Uncovering Patterns in Dairy Cow Behaviour: A Deep Learning Approach with Tri-Axial Accelerometer Data
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
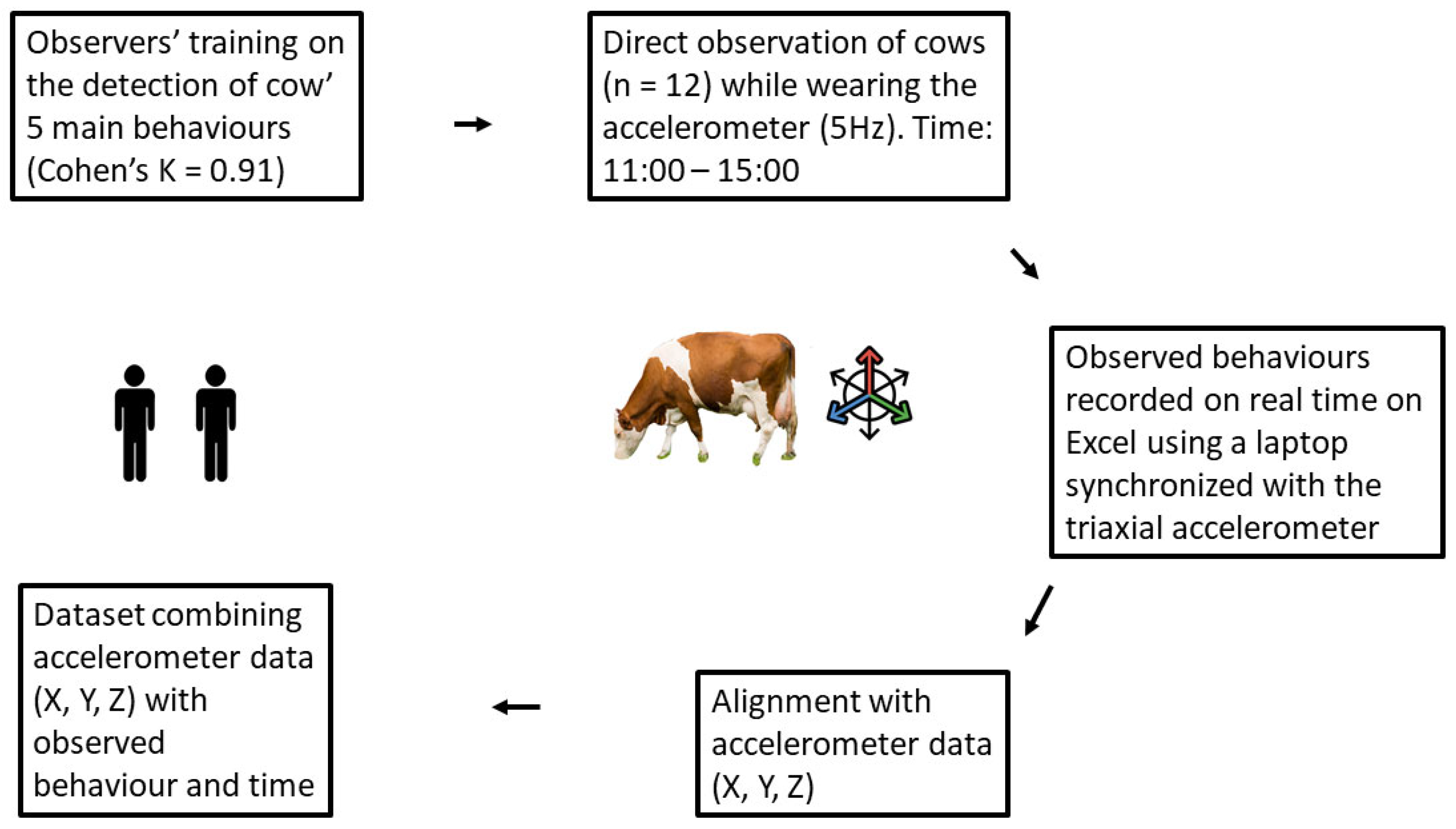
2.2. Data Collection
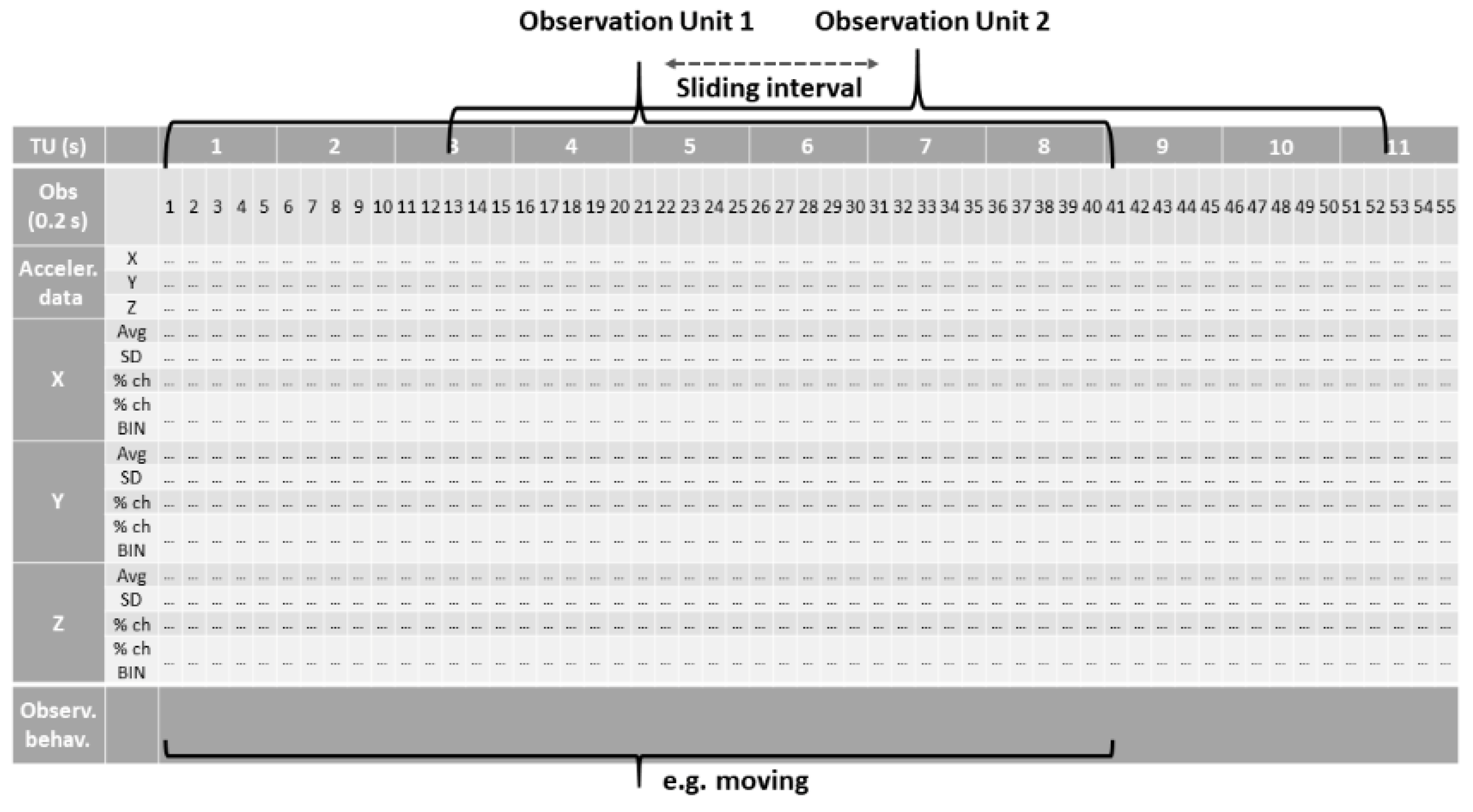
2.3. Dataset Preparation
2.4. Data Modelling
- –
- Convolution is a process in which a small matrix (the kernel or filter) is slid across the input dataset and is transformed on the basis of the filter values. As reported in Table 3, in the Conv1d_1, Conv1d_2 and Conv1d_3 layers, the filters were set at 128, 64 and 32, respectively. For all three layers, the kernel size was set at 3, and the activation function used was the rectified linear unit (RELU). We set padding = ‘valid’ so that the size of the feature maps would gradually decrease as it went through the convolutional layers, which is the default setting option in Keras. Otherwise, ‘Zero Padding’ means filling two edges of each layer’s inputs with zero to keep the size of the inputs and outputs the same. The stride parameter is the number of pixels that a filter moves by once it is inside an input. If it is one, the filter moves right, one pixel at a time. We made the stride parameters equal to one for the convolutional layers and to the same value as the pool size for the pooling layers. If the values of the stride and pooling kernel size are the same, each kernel is prevented from being overlapped.
