Natural Evolution of Porcine Epidemic Diarrhea Viruses Isolated from Maternally Immunized Piglets
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Cell Culture and Clinical Samples
2.2. Multiple Detection of Virus
2.3. Histological Analysis of the Materials
2.4. Sequence Analysis and Phylogenetic Construction
2.5. Virus Isolation and Purification
2.6. Selection of Susceptible Cells
2.7. Tissue Culture Infection Dose (TCID50) Measurement
2.8. Growth Dynamic Curve of Different Cells Infected by the Virus
2.9. Indirect Immunofluorescence Assay (IFA)
2.10. Analysis of S Gene Recombination
3. Results
3.1. PEDV Virus Screening
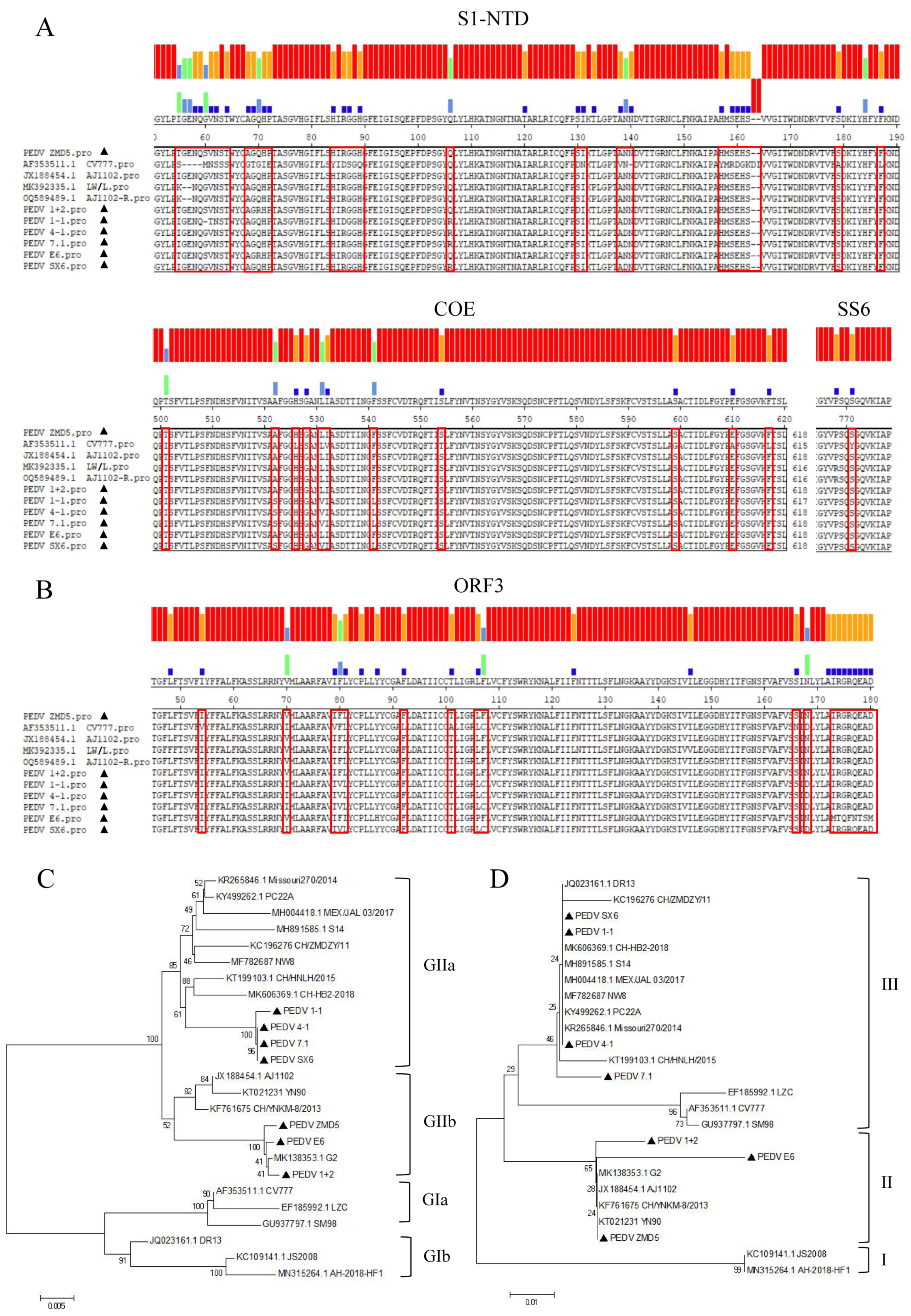
3.2. S, ORF3 Amino Acid Sequence Alignment and Evolutionary Analysis
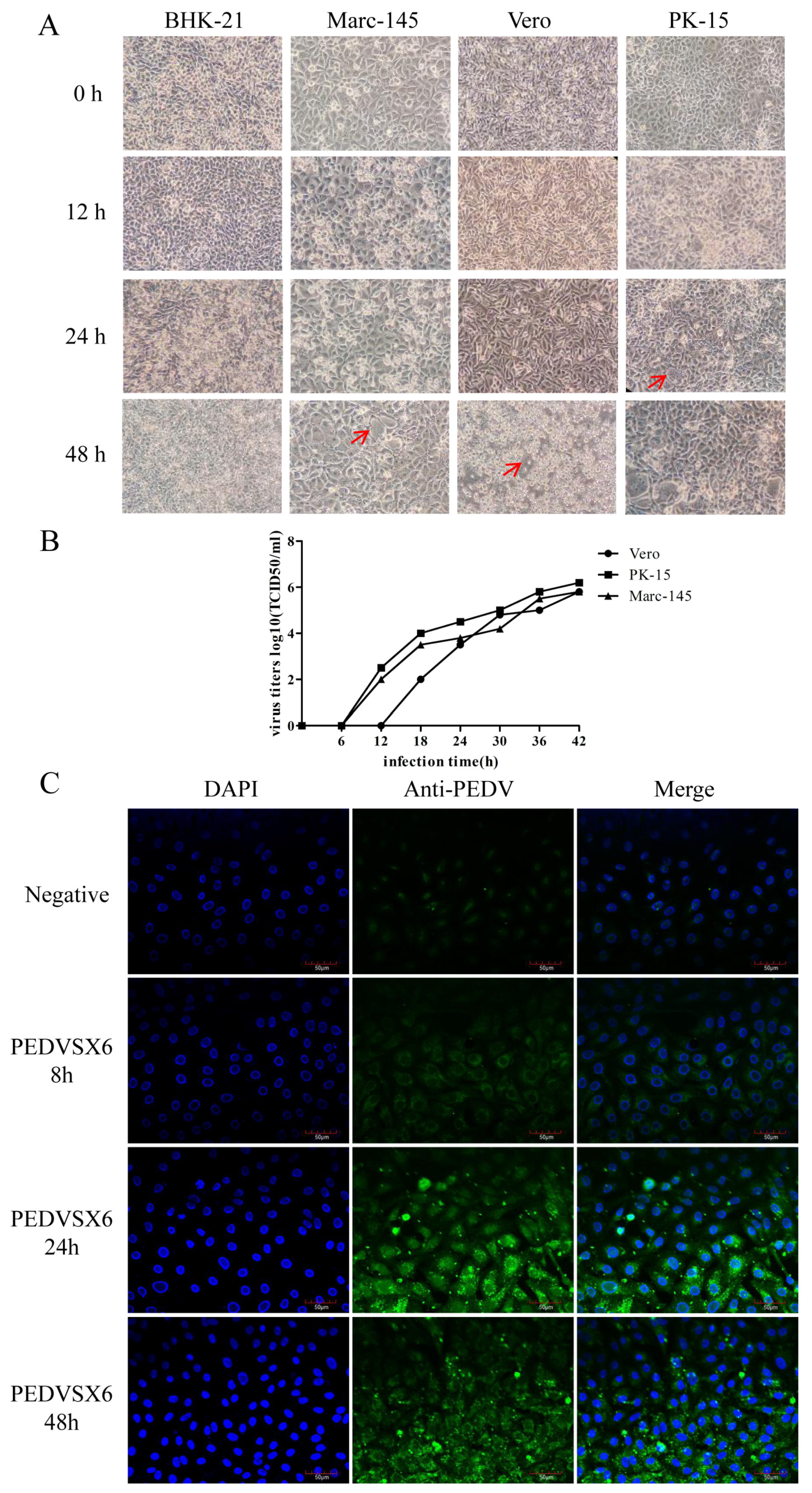
3.3. Isolation and Identification of PEDV Virus
3.4. Analysis of S Gene Recombination
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.Q.; Cai, R.J.; Chen, Y.Q.; Liang, P.S.; Chen, D.K.; Song, C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Dickerman, A.W.; Piñeyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013, 4, e00737-13. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Wang, Q.; Scheuer, K.A.; Lu, Z.; Zhang, Y.; Saif, L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014, 20, 662–665. [Google Scholar] [CrossRef]
- Yang, D.; Su, M.; Li, C.; Zhang, B.; Qi, S.; Sun, D.; Yin, B. Isolation and characterization of a variant subgroup GII-a porcine epidemic diarrhea virus strain in China. Microb. Pathog. 2020, 140, 103922. [Google Scholar] [CrossRef]
- Cui, J.T.; Qiao, H.; Hou, C.Y.; Zheng, H.H.; Li, X.S.; Zheng, L.L.; Chen, H.Y. Characteristics of the spike and ORF3 genes of porcine epidemic diarrhea virus in Henan and Shanxi provinces of China. Arch. Virol. 2020, 165, 2323–2333. [Google Scholar] [CrossRef]
- Chen, P.; Wang, K.; Hou, Y.; Li, H.; Li, X.; Yu, L.; Jiang, Y.; Gao, F.; Tong, W.; Yu, H.; et al. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011–2017. Infect. Genet. Evol. 2019, 69, 153–165. [Google Scholar] [CrossRef]
