Characterization of Two Mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with Phylogenetic Relationships and Selection Pressure Analyses of Hylidae
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Primer Design, PCR Amplification, and Sequencing
2.3. Mitogenome Annotation and Sequence Analyses
2.4. Genetic Distance
2.5. Phylogenetic Analyses
2.6. Detecting Selective Pressure
3. Results
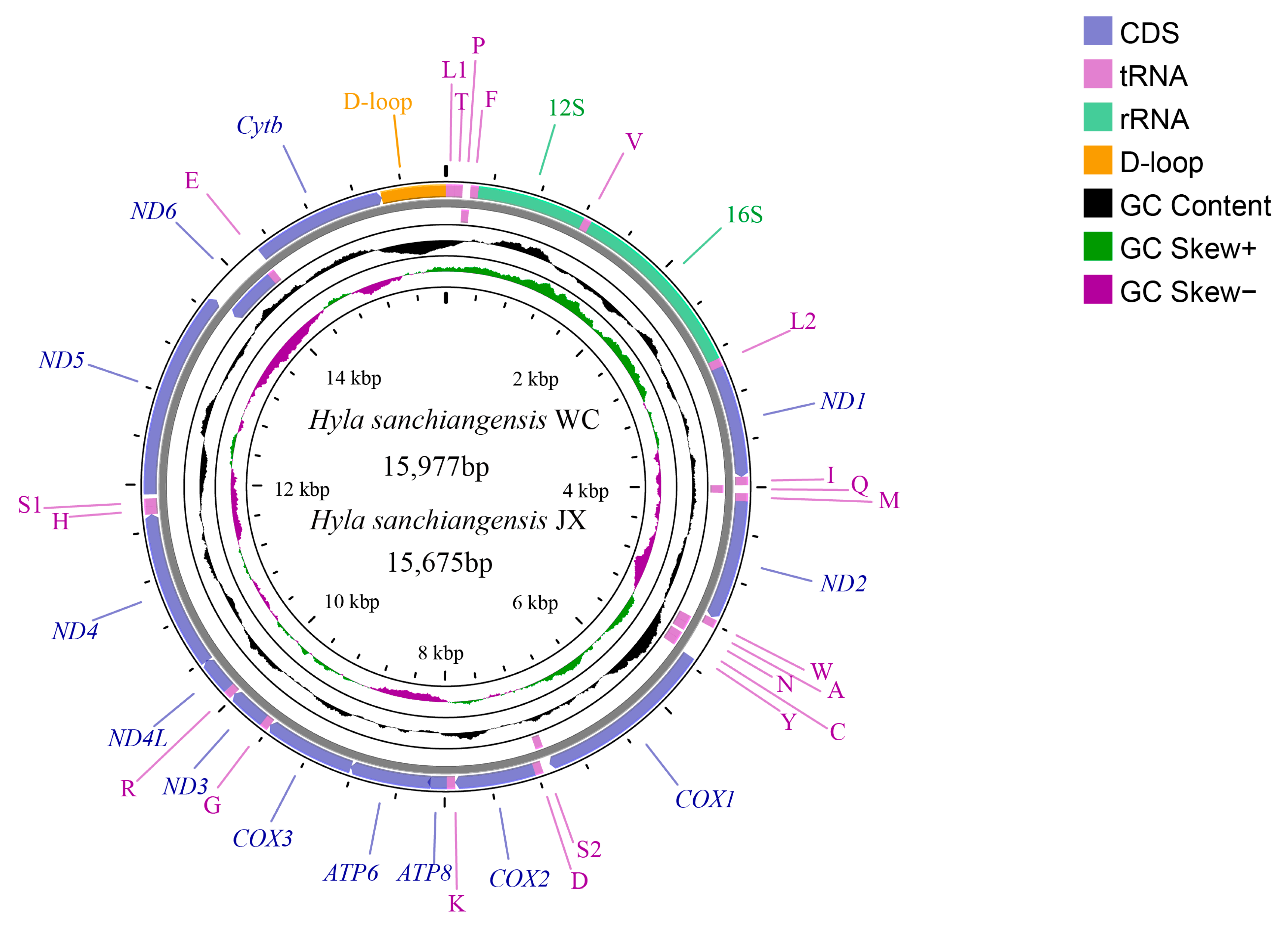
3.1. Mitogenome Organization and Structure
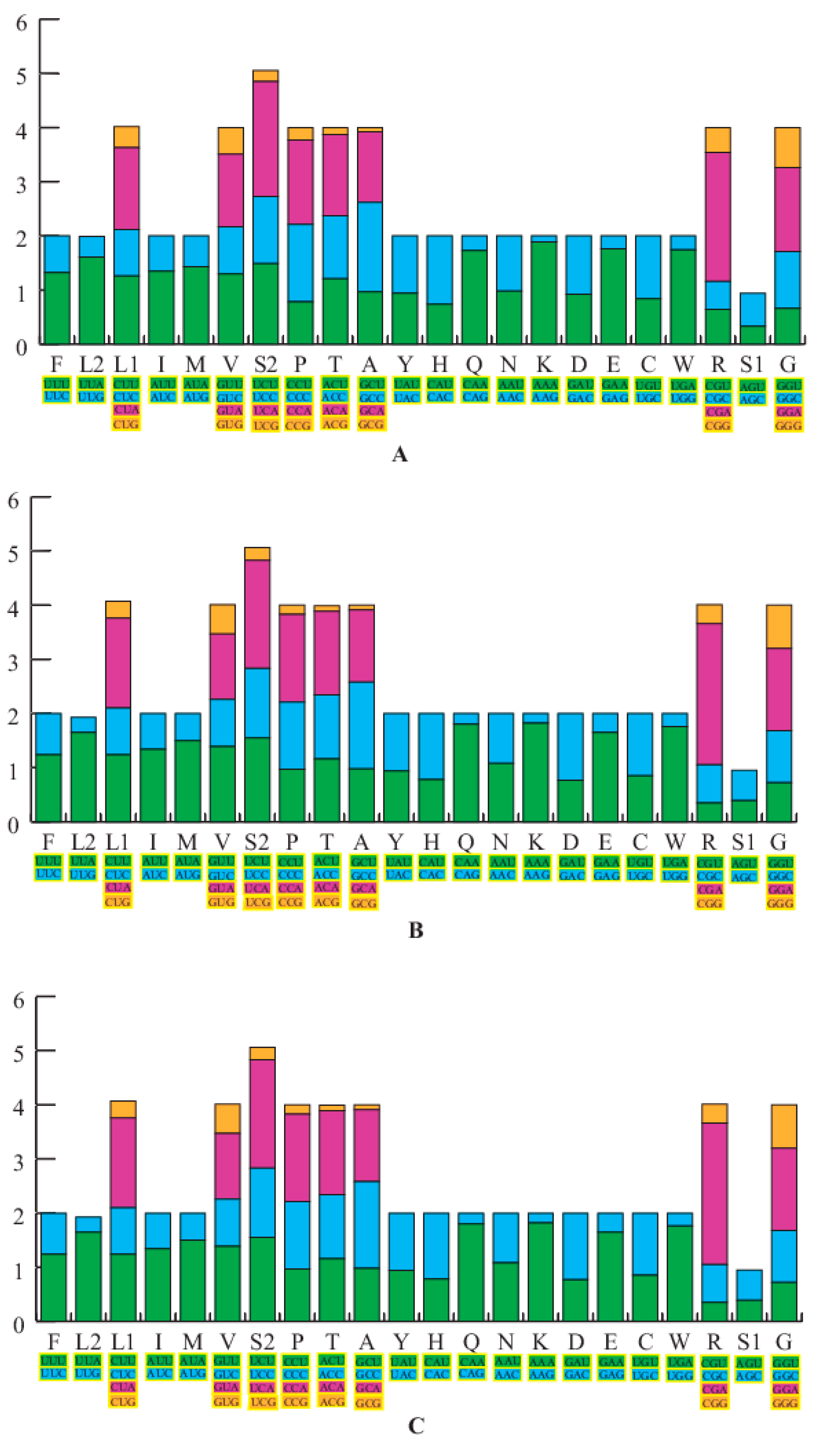
3.2. Protein-Coding Genes and Codon Usages
3.3. Ribosomal and Transfer RNAs