- –
- The dropout layer randomly selects neurons that are ignored during training. This helps to prevent overfitting. To accomplish this, a rate frequency is adopted at each step. In this model, the rate was set to 0.3.
- –
- Max pooling was used to reduce the size of the tensor and to accelerate calculations. It downsamples the input representation by calculating the largest value over the window as defined by pool size, which in our case was set to 2. We maintained stride and padding parameters equal to those of the convolution layers.
- –
- The flattened layer reduces the data into an array so that the CNN can read it by removing every dimension but one. As reported in Table 3, the output shape of the layer is 544, which is equal to 17 times 32, the two dimensions of the previous layer.
- –
- The dense layer consists of a finite number of neurons (mathematical functions) that receive one vector as input and return another vector as output. The first dense layer was made of 100 neurons with a ‘RELU’ activation function and was connected to the last dense layer with a softmax activation function and a length of 5, which is equal to the number of activities to be classified by the model. The model was deployed in Python using Keras [28] with a TensorFlow backend.
- –
- It is noteworthy that the final layer’s output shape is 5, given that there are 5 behaviours to classify.
| Layer (Type) | Output Shape | Parameters | Hyperparameters |
|---|---|---|---|
| Conv1d_1 (Conv1D) | (None, 38, 128) | 5888 | filter = 120, kernel_size = 3, strides = 1, padding = ‘valid’ |
| Conv1d_2 (Conv1D) | (None, 36, 64) | 24,640 | filter = 64, kernel_size = 3, strides = 1, padding = ‘valid’ |
| Conv1d_3 (Conv1D) | (None, 34, 32) | 6176 | filter = 32, kernel_size = 3, strides = 1, padding = ‘valid’ |
| Dropout_1 (Dropout) | (None, 34, 32) | 0 | rate = 0.3 |
| Max_pooling1d_1 (Max-pooling) | (None, 17, 32) | 0 | pool_size = 2, strides = None, padding = ‘valid’ |
| Flatten_1 (Flatten) | (None, 544) | 0 | - |
| Dense_1 (Dense) | (None, 100) | 54,500 | units = 100, activation= RELU |
| Dense_2 (Dense) | (None, 5) | 505 | units = 5, activation = RELU |
| Total parameters: 91,709 | |||
| Trainable parameters: 91,709 | |||
| Non-trainable parameters: 0 | |||
| Parameter | Value |
|---|---|
| GPU | Nvidia K80/T4 |
| GPU memory | 12 GB/16 GB |
| GPU memory Clock | 0.82 GHz/1.59 GHz |
| Performance | 4.1 TFLOPS/8.1 TFLOPS |
| Support mixed precision | No/Yes |
| GPU release year | 2014/2018 |
| No. CPU cores | 2 |
| Available RAM | 12 GB (upgradable to 26.75 GB) |
2.5. Model Assessment
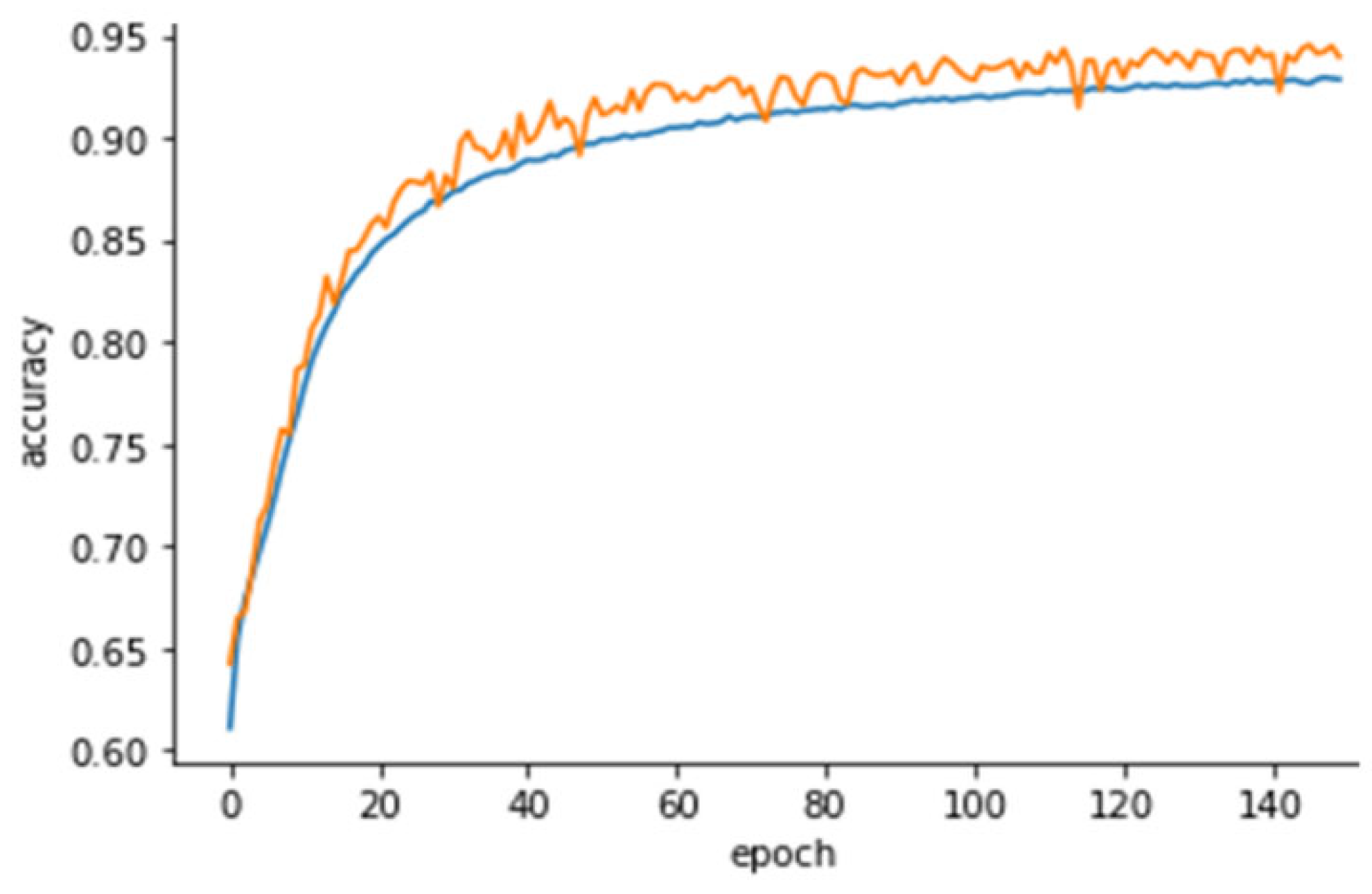
