Program, N.C.S.; Zhao, S.; Bailey, J.A.; Sahinalp, S.C.; Alkan, C.; Tuzun, E.; Green, E.D.; Eichler, E.E. Analysis of Primate Genomic Variation Reveals a Repeat-Driven Expansion of the Human Genome. Genome Res. 2003, 13, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Honey, J.G. Family Camelidae. In The Evolution of Artiodactyls; Johns Hopkins University Press: Baltimore, MD, USA, 2007; pp. 177–188. [Google Scholar]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; AbuTarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef] [PubMed]
- Bibi, F.; Bukhsianidze, M.; Gentry, A.W.; Geraads, D.; Kostopoulos, D.S.; Vrba, E.S. The fossil record and evolution of Bovidae: State of the field. Palaeontol. Electron. 2009, 12, 10A. [Google Scholar]
- Vrba, E.S. Phylogenetic analysis and classification of fossil and recent Alcelaphini Mammalia: Bovidae. Biol. J. Linn. Soc. 1979, 11, 207–228. [Google Scholar] [CrossRef]
- Groenen, M.; Dijkhof, R.; Verstege, A.; van der Poel, J. The complete sequence of the gene encoding bovine α2-casein. Gene 1993, 123, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Parveen, S.; Zhu, P.; Shafique, L.; Lan, H.; Xu, D.; Ashraf, S.; Ashraf, S.; Sherazi, M.; Liu, Q. Molecular Characterization and Phylogenetic Analysis of Casein Gene Family in Camelus ferus. Genes 2023, 14, 256. [Google Scholar] [CrossRef]
- Pauciullo, A.; Gauly, M.; Cosenza, G.; Wagner, H.; Erhardt, G. Lama glama αS1-casein: Identification of new polymorphisms in the CSN1S1 gene. J. Dairy Sci. 2017, 100, 1282–1289. [Google Scholar] [CrossRef]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef]
- Cosenza, G.; Pauciullo, A.; Macciotta, N.P.P.; Apicella, E.; Steri, R.; La Battaglia, A.; Jemma, L.; Coletta, A.; Di Berardino, D.; Ramunno, L. Mediterranean river buffalo CSN1S1 gene: Search for polymorphisms and association studies. Anim. Prod. Sci. 2015, 55, 654–660. [Google Scholar] [CrossRef]
- Pauciullo, A.; Martorello, S.; Carku, K.; Versace, C.; Coletta, A.; Cosenza, G. A novel duplex ACRS-PCR for composite CSN1S1–CSN3 genotype discrimination in domestic buffalo. Ital. J. Anim. Sci. 2021, 20, 1264–1269. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Rando, A.; Pauciullo, A.; Illario, R.; Gallo, D.; Di Berardino, D.; Masina, P. Comparative analysis of gene sequence of goat CSN1S1 F and N alleles and characterization of CSN1S1 transcript variants in mammary gland. Gene 2005, 345, 289–299. [Google Scholar] [CrossRef]