- Wen, Z.; Li, J.; Zhang, Y.; Zhou, Q.; Gong, L.; Xue, C.; Cao, Y. Genetic epidemiology of porcine epidemic diarrhoea virus circulating in China in 2012–2017 based on spike gene. Transbound. Emerg. Dis. 2018, 65, 883–889. [Google Scholar] [CrossRef]
- Kocherhans, R.; Bridgen, A.; Ackermann, M.; Tobler, K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus. Genes 2001, 23, 137–144. [Google Scholar] [CrossRef]
- Sun, M.; Ma, J.; Wang, Y.; Wang, M.; Song, W.; Zhang, W.; Lu, C.; Yao, H. Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J. Clin. Microbiol. 2015, 53, 1484–1492. [Google Scholar] [CrossRef]
- Wrapp, D.; McLellan, J.S. The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epide-mic Diarrhea Virus Spike Protein in the Prefusion Conformation. J. Virol. 2019, 93, e00923-19. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.-T.K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019, 5, eaav4580. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, C.; Shi, H.; Qiu, H.; Liu, S.; Chen, X.; Zhang, Z.; Feng, L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010, 155, 1471–1476. [Google Scholar] [CrossRef]
- Sun, M.; Ma, J.; Yu, Z.; Pan, Z.; Lu, C.; Yao, H. Identification of two mutation sites in spike and envelope proteins mediating optimal cellular infection of porcine epidemic diarrhea virus from different pathways. Vet. Res. 2017, 48, 44. [Google Scholar] [CrossRef]
- Nandre, R.M.; Chaudhari, A.A.; Matsuda, K.; Lee, J.H. Immunogenicity of a Salmonella Enteritidis mutant as vaccine candidate and its protective efficacy against salmonellosis in chickens. Vet. Immunol. Immunopathol. 2011, 144, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Ming, K.; Chen, Y.; Yao, F.; Shi, J.; Yang, J.; Du, H.; Wang, X.; Wang, Y.; Liu, J. Phosphorylated Codonopsis pilosula polysaccharide could inhibit the virulence of duck hepatitis A virus compared with Codonopsis pilosula polysaccharide. Int. J. Biol. Macromol. 2017, 94 Pt A, 28–35. [Google Scholar] [CrossRef]
- Wen, F.; Yang, J.; Li, A.; Gong, Z.; Yang, L.; Cheng, Q.; Wang, C.; Zhao, M.; Yuan, S.; Chen, Y.; et al. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS ONE 2021, 16, e0253622. [Google Scholar] [CrossRef]