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Britt, J.H.; Cushman, R.A.; Dechow, C.D.; Dobson, H.; Humblot, P.; Hutjens, M.F.; Jones, G.A.; Mitloehner, F.M.; Ruegg, P.L.; Sheldon, I.M.; et al. Review: Perspective on High-Performing Dairy Cows and Herds. Animal 2021, 15, 100298. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive Analysis of Machine Learning Models for Prediction of Sub-Clinical Mastitis: Deep Learning and Gradient-Boosted Trees Outperform Other Models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef] [PubMed]
- Yunta, C.; Guasch, I.; Bach, A. Short Communication: Lying Behavior of Lactating Dairy Cows Is Influenced by Lameness Especially around Feeding Time. J. Dairy Sci. 2012, 95, 6546–6549. [Google Scholar] [CrossRef]
- Wagner, N.; Antoine, V.; Mialon, M.-M.; Lardy, R.; Silberberg, M.; Koko, J.; Veissier, I. Machine Learning to Detect Behavioural Anomalies in Dairy Cows under Subacute Ruminal Acidosis. Comput. Electron. Agric. 2020, 170, 105233. [Google Scholar] [CrossRef]
- Dantzer, R.; Kelley, K.W. Twenty Years of Research on Cytokine-Induced Sickness Behavior. Brain Behav. Immun. 2007, 21, 153–160. [Google Scholar] [CrossRef]
- Stangaferro, M.L.; Wijma, R.; Caixeta, L.S.; Al-Abri, M.A.; Giordano, J.O. Use of Rumination and Activity Monitoring for the Identification of Dairy Cows with Health Disorders: Part II. Mastitis. J. Dairy Sci. 2016, 99, 7411–7421. [Google Scholar] [CrossRef]
- De Boyer des Roches, A.; Faure, M.; Lussert, A.; Herry, V.; Rainard, P.; Durand, D.; Foucras, G. Behavioral and Patho-Physiological Response as Possible Signs of Pain in Dairy Cows during Escherichia Coli Mastitis: A Pilot Study. J. Dairy Sci. 2017, 100, 8385–8397. [Google Scholar] [CrossRef]
- Norring, M.; Häggman, J.; Simojoki, H.; Tamminen, P.; Winckler, C.; Pastell, M. Short Communication: Lameness Impairs Feeding Behavior of Dairy Cows. J. Dairy Sci. 2014, 97, 4317–4321. [Google Scholar] [CrossRef]
- Riaboff, L.; Poggi, S.; Madouasse, A.; Couvreur, S.; Aubin, S.; Bédère, N.; Goumand, E.; Chauvin, A.; Plantier, G. Development of a Methodological Framework for a Robust Prediction of the Main Behaviours of Dairy Cows Using a Combination of Machine Learning Algorithms on Accelerometer Data. Comput. Electron. Agric. 2020, 169, 105179. [Google Scholar] [CrossRef]
- Abeni, F.; Galli, A. Monitoring Cow Activity and Rumination Time for an Early Detection of Heat Stress in Dairy Cow. Int. J. Biometeorol. 2017, 61, 417–425. [Google Scholar] [CrossRef]
- Marchesini, G.; Cortese, M.; Mottaran, D.; Ricci, R.; Serva, L.; Contiero, B.; Segato, S.; Andrighetto, I. Effects of Axial and Ceiling Fans on Environmental Conditions, Performance and Rumination in Beef Cattle during the Early Fattening Period. Livest. Sci. 2018, 214, 225–230. [Google Scholar] [CrossRef]
