Genome-Wide Association Study Identifies the Crucial Candidate Genes for Teat Number in Crossbred Commercial Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animal Samples and Genotyping and Quality Control
2.3. Population Structure
2.4. Genome-Wide Association Study
2.5. Functional Candidate Genes Search
3. Results
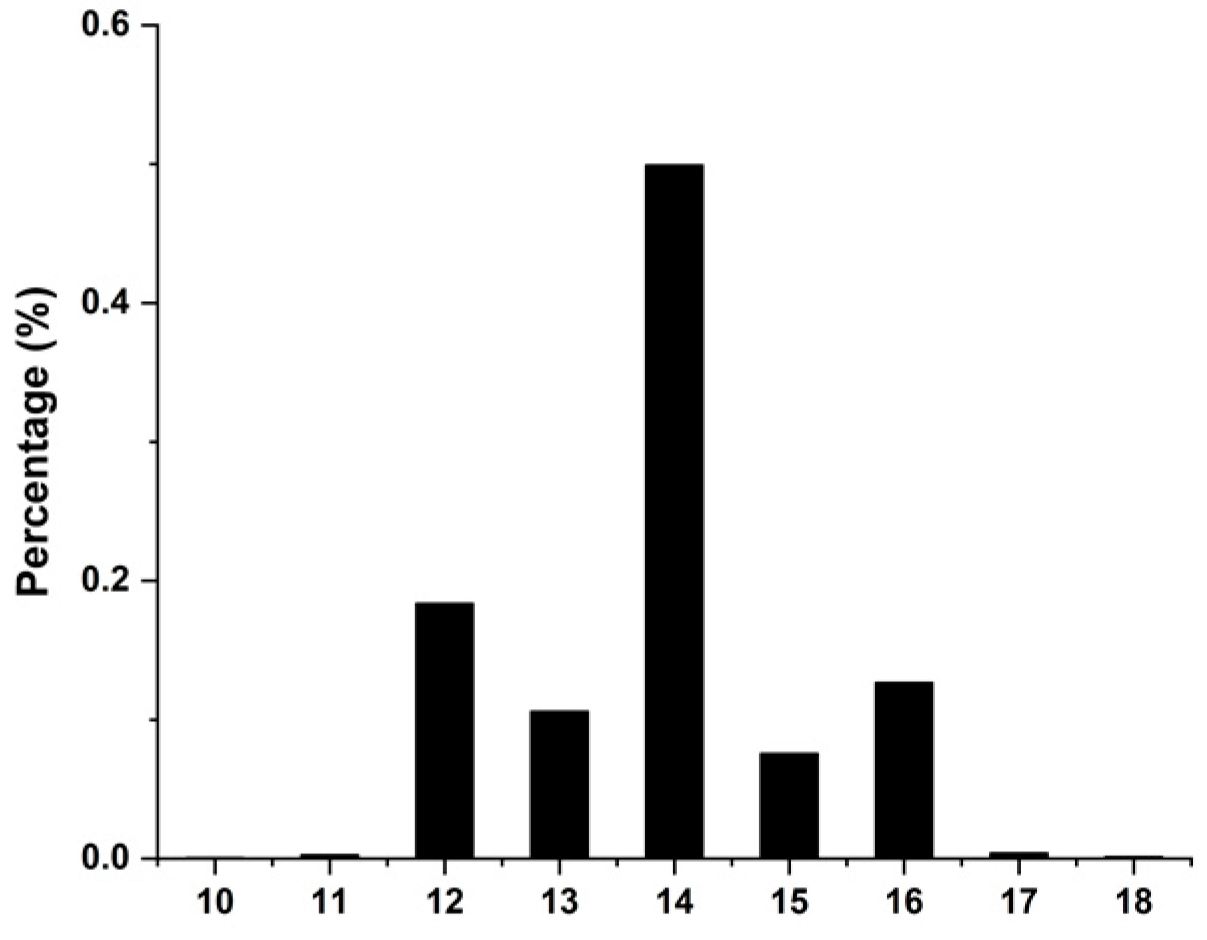
3.1. Phenotype Statistic
3.2. Genome-Wide Association Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.; Chen, C.; Yang, B.; Guo, Y.; Ai, H.; Ren, J.; Peng, Z.; Tu, Z.; Yang, X.; Meng, Q.; et al. A systems genetics study of swine illustrates mechanisms underlying human phenotypic traits. BMC Genom. 2015, 16, 88. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, G.A.; Nonneman, D.J. Genetic analysis of teat number in pigs reveals some developmental pathways independent of vertebra number and several loci which only affect a specific side. Genet. Sel. Evol. 2017, 49, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Ding, R.; Peng, L.; Wu, J.; Ye, Y.; Zhou, S.; Wang, X.; Quan, J.; Zheng, E.; Cai, G.; et al. Genome-wide association analyses identify known and novel loci for teat number in Duroc pigs using single-locus and multi-locus models. BMC Genom. 2020, 21, 344. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Zhang, T.; Zhang, L.; Yan, H.; Liu, X.; Wang, L. Genotyping by sequencing reveals a new locus for pig teat number. Anim. Genet. 2017, 48, 470–472. [Google Scholar] [CrossRef]
- Tomiyama, M.; Kanetani, T.; Tatsukawa, Y.; Mori, H.; Oikawa, T. Genetic parameters for preweaning and early growth traits in Berkshire pigs when creep feeding is used. J. Anim. Sci. 2010, 88, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, X.; Tan, Z.; Xing, K.; Yang, T.; Wang, Y.; Sun, D.; Wang, C. Genome-wide association study for reproductive traits in a Large White pig population. Anim. Genet. 2018, 49, 127–131. [Google Scholar] [CrossRef]
- Akanno, E.C.; Chen, L.; Abo-Ismail, M.K.; Crowley, J.J.; Wang, Z.; Li, C.; Basarab, J.A.; MacNeil, M.D.; Plastow, G.S. Genome-wide association scan for heterotic quantitative trait loci in multi-breed and crossbred beef cattle. Genet. Sel. Evol. 2018, 50, 48. [Google Scholar] [CrossRef]
- Georges, M. Mapping, fine mapping, and molecular dissection of quantitative trait Loci in domestic animals. Annu. Rev. Genom. Hum. Genet. 2007, 8, 131–162. [Google Scholar] [CrossRef]