- Zhang, H.; Han, F.; Yan, X.; Liu, L.; Shu, X.; Hu, H. Prevalence and phylogenetic analysis of spike gene of porcine epidemic diarrhea virus in Henan province, China in 2015–2019. Infect. Genet. Evol. 2021, 88, 104709. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423. [Google Scholar] [CrossRef]
- Jang, G.; Lee, S.; Lee, C. Assessing the risk of recurrence of porcine epidemic diarrhea virus in affected farms on Jeju Island, South Korea. J. Vet. Sci. 2021, 22, e48. [Google Scholar] [CrossRef]
- Guo, J.; Fang, L.; Ye, X.; Chen, J.; Xu, S.; Zhu, X.; Miao, Y.; Wang, D.; Xiao, S. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound. Emerg. Dis. 2019, 66, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Zhou, J.; Wang, X.; Ma, L.; Li, J.; Yang, L.; Yuan, H.; Pang, D.; Ouyang, H. Porcine Epidemic Diarrhea Virus: An Updated Overview of Virus Epidemiology, Virulence Variation Patterns and Virus-Host Interactions. Viruses 2022, 14, 2434. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, Z.; Li, Y.; Gao, S.; Xiao, S. Porcine epidemic diarrhea virus: Molecular mechanisms of attenuation and vaccines. Microb. Pathog. 2020, 149, 104553. [Google Scholar] [CrossRef]
- Chang, S.H.; Bae, J.L.; Kang, T.J.; Kim, J.; Chung, G.H.; Lim, C.W.; Laude, H.; Yang, M.S.; Jang, Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002, 14, 295–299. [Google Scholar] [PubMed]
- Sun, D.; Feng, L.; Shi, H.; Chen, J.; Cui, X.; Chen, H.; Liu, S.; Tong, Y.; Wang, Y.; Tong, G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008, 131, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Terada, Y.; Enjuanes, L.; Ohashi, S.; Kamitani, W. S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets. Viruses 2018, 10, 467. [Google Scholar] [CrossRef]
- Wang, K.; Lu, W.; Chen, J.; Xie, S.; Shi, H.; Hsu, H.; Yu, W.; Xu, K.; Bian, C.; Fischer, W.B.; et al. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012, 586, 384–391. [Google Scholar] [CrossRef]
- Liang, W.; Zhou, D.; Geng, C.; Yang, K.; Duan, Z.; Guo, R.; Liu, W.; Yuan, F.; Liu, Z.; Gao, T.; et al. Isolation and evolutionary analyses of porcine epidemic diarrhea virus in Asia. PeerJ 2020, 8, e10114. [Google Scholar] [CrossRef]