- Marchesini, G.; Mottaran, D.; Contiero, B.; Schiavon, E.; Segato, S.; Garbin, E.; Tenti, S.; Andrighetto, I. Use of Rumination and Activity Data as Health Status and Performance Indicators in Beef Cattle during the Early Fattening Period. Vet. J. 2018, 231, 41–47. [Google Scholar] [CrossRef]
- Cabrera, V.E.; Barrientos-Blanco, J.A.; Delgado, H.; Fadul-Pacheco, L. Symposium Review: Real-Time Continuous Decision Making Using Big Data on Dairy Farms. J. Dairy Sci. 2020, 103, 3856–3866. [Google Scholar] [CrossRef]
- Borchers, M.R.; Chang, Y.M.; Tsai, I.C.; Wadsworth, B.A.; Bewley, J.M. A Validation of Technologies Monitoring Dairy Cow Feeding, Ruminating, and Lying Behaviors. J. Dairy Sci. 2016, 99, 7458–7466. [Google Scholar] [CrossRef]
- Benaissa, S.; Tuyttens, F.A.M.; Plets, D.; de Pessemier, T.; Trogh, J.; Tanghe, E.; Martens, L.; Vandaele, L.; Van Nuffel, A.; Joseph, W.; et al. On the Use of On-Cow Accelerometers for the Classification of Behaviours in Dairy Barns. Res. Vet. Sci. 2019, 125, 425–433. [Google Scholar] [CrossRef]
- Riaboff, L.; Shalloo, L.; Smeaton, A.F.; Couvreur, S.; Madouasse, A.; Keane, M.T. Predicting livestock behaviour using accelerometers: A systematic review of processing techniques for ruminant behaviour prediction from raw accelerometer data. Comput. Electron. Agric. 2022, 192, 106610. [Google Scholar] [CrossRef]
- Santos, A.D.S.; de Medeiros, V.W.C.; Gonçalves, G.E. Monitoring and classification of cattle behavior: A survey. Smart Agric. Technol. 2023, 3, 100091. [Google Scholar] [CrossRef]
- Peng, Y.; Kondo, N.; Fujiura, T.; Suzuki, T.; Wulandari; Yoshioka, H.; Itoyama, E. Classification of multiple cattle behavior patterns using a recurrent neural network with long short-term memory and inertial measurement units. Comput. Electron. Agric. 2019, 157, 247–253. [Google Scholar] [CrossRef]
- Peng, Y.; Kondo, N.; Fujiura, T.; Suzuki, T.; Ouma, S.; Wulandari; Yoshioka, H.; Itoyama, E. Dam behavior patterns in Japanese black beef cattle prior to calving: Automated detection using LSTM-RNN. Comput. Electron. Agric. 2020, 169, 105178. [Google Scholar] [CrossRef]
- Awais, M.; Chiari, L.; Ihlen, E.A.F.; Helbostad, J.L.; Palmerini, L. Classical Machine Learning Versus Deep Learning for the Older Adults Free-Living Activity Classification. Sensors 2021, 21, 4669. [Google Scholar] [CrossRef]
- Li, G.; Erickson, G.E.; Xiong, Y. Individual Beef Cattle Identification Using Muzzle Images and Deep Learning Techniques. Animals 2022, 12, 1453. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, M.; Peng, Z.; Liu, M.; Wang, M.; Peng, Y. Recognising Cattle Behaviour with Deep Residual Bidirectional LSTM Model Using a Wearable Movement Monitoring Collar. Agriculture 2022, 12, 1237. [Google Scholar] [CrossRef]