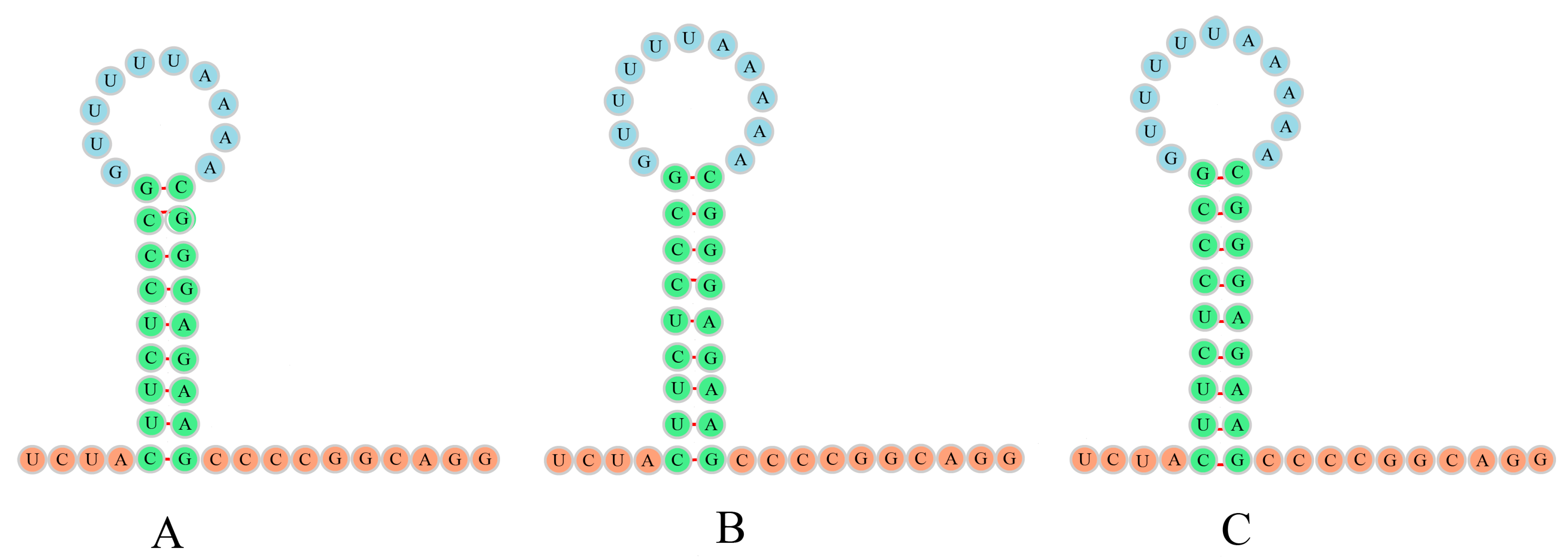
3.4. Intergenic Regions and L-Strand Origin of Replication
3.5. Genetic Distance
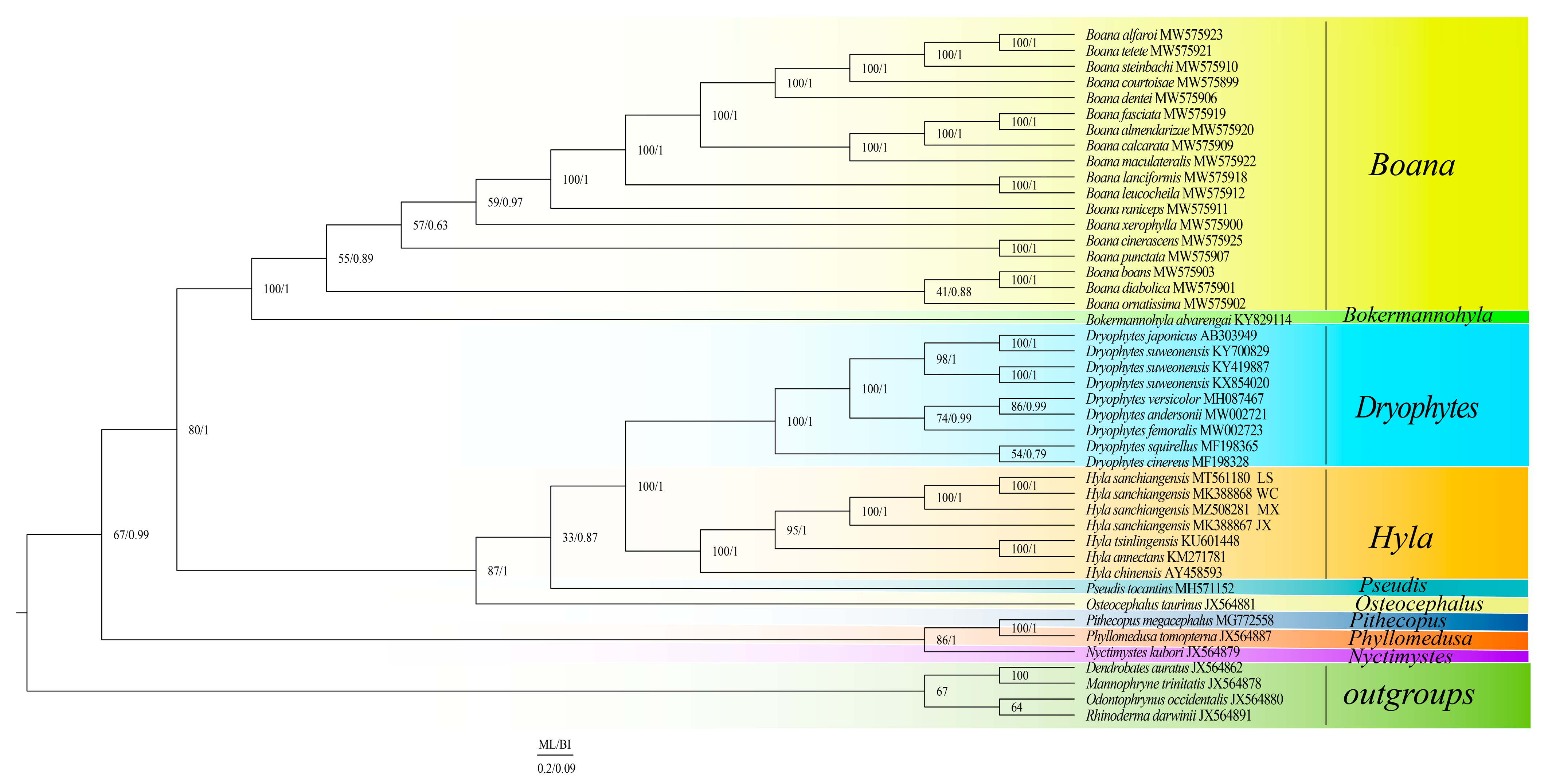
3.6. Phylogeny of Hylidae
3.7. Detecting Selective Pressure
4. Discussion
4.1. Mitogenome Structure
4.2. Genetic Distance and Phylogeny of Hylidae
4.3. Detecting Selective Pressure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeisset, I.; Beebee, T.J.C. Amphibian phylogeography: A model for understanding historical aspects of species distributions. Heredity 2008, 101, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, F.; Milinkovitch, M.C. Amphibians as indicators of early tertiary “out-of-India” dispersal of vertebrates. Science 2001, 292, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Wang, K. AmphibiaChina: An online database of Chinese Amphibians. Zool. Res. 2016, 37, 57–59. [Google Scholar]
- Li, J.T.; Wang, J.S.; Nian, H.H.; Litvinchuk, S.N.; Wang, J.; Li, Y.; Rao, D.Q.; Klaus, S. Amphibians crossing the Bering Land Bridge: Evidence from holarctic treefrogs (Hyla, Hylidae, Anura). Mol. Phylogenet. Evol. 2015, 87, 80–90. [Google Scholar] [CrossRef]
- Wiens, J.J.; Graham, C.H.; Moen, D.S.; Smith, S.A.; Reeder, T.W. Evolutionary and ecological causes of the latitudinal diversity gradient in hylid frogs: Treefrog trees unearth the roots of high tropical diversity. Am. Nat. 2006, 168, 579–596. [Google Scholar] [CrossRef]
- Duellman, W.E.; Marion, A.B.; Hedges, S.B. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa 2016, 4104, 1–109. [Google Scholar] [CrossRef]
- Frost, D.R. Amphibian Species of the World: An Online Reference, Version 5.4. 2010. Available online: http://research.amnh.org/vz/herpetology/amphibia (accessed on 26 August 2022).