- Feng, B.; Li, C.; Qiu, Y.; Qi, W.; Qiu, M.; Li, J.; Lin, H.; Zheng, W.; Zhu, J.; Chen, N. Genomic Characterizations of Porcine Epidemic Diarrhea Viruses (PEDV) in Diarrheic Piglets and Clinically Healthy Adult Pigs from 2019 to 2022 in China. Animals 2023, 13, 1562. [Google Scholar] [CrossRef]
- Wang, D.; Ge, X.; Chen, D.; Li, J.; Cai, Y.; Deng, J.; Zhou, L.; Guo, X.; Han, J.; Yang, H. The S Gene Is Necessary but Not Sufficient for the Virulence of Porcine Epidemic Diarrhea Virus Novel Variant Strain BJ2011C. J. Virol. 2018, 92, e00603–e00618. [Google Scholar] [CrossRef]
- Park, S.-J.; Moon, H.J.; Luo, Y.; Kim, H.-K.; Kim, E.M.; Yang, J.S.; Song, D.S.; Kang, B.K.; Lee, C.S.; Park, B.K. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus. Genes 2008, 36, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zeng, L.; Yang, J.; Yu, F.; Ge, J.; Guo, Q.; Gao, X.; Song, T. Sequence heterogeneity of the ORF3 gene of porcine epidemic diarrhea viruses field samples in Fujian, China, 2010–2012. Viruses 2013, 5, 2375–2383. [Google Scholar] [CrossRef] [PubMed]


| Name of Sample | Time of Collection | PEDV Vaccination Status (Sows) | PEDV Positive or Not |
|---|---|---|---|
| HB-lf-01 | November 2019 | Inactivated vaccine CV777 | Negative |
| HB-lf-02 | November 2019 | Inactivated vaccine CV777 | Negative |
| HB-lf-03 | November 2019 | Inactivated vaccine CV777 | Negative |
| Ll | November 2019 | Duplex live attenuated vaccine AJ1102 | Negative |
| ZMD1 | November 2019 | Live attenuated triple vaccine CV777 | Negative |
| ZMD2 | November 2019 | Live attenuated triple vaccine CV777 | Negative |
| E1-4 | December 2019 | Inactivated vaccine CV777 | Negative |
| E5 | December 2019 | Inactivated vaccine CV777 | Negative |
| E6 | December 2019 | Inactivated vaccine CV777 | Positive |
| 1-1 | December 2019 | Inactivated vaccine CHYJ | Positive |
| 2-1 | December 2019 | Inactivated vaccine CHYJ | Negative |
| 3-1 | December 2019 | Inactivated vaccine CHYJ | Negative |
| 4-1 | December 2019 | Inactivated vaccine CHYJ | Negative |