- Andersson, L. Molecular consequences of animal breeding. Curr. Opin. Genet. Dev. 2013, 23, 295–301. [Google Scholar] [CrossRef]
- Hong, Y.F.; Ye, J.; Dong, L.S.; Li, Y.L.; Yan, L.M.; Cai, G.Y.; Liu, D.W.; Tan, C.; Wu, Z.F. Genome-Wide Association Study for Body Length, Body Height, and Total Teat Number in Large White Pigs. Front. Genet. 2021, 12, 650370. [Google Scholar] [CrossRef]
- Mikawa, S.; Sato, S.; Nii, M.; Morozumi, T.; Yoshioka, G.; Imaeda, N.; Yamaguchi, T.; Hayashi, T.; Awata, T. Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet. 2011, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Duijvesteijn, N.; Veltmaat, J.M.; Knol, E.F.; Harlizius, B. High-resolution association mapping of number of teats in pigs reveals regions controlling vertebral development. BMC Genom. 2014, 15, 542. [Google Scholar] [CrossRef] [PubMed]
- van Son, M.; Lopes, M.S.; Martell, H.J.; Derks, M.; Gangsei, L.E.; Kongsro, J.; Wass, M.N.; Grindflek, E.H.; Harlizius, B. A QTL for Number of Teats Shows Breed Specific Effects on Number of Vertebrae in Pigs: Bridging the Gap Between Molecular and Quantitative Genetics. Front. Genet. 2019, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Veltmaat, J.M. Prenatal Mammary Gland Development in the Mouse: Research Models and Techniques for Its Study from Past to Present. Methods Mol. Biol. 2017, 1501, 21–76. [Google Scholar] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Gao, G.X.; Zhou, Y.; Guo, C.X.; Li, B.; El-Ashram, S.; Li, Z.L. Genome-wide association studies uncover genes associated with litter traits in the pig. Animal 2022, 16, 100672. [Google Scholar] [CrossRef]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 2009, 3, 281–290. [Google Scholar] [CrossRef]
- Fettelschoss, V.; Burda, P.; Sagne, C.; Coelho, D.; De Laet, C.; Lutz, S.; Suormala, T.; Fowler, B.; Pietrancosta, N.; Gasnier, B.; et al. Clinical or ATPase domain mutations in ABCD4 disrupt the interaction between the vitamin B (12)-trafficking proteins ABCD4 and LMBD1. J. Biol. Chem. 2017, 292, 11980–11991. [Google Scholar] [CrossRef]