- Fitak, R.R.; Mohandesan, E.; Corander, J.; Burger, P.A. The de novo genome assembly and annotation of a female domestic dromedary of North African origin. Mol. Ecol. Resour. 2016, 16, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Fitak, R.R.; Mohandesan, E.; Corander, J.; Yadamsuren, A.; Chuluunbat, B.; Abdelhadi, O.; Raziq, A.; Nagy, P.; Walzer, C.; Faye, B.; et al. Genomic signatures of domestication in Old World camels. Commun. Biol. 2020, 3, 316. [Google Scholar] [CrossRef] [PubMed]

| Position | Nucleotide | Bactrian Present Study (OQ730238) | Bactrian Genome (NW_011517196) | Nucleotide | Dromedary Present Study (OQ730239) | Dromedary Genome (NW_011591251) |
|---|---|---|---|---|---|---|
| Promoter | 311 | R | G | 311 | G | G |
| 674 | R | G | 674 | G | G | |
| Intron 1 | 845 | Y | T | 845 | T | T |
| 865 | W | T | 865 | T | T | |
| 905 | Y | C | 905 | C | C | |
| 1086 | R | A | 1086 | A | A | |
| 1197 | R | G | 1197 | G | G | |
| 1294 | R | G | 1294 | G | G | |
| 1399 | R | G | 1399 | G | G | |
| 1568 | T | T | 1568 | K | T | |
| Intron 3 | 2848 | R | A | 2857 | A | A |
| 2854 | G | G | 2863 | R | G | |
| Intron 5 | 3530 | Y | T | 3539 | T | T |
| 3587 | W | A | 3596 | A | A | |
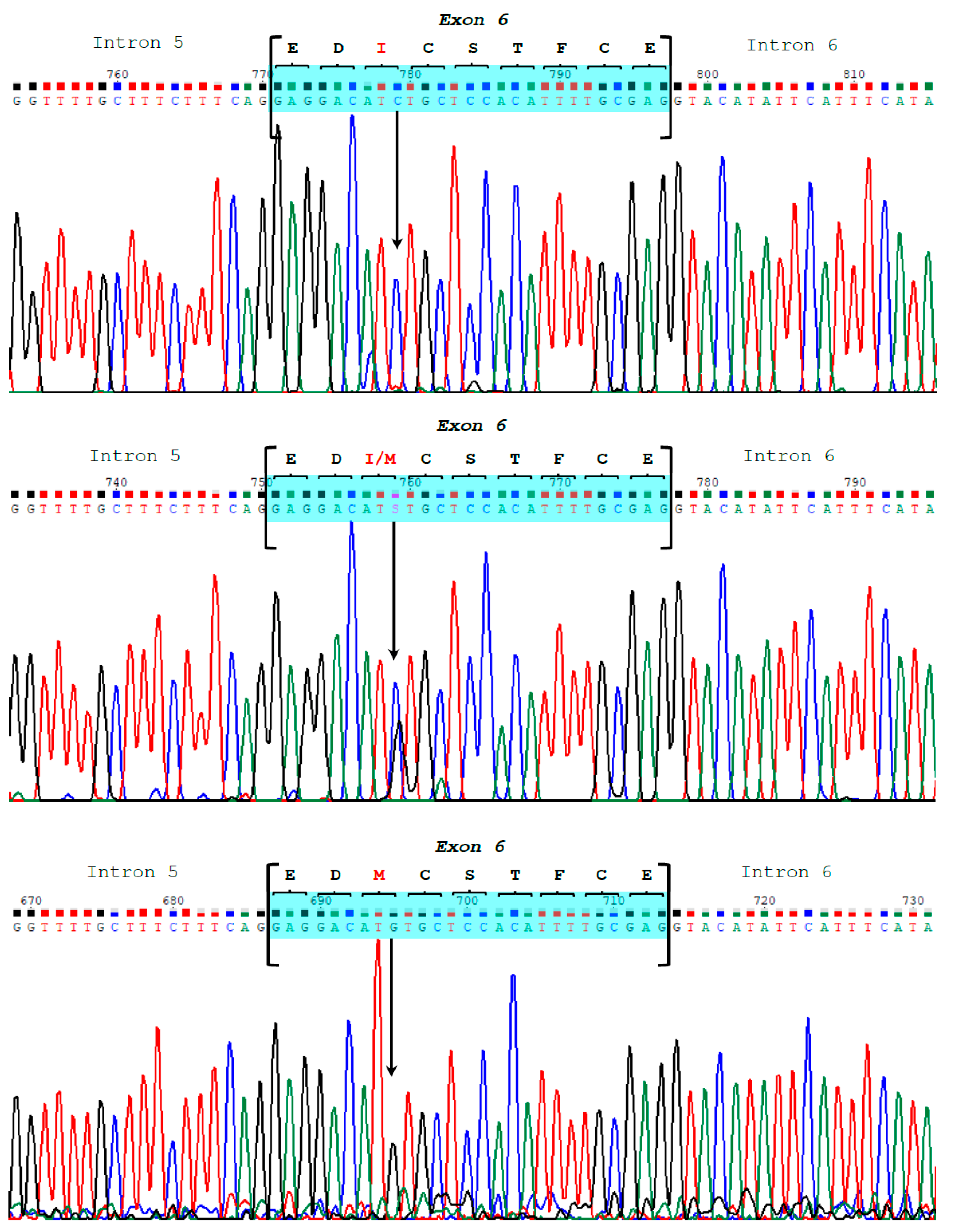
| Exon 6 | 3639 | S | C | 3648 | C | C |
| Intron 6 | 3757 | G | G | 3766 | R | G |
| 4429 | C | C | 4438 | Y | C | |
| Intron 7 | 5412 | R | G | 5421 | G | G |
| 5798 | G | G | 5807 | K | G | |
| Intron 8 | 6028 | A | A | 6037 | R | A |
| 6281 | G | G | 6290 | R | G | |
| 7198 | A | A | 7236 | M | A | |
| Intron 9 | 7942 | G | G | 7980 | K | G |
| 8139 | S | C | 8177 | C | C | |
| 8169 | W | T | 8207 | T | T | |
| Intron 10 | 8623 | C | T | 8659 | Y | T |
| 8767 | A | A | 8793 | R | A | |
| Intron 11 | 9662 | T | T | 9698 | Y | T |
| Intron 12 | 9883 | T | T | 9919 | W | T |
| Intron 14 | 11329 | A | A | 11368 | M | A |
| 12073 | Y | C | 12112 | C | C | |
| Intron 15 | 12261 | A | A | 12300 | W | A |
| Intron 16 | 13842 | Y | T | 13881 | T | T |
| Exon 17 | 15069 | T | T | 15110 | K | T |
| Total | 18 | 16 |
| Transcription Factor | Consensus Motif | Signal Sequence | Strand | Score | C. bactrianus | C. dromedarius |
|---|---|---|---|---|---|---|
| AP-1 | NNTGACTCANN | CCTGACTCCCT | + | 0.913 | −677/−667 | −677/−667 |
| TEC1 | TNCATTCYWW | TTCATTCCAT | + | 0.985 | −620/−629 | - |
| AP-4 | NNCAGCTGNN | CACAGCTGGT | + | 0.989 | −591/−582 | −591/−582 |
| Oct-1 | CWNAWTKWSATRYN | CACAATTAAATATG | + | 0.946 | −573/−560 | −573/−560 |