| 5-1 | December 2019 | Inactivated vaccine CHYJ | Positive |
| 6-1 | December 2019 | Inactivated vaccine CHYJ | Negative |
| 1 + 2 | December 2019 | Duplex live attenuated vaccine LW/L | Positive |
| JZ-p1 | December 2019 | Live attenuated triple vaccine CV777 | Negative |
| JZ-p2 | December 2019 | Live attenuated triple vaccine CV777 | Negative |
| 7.1 | January 2020 | Inactivated vaccine CV777 | Positive |
| 7.2 | January 2020 | Inactivated vaccine CV777 | Positive |
| 7.3 | January 2020 | Inactivated vaccine CV777 | Negative |
| ZMD3 | January 2020 | Live attenuated triple vaccine CV777 | Positive |
| ZMD4 | January 2020 | Live attenuated triple vaccine CV777 | Positive |
| ZMD5 | January 2020 | Live attenuated triple vaccine CV777 | Positive |
| C1 | January 2020 | Duplex live attenuated vaccine SCSZ-1 | Positive |
| C2 | January 2020 | Duplex live attenuated vaccine SCSZ-1 | Negative |
| C3 | January 2020 | Duplex live attenuated vaccine SCSZ-1 | Negative |
| C4 | January 2020 | Duplex live attenuated vaccine SCSZ-1 | Negative |
| 4-1 | November 2020 | Duplex live attenuated vaccine AJ1102 | Positive |
| 3-2 | November 2020 | Duplex live attenuated vaccine AJ1102 | Negative |
| 2-2 | November 2020 | Duplex live attenuated vaccine AJ1102 | Negative |
| 1-1 | November 2020 | Duplex live attenuated vaccine AJ1102 | Negative |
| E7-8 | November 2020 | Inactivated vaccine CV777 | Negative |
| E9 | November 2020 | Inactivated vaccine CV777 | Positive |
| HN-01 | December 2020 | Inactivated vaccine CV777 | Negative |
| HN-02 | December 2020 | Inactivated vaccine CV777 | Positive |
| HN-03 | December 2020 | Inactivated vaccine CV777 | Positive |
| HN-04 | December 2020 | Inactivated vaccine CV777 | Positive |
| HN-05 | December 2020 | Inactivated vaccine CV777 | Negative |
| J1 | December 2020 | Live attenuated triple vaccine CV777 | Negative |
| J2 | December 2020 | Live attenuated triple vaccine CV777 | Negative |
| J3 | December 2020 | Live attenuated triple vaccine CV777 | Positive |
| SX1-3 | March 2021 | Duplex live attenuated vaccine AJ1102 | Negative |
| SX4-5 | March 2021 | Duplex live attenuated vaccine AJ1102 | Positive |