- Ray, J.G.; Blom, H.J. Vitamin B12 insufficiency and the risk of fetal neural tube defects. QJM Int. J. Med. 2003, 96, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Groenen, P.M.; van Rooij, I.A.; Peer, P.G.; Gooskens, R.H.; Zielhuis, G.A.; Steegers-Theunissen, R.P. Marginal maternal vitamin B12 status increases the risk of offspring with spina bifida. Am. J. Obstet. Gynecol. 2004, 191, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, H.; Zhang, Z.; Gao, J.; Yang, J.; Wu, Z.; Fan, Y.; Xing, Y.; Li, L.; Xiao, S.; et al. VRTN is Required for the Development of Thoracic Vertebrae in Mammals. Int. J. Biol. Sci. 2018, 14, 667–681. [Google Scholar] [CrossRef]
- Spina, E.; Cowin, P. Embryonic mammary gland development. Semin. Cell Dev. Biol. 2021, 114, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.W. Cooperation of signalling pathways in embryonic mammary gland development. Nat. Rev. Genet. 2007, 8, 963–972. [Google Scholar] [CrossRef]
- Capuco, A.V.; Ellis, S.E. Comparative aspects of mammary gland development and homeostasis. Annu. Rev. Anim. Biosci. 2013, 1, 179–202. [Google Scholar] [CrossRef]
- Yang, J.; Huang, L.; Yang, M.; Fan, Y.; Li, L.; Fang, S.; Deng, W.; Cui, L.; Zhang, Z.; Ai, H.; et al. Possible introgression of the VRTN mutation increasing vertebral number, carcass length and teat number from Chinese pigs into European pigs. Sci. Rep. 2016, 6, 19240. [Google Scholar] [CrossRef]
- Schaid, D.J.; Chen, W.; Larson, N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018, 19, 491–504. [Google Scholar] [CrossRef]
- Hiremath, B.; Subramaniam, N.; Chandrashekhar, N. Giant accessory breast: A rare occurrence reported, with a review of the literature. BMJ Case Rep. 2015, 2015, bcr2015210918. [Google Scholar] [CrossRef]
- Chomwisarutkun, K.; Murani, E.; Brunner, R.; Ponsuksili, S.; Wimmers, K. QTL region-specific microarrays reveal differential expression of positional candidate genes of signaling pathways associated with the liability for the inverted teat defect. Anim. Genet. 2013, 44, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Kessaris, N.; Jamen, F.; Rubin, L.L.; Richardson, W.D. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development 2004, 131, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, S.V.; Kniazev, S.P.; Ermolaev, V.I. Model of genetic control of the number and location of nipples in domestic pig. Russ. J. Genet. 2012, 48, 1128–1140. [Google Scholar] [CrossRef]