- Li, G.; Huang, Y.; Chen, Z.; Chesser, G.D.; Purswell, J.L.; Linhoss, J.; Zhao, Y. Practices and Applications of Convolutional Neural Network-Based Computer Vision Systems in Animal Farming: A Review. Sensors 2021, 21, 1492. [Google Scholar] [CrossRef] [PubMed]
- Mathis, M.W.; Mathis, A. Deep Learning Tools for the Measurement of Animal Behavior in Neuroscience. Curr. Opin. Neurobiol. 2020, 60, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nunavath, V.; Johansen, S.; Johannessen, T.S.; Jiao, L.; Hansen, B.H.; Berntsen, S.; Goodwin, M. Deep Learning for Classifying Physical Activities from Accelerometer Data. Sensors 2021, 21, 5564. [Google Scholar] [CrossRef]
- Balasso, P.; Marchesini, G.; Ughelini, N.; Serva, L.; Andrighetto, I. Machine Learning to Detect Posture and Behavior in Dairy Cows: Information from an Accelerometer on the Animal’s Left Flank. Animals 2021, 11, 2972. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python, 2nd ed.; Manning Publications: Shelter Island, NY, USA, 2021; ISBN 978-1-63835-009-5. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Regularization for Deep Learning. In Deep Learning; MIT Press: Cambridge, UK, 2016; pp. 241–249. [Google Scholar]
- Sokolova, M.; Lapalme, G. A Systematic Analysis of Performance Measures for Classification Tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Wang, J.; He, Z.; Zheng, G.; Gao, S.; Zhao, K. Development and Validation of an Ensemble Classifier for Real-Time Recognition of Cow Behavior Patterns from Accelerometer Data and Location Data. PLoS ONE 2018, 13, e0203546. [Google Scholar] [CrossRef]
- Con, D.; van Langenberg, D.R.; Vasudevan, A. Deep Learning vs Conventional Learning Algorithms for Clinical Prediction in Crohn’s Disease: A Proof-of-Concept Study. World J. Gastroenterol. 2021, 27, 6476–6488. [Google Scholar] [CrossRef]
- Cabezas, J.; Yubero, R.; Visitación, B.; Navarro-García, J.; Algar, M.J.; Cano, E.L.; Ortega, F. Analysis of Accelerometer and GPS Data for Cattle Behaviour Identification and Anomalous Events Detection. Entropy 2022, 24, 336. [Google Scholar] [CrossRef]
- Vázquez Diosdado, J.A.; Barker, Z.E.; Hodges, H.R.; Amory, J.R.; Croft, D.P.; Bell, N.J.; Codling, E.A. Classification of Behaviour in Housed Dairy Cows Using an Accelerometer-Based Activity Monitoring System. Anim. Biotelem. 2015, 3, 15. [Google Scholar] [CrossRef]