| Pit-1a | NNGAATATKCANNNN | AATATGAATATTATT | − | 0.944 | −565/−551 | −565/−551 |
| C/EBP-β | RNRTKNNGMAAKNN | AAGTTAAGAAAGTA | + | 0.908 | −527/−514 | −527/−514 |
| AP-4 | NNCAGCTGNN | GAGAGCTGAG | − | 0.934 | −482/−473 | −482/−473 |
| C/EBP-β * | RNRTKNNGMAAKNN | GACTTGCATAAGACT | − | 0.909 | −453/−439 | - |
| YY1 | CCATNTWNNNW | CCATATTTTTA | + | 0.899 | −436/−426 | −436/−426 |
| Pit-1a | TGAATAWNWA | TGAATATGAA | + | 0.859 | −404/−395 | −404/−395 |
| Oct-1 | CWNAWTKWSATRYN | AATATGAAAAATGT | − | 0.847 | −402/−389 | −402/−389 |
| Oct-1 | NNNRTAATNANNN | GTATTAATGAAAT | + | 0.870 | −378/−366 | −378/−366 |
| Oct-1 | CWNAWTKWSATRYN | CACATCCAAAATAT | − | 0.890 | −356/−343 | −356/−343 |
| C/EBP-α | NNTKTGGWNANNN | TATTTGTTTAAAG | + | 0.901 | −333/−321 | −333/−321 |
| STAT5A | TTCCCRKAA | TTCTAGGAA | − | 0.956 | −290/−282 | −290/−282 |
| Hfh1 | NAWTGTTTATWT | AAAAAAAAAATC | − | 0.924 | −283/−272 | −283/−272 |
| C/EBP-α | TRRCCAATSRN | GAACCACACAG | + | 0.799 | −269/−259 | −269/−259 |
| HNF-3/FOXA1 | NNNTRTTTRYTY | CTATAAATAATT | − | 0.861 | −222/−211 | −222/−211 |
| GR | NTGCGTRGGCGK | ATTCCTACACAC | + | 0.787 | −213/−202 | −213/−202 |
| TATA box | NCTATAAAAR | ACTATAAAAT | + | 0.964 | −203/−194 | −203/−194 |
| STATx | TTCCCRKAA | TTCTTATAA | + | 0.905 | −187/−179 | −187/−179 |
| STAT5A | TTCCCRKAA | TCCTTGGAA | + | 0.813 | −147/−139 | −147/−139 |
| STAT5A | TTCCCRKAA | TTCTTAGAA | + | 0.912 | −91/−83 | −91/−83 |
| TATA box | WTATAAAW | ATTTAAAT | + | 0.856 | −24/−16 | −24/−16 |
| Total elements | 24 | 22 | ||||
| Genotype Distribution | Allelic Frequency | |||||
|---|---|---|---|---|---|---|
| TT | GT | GG | Total | T | G | |
| Observed | 83 | 61 | 13 | 157 | 0.723 | 0.277 |
| Expected | 82.05 | 62.89 | 12.05 | |||
| Camelus bactrianus CSN1S2 GenBank ID: OQ730238 | Camelus dromedarius CSN1S2 GenBank ID: OQ730239 | Bos taurus CSN1S2 GenBank ID: M94327.1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Name | Strand | Position | Name | Strand | Position | Name | Strand | ||||
| Intron | Nucleotide | Intron | Nucleotide | Intron | Nucleotide | |||||||
| LINEs | ||||||||||||
| LINE 1 | ||||||||||||
| 10 | 8635/8784 | G:L1MA10 | + | 10 | 8613/8834 | G:L1MA10 | + | |||||
| LINE 2 | ||||||||||||
| 1 | 1270/1320 | A: L2a | + | 1 | 1270/1319 | A: L2a | + | 1 | 3953/4000 | A:L2a | + | |
| 6 | 4198/4270 | B:L2a | − | 6 | 4207/4279 | B:L2a | − | 6 | 8069/8151 | B:L2a | − | |
| 11 | 9528/9624 | H:L2a | − | 11 | 9564/9659 | H:L2a | − | 12 | 12212/12306 | H:L2a | − | |
| DNA elements | ||||||||||||