| SX6 | March 2021 | Duplex live attenuated vaccine AJ1102 | Positive |
| SX7-9 | April 2021 | Duplex live attenuated vaccine AJ1102 | Negative |
| SX10 | April 2021 | Duplex live attenuated vaccine AJ1102 | Positive |
| Strain | GenBank Entry Number | Origin Country | Collection Year |
|---|---|---|---|
| CV777 | AF353511.1 | Belgium | 1978 |
| LZC | EF185992.1 | China | 2006 |
| DR13 | JQ023161.1 | Korea | 1999 |
| AJ1102 | JX188454.1 | China | 2012 |
| PC22A | KY499262.1 | USA | 2017 |
| Missouri270/2014 | KR265846.1 | USA | 2016 |
| S14 | MH891585.1 | Korea | 2019 |
| MEX/JAL/03/2017 | MH004418.1 | Mexico | 2017 |
| AH-2018-HF1 | MN315264.1 | China | 2018 |
| G2 | MK138353.1 | China | 2018 |
| CH-HB2-2018 | MK606369.1 | China | 2018 |
| CH/HNLH/2015 | KT199103.1 | China | 2015 |
| SM98 | GU937797.1 | Korea | 2011 |
| JS2008 | KC109141.1 | China | 2013 |
| YN90 | KT021231 | China | 2015 |
| CH/ZMDZY/11 | KC196276 | China | 2013 |
| NW8 | MF782687 | China | 2017 |
| CH/YNKM-8/2013 | KF761675 | China | 2013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.; Jiang, F.; Wang, S.; Wu, H.; Liu, Y.; Wang, B.; Hou, W.; Yu, X.; Wang, H. Natural Evolution of Porcine Epidemic Diarrhea Viruses Isolated from Maternally Immunized Piglets. Animals 2023, 13, 1766. https://doi.org/10.3390/ani13111766
Ge Y, Jiang F, Wang S, Wu H, Liu Y, Wang B, Hou W, Yu X, Wang H. Natural Evolution of Porcine Epidemic Diarrhea Viruses Isolated from Maternally Immunized Piglets. Animals. 2023; 13(11):1766. https://doi.org/10.3390/ani13111766
Chicago/Turabian StyleGe, Yufang, Feiyang Jiang, Sibei Wang, Heqiong Wu, Yuan Liu, Bin Wang, Wei Hou, Xiuju Yu, and Haidong Wang. 2023. "Natural Evolution of Porcine Epidemic Diarrhea Viruses Isolated from Maternally Immunized Piglets" Animals 13, no. 11: 1766. https://doi.org/10.3390/ani13111766
APA StyleGe, Y., Jiang, F., Wang, S., Wu, H., Liu, Y., Wang, B., Hou, W., Yu, X., & Wang, H. (2023). Natural Evolution of Porcine Epidemic Diarrhea Viruses Isolated from Maternally Immunized Piglets. Animals, 13(11), 1766. https://doi.org/10.3390/ani13111766