- Pavlovic, D.; Czerkawski, M.; Davison, C.; Marko, O.; Michie, C.; Atkinson, R.; Crnojevic, V.; Andonovic, I.; Rajovic, V.; Kvascev, G.; et al. Behavioural Classification of Cattle Using Neck-Mounted Accelerometer-Equipped Collars. Sensors 2022, 22, 2323. [Google Scholar] [CrossRef]
- Roland, L.; Schweinzer, V.; Kanz, P.; Sattlecker, G.; Kickinger, F.; Lidauer, L.; Berger, A.; Auer, W.; Mayer, J.; Sturm, V.; et al. Technical Note: Evaluation of a Triaxial Accelerometer for Monitoring Selected Behaviors in Dairy Calves. J. Dairy Sci. 2018, 101, 10421–10427. [Google Scholar] [CrossRef]
- Martiskainen, P.; Järvinen, M.; Skön, J.-P.; Tiirikainen, J.; Kolehmainen, M.; Mononen, J. Cow Behaviour Pattern Recognition Using a Three-Dimensional Accelerometer and Support Vector Machines. Appl. Anim. Behav. Sci. 2009, 119, 32–38. [Google Scholar] [CrossRef]
- Benaissa, S.; Tuyttens, F.A.M.; Plets, D.; Martens, L.; Vandaele, L.; Joseph, W.; Sonck, B. Improved Cattle Behaviour Monitoring by Combining Ultra-Wideband Location and Accelerometer Data. Animal 2023, 17, 100730. [Google Scholar] [CrossRef]
- Cook, N.B. Symposium Review: The Impact of Management and Facilities on Cow Culling Rates. J. Dairy Sci. 2020, 103, 3846–3855. [Google Scholar] [CrossRef]


| Behaviour | Definition 1 |
|---|---|
| Standing still | Cows stand still without moving their legs or showing any sign of activity |
| Feeding | Cows ingest feed and chew it at the feed bunk |
| Moving (walking or moving slightly) | Cows walk across the pen or, while standing, perform other behaviours other than those described here, such as agonistic behaviours and drinking, which are characterized by at least one step every 10 s |
| Ruminating | Cows chew a bolus, a process which begins upon regurgitating the bolus and ends when the bolus is swallowed, in either a standing or lying position |
| Resting | Cows lie on the floor, neither moving nor ruminating |
| Behaviour | Precision | Recall | F1 Score | Number of Observation Units |
|---|---|---|---|---|
| Feeding | 0.96 (0.89) | 0.96 (0.91) | 0.96 (0.90) | 8192 |
| Moving | 0.94 (0.86) | 0.94 (0.89) | 0.94 (0.88) | 8857 |
| Resting | 0.99 (0.98) | 0.99 (0.96) | 0.99 (0.97) | 13,141 |
| Ruminating | 0.99 (0.88) | 0.96 (0.92) | 0.97 (0.90) | 5001 |
| Standing still | 0.93 (0.88) | 0.93 (0.83) | 0.93 (0.85) | 7144 |
| Metrics | ||||
| Accuracy | 0.96 (0.91) | 42,335 | ||
| Macro average | 0.96 (0.90) | 0.96 (0.90) | 0.96 (0.90) | 42,335 |
| Weighted average | 0.96 (0.91) | 0.96 (0.91) | 0.96 (0.91) | 42,335 |
| Predicted | Actual | ||||
|---|---|---|---|---|---|
| Feeding | Moving | Resting | Ruminating | Standing Still | |
| Feeding | 7896 | 147 | 12 | 1 | 136 |
| Moving | 185 | 8352 | 21 | 7 | 292 |
| Resting | 12 | 18 | 13,058 | 43 | 10 |