| hAT-Charlie | 7 | 5319/5370 | C:MER5A | + | 7 | 5328/5379 | C:MER5A | + | ||||
| SINEs | ||||||||||||
| MIRs | ||||||||||||
| 8 | 6562/6785 | D:MIRb | + | 8 | 6572/6794 | D:MIRb | + | |||||
| 8 | 7330/7385 | E:MIRb | − | 8 | 7368/7423 | E:MIRb | − | |||||
| 9 | 8093/8232 | F:MIRc | − | 9 | 8131/8270 | F:MIRc | − | 11 | 10742/10888 | F:MIRc | − | |
| 14 | 11593/11775 | I:MIRb | − | 14 | 11632/11814 | I:MIRb | − | 15 | 14773/15100 | I:MIRb | − | |
| 16 | 13828/13938 | L:MIRc | − | 16 | 13867/14030 | L:MIRc | − | 17 | 19404/19481 | L:MIRc | − | |
| Alu/B1 | 2 | 4894/5086 | M:BMF | + | ||||||||
| 8 | 9534/9772 | N:ARS2 | + | |||||||||
| 12 | 11563/11967 | O:BDF | + | |||||||||
| 13 | 13489/13703 | P:BMF | + | |||||||||
| 15 | 14664/14947 | Q:BDF | + | |||||||||
| 17 | 16405/16608 | R:BMF | + | |||||||||
| 17 | 17300/17827 | S:ARS2 | − | |||||||||
| 17 | 19128/19403 | T:BMF | + | |||||||||
| miRNA Name | miRNA Sequence | Seed Location | Custom Target Sequence | Target Score | |
|---|---|---|---|---|---|
| T/T | G/G | ||||
| hsa-miR-298 | AGCAGAAGCAGGGAGGUUCUCCCA | 20 | CTTCTGCA | 93 | 87 |
| hsa-miR-4418 | CACUGCAGGACUCAGCAG | 23 | CTGCAGT | 83 | 76 |
| hsa-miR-3158-5p | CCUGCAGAGAGGAAGCCCUUC | 22 | TCTGCAG | 81 | 65 |
| hsa-miR-548av-3p | AAAACUGCAGUUACUUUUGC | 25 | GCAGTTT | 66 | - |
| hsa-miR-4662a-3p | AAAGAUAGACAAUUGGCUAAAU | 16 | CTATCTT | 52 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauciullo, A.; Versace, C.; Gaspa, G.; Letaief, N.; Bedhiaf-Romdhani, S.; Fulgione, A.; Cosenza, G. Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification. Animals 2023, 13, 2805. https://doi.org/10.3390/ani13172805
Pauciullo A, Versace C, Gaspa G, Letaief N, Bedhiaf-Romdhani S, Fulgione A, Cosenza G. Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification. Animals. 2023; 13(17):2805. https://doi.org/10.3390/ani13172805
Chicago/Turabian StylePauciullo, Alfredo, Carmine Versace, Giustino Gaspa, Neyrouz Letaief, Sonia Bedhiaf-Romdhani, Andrea Fulgione, and Gianfranco Cosenza. 2023. "Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification" Animals 13, no. 17: 2805. https://doi.org/10.3390/ani13172805
APA StylePauciullo, A., Versace, C., Gaspa, G., Letaief, N., Bedhiaf-Romdhani, S., Fulgione, A., & Cosenza, G. (2023). Sequencing and Characterization of αs2-Casein Gene (CSN1S2) in the Old-World Camels Have Proven Genetic Variations Useful for the Understanding of Species Diversification. Animals, 13(17), 2805. https://doi.org/10.3390/ani13172805