- Fernandez, A.; Toro, M.; Rodriguez, C.; Silio, L. Heterosis and epistasis for teat number and fluctuating asymmetry in crosses between Jiaxing and Iberian pigs. Heredity 2004, 93, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, W.; Fu, Y.; Fang, X.; Ren, S.; Ren, J. Genome-wide detection of genetic loci and candidate genes for teat number and body conformation traits at birth in Chinese Sushan pigs. Anim. Genet. 2019, 50, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.S.; Bastiaansen, J.W.; Harlizius, B.; Knol, E.F.; Bovenhuis, H. A genome-wide association study reveals dominance effects on number of teats in pigs. PLoS ONE 2014, 9, e105867. [Google Scholar] [CrossRef]
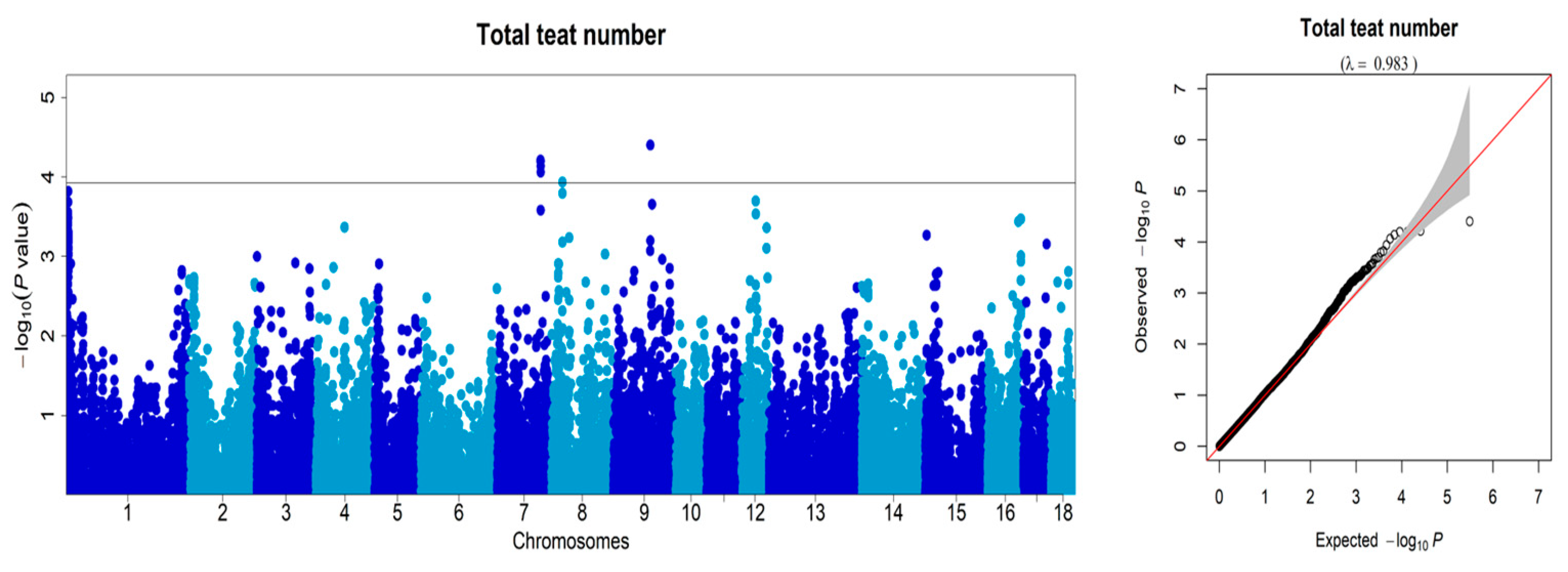
| chr | SNP ID | Position (bp) | p-Value | Gene | Region |
|---|---|---|---|---|---|
| 9 | ALGA0054033 | 82,853,264 | 3.94 × 10−5 | / | / |
| 7 | Affx-114687136 | 97,568,284 | 6.12 × 10−5 | ABCD4 | 3′ UTR |
| 7 | Affx-115258151 | 97,595,573 | 6.29 × 10−5 | ABCD4 | 5′ UTR |
| 7 | Affx-114892585 | 97,575,068 | 6.32 × 10−5 | ABCD4 | Intron 7-8 |
| 7 | WU_10.2_7_103460706 | 97,617,907 | 7.26 × 10−5 | VRTN | Intron 1-2 |
| 7 | WU_10.2_7_103232787 | 97,584,287 | 8.67 × 10−5 | ABCD4 | Intron 1-2 |
| 8 | ASGA0094767 | 24,709,455 | 1.16 × 10−4 | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Li, X.; Zhuang, Z.; Zhou, S.; Wu, J.; Xu, C.; Ruan, D.; Qiu, Y.; Zhao, H.; Zheng, E.; et al. Genome-Wide Association Study Identifies the Crucial Candidate Genes for Teat Number in Crossbred Commercial Pigs. Animals 2023, 13, 1880. https://doi.org/10.3390/ani13111880
Yang L, Li X, Zhuang Z, Zhou S, Wu J, Xu C, Ruan D, Qiu Y, Zhao H, Zheng E, et al. Genome-Wide Association Study Identifies the Crucial Candidate Genes for Teat Number in Crossbred Commercial Pigs. Animals. 2023; 13(11):1880. https://doi.org/10.3390/ani13111880
Chicago/Turabian StyleYang, Lijuan, Xuehua Li, Zhanwei Zhuang, Shenping Zhou, Jie Wu, Cineng Xu, Donglin Ruan, Yibin Qiu, Hua Zhao, Enqin Zheng, and et al. 2023. "Genome-Wide Association Study Identifies the Crucial Candidate Genes for Teat Number in Crossbred Commercial Pigs" Animals 13, no. 11: 1880. https://doi.org/10.3390/ani13111880
APA StyleYang, L., Li, X., Zhuang, Z., Zhou, S., Wu, J., Xu, C., Ruan, D., Qiu, Y., Zhao, H., Zheng, E., Cai, G., Wu, Z., & Yang, J. (2023). Genome-Wide Association Study Identifies the Crucial Candidate Genes for Teat Number in Crossbred Commercial Pigs. Animals, 13(11), 1880. https://doi.org/10.3390/ani13111880