| Ruminating | 20 | 23 | 96 | 4793 | 69 |
| Standing still | 138 | 337 | 9 | 19 | 6641 |
| Study | Year | Behaviour | N | h | Acceler. | Other Sensors | Sensor Location | Sampling Rate (Hz) | Models | Overall Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| Present | 2023 | F, M, R, Ru, Ss | 12 | 27 | Single | - | Left flank | 5 | CNN, KNN, LSTM, RF, SVM, XGB | 0.96 |
| [9] | 2020 | G, R, Ru, W | 86 | 57 | Single | - | Neck | 59.5 | ADA, RF, SVM, XGB, | 0.98 |
| [15] | 2019 | F, L, S | 16 | 96 | Multiple | - | Leg and neck | 1 | KNN, NB, SVM | 0.98 |
| [18] | 2019 | C, F, H, L, Li, M, Ru | 6 | 68 | Single | IMU | Neck | 20 | CNN, LSTM-RNN | 0.89 |
| [19] | 2020 | Cb | 3 | 150 | Single | IMU | Neck | 20 | LSTM-RNN | 0.80 |
| [22] | 2022 | F, L, Li, Ri, Ru | 12 | 1066 | Single | IMU | Neck | 10 | Bid. LSTM, LSTM-RNN | 0.95 |
| [26] | 2022 | F, L, M, R, Ru, S, Ss | 12 | 27 | Single | - | Left flank | 5 | KNN, RF, SVM, XGB | 0.76 |
| [31] | 2018 | C, F, L, S, W | 5 | 200 | Multiple | RFL | Legs and neck | 1 | MBP-ADAB | 0.73 |
| [33] | 2022 | G, L, O, Ru, Ss | 10 | 50 | Single | GPS | Neck | 10 | RF | 0.88–0.93 |
| [34] | 2015 | C, F, L, S, W | 6 | 33 | Single | - | Neck | 50 | DT, HMM, K-means, SVM | 0.88, 0.82 ** |
| [35] | 2022 | F, O, R | 18 | 60 | Single | PC | Neck and halter | 10 | HMM, LDA, PLSDA | 0.83 |
| [36] | 2018 | F, L, Ru, W | 15 | 60 | Single | - | Ear | 10 | HMM | 0.71 |
| [37] | 2009 | C, F, L Ru, S | 30 | 95.5 | Single | - | Neck | 10 | SVM | 0.78 |
| [38] | 2023 | D, F, R, Ru | 30 | 156 | Single | UWB | Neck | 2 | DT, HMM, K-means, SVM | 0.99 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasso, P.; Taccioli, C.; Serva, L.; Magrin, L.; Andrighetto, I.; Marchesini, G. Uncovering Patterns in Dairy Cow Behaviour: A Deep Learning Approach with Tri-Axial Accelerometer Data. Animals 2023, 13, 1886. https://doi.org/10.3390/ani13111886
Balasso P, Taccioli C, Serva L, Magrin L, Andrighetto I, Marchesini G. Uncovering Patterns in Dairy Cow Behaviour: A Deep Learning Approach with Tri-Axial Accelerometer Data. Animals. 2023; 13(11):1886. https://doi.org/10.3390/ani13111886
Chicago/Turabian StyleBalasso, Paolo, Cristian Taccioli, Lorenzo Serva, Luisa Magrin, Igino Andrighetto, and Giorgio Marchesini. 2023. "Uncovering Patterns in Dairy Cow Behaviour: A Deep Learning Approach with Tri-Axial Accelerometer Data" Animals 13, no. 11: 1886. https://doi.org/10.3390/ani13111886
APA StyleBalasso, P., Taccioli, C., Serva, L., Magrin, L., Andrighetto, I., & Marchesini, G. (2023). Uncovering Patterns in Dairy Cow Behaviour: A Deep Learning Approach with Tri-Axial Accelerometer Data. Animals, 13(11), 1886. https://doi.org/10.3390/ani13111886