- Pope, C.H. Four new frogs from Fukien Province, China. In American Museum Novitates; No. 352; The American Museum of National History: New York City, NY, USA, 1929. [Google Scholar]
- Kishida, O.; Nishimura, K. Multiple inducible defences against multiple predators in the anuran tadpole. Rana pirica. Evol. Ecol. Res. 2005, 7, 619–631. [Google Scholar]
- Ghioca-Robrecht, D.; Smith, L.; Densmore, L. Ecological correlates of trophic polyphenism in spadefoot tadpoles inhabiting playas. Can. J. Zool. 2009, 87, 229–238. [Google Scholar] [CrossRef]
- Székely, P.; Cogălniceanu, D.; Tudor, M. Effect of habitat drying on the development of the Eastern spadefoot toad (Pelobates syriacus) tadpoles. Amphib. Reptilia 2010, 31, 425–434. [Google Scholar] [CrossRef]
- Ledón-Rettig, C.C.; Pfennig, D.W. Emerging model systems in eco-evo-devo: The environmentally responsive spadefoot toad. Evol. Dev. 2011, 13, 391–400. [Google Scholar] [CrossRef]
- Johnson, J.B.; Saenz, D.; Adams, C.K.; Hibbitts, T.J. Naturally occurring variation in tadpole morphology and performance linked to predator regime. Ecol. Evol. 2015, 5, 2991–3002. [Google Scholar] [CrossRef] [PubMed]
- Van Buskirk, J. Spatially heterogeneous selection in nature favors phenotypic plasticity in anuran larvae. Evolution 2017, 71, 1670–1685. [Google Scholar] [CrossRef]
- Touchon, J.C.; Robertson, J.M. You cannot have it all: Heritability and constraints of predator-induced developmental plasticity in a Neotropical treefrog. Evolution 2018, 72, 2758–2772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zheng, R.Q. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and phylogenetic relationship of Dicroglossidae. BMC Evol. Biol. 2018, 18, 26. [Google Scholar] [CrossRef]
- Cai, Y.T.; Li, Q.; Zhang, J.Y.; Storey, K.B.; Yu, D.N. Characterization of the mitochondrial genomes of two toads, Anaxyrus americanus (Anura: Bufonidae) and Bufotes pewzowi (Anura: Bufonidae), with phylogenetic and selection pressure analyses. PeerJ 2020, 8, e8901. [Google Scholar] [CrossRef] [PubMed]
- Roe, B.A.; Ma, D.P.; Wilson, R.; Wong, J. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J. Biol. Chem. 1985, 260, 9759–9774. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Wu, X.B.; Wang, Y.Q.; Zhou, K.Y.; Zhu, W.Q.; Nie, J.S.; Wang, C.L. Complete mitochondrial DNA sequence of Chinese alligator, Alligator sinensis, and phylogeny of crocodiles. Chin. Sci. Bull. 2003, 48, 2050–2054. [Google Scholar] [CrossRef]
- Sano, N.; Kurabayashi, A.; Fujii, T.; Yonekawa, H.; Sumida, M. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of the bell-ring frog, Buergeria buergeri (family Rhacophoridae). Genes Genet. Syst. 2004, 79, 151–163. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Usuki, C.; Mikami, N.; Fujii, T.; Yonekawa, H.; Sumida, M.; Hasegawa, M. Complete nucleotide sequence of the mitochondrial genome of a Malagasy poison frog Mantella madagascariensis: Evolutionary implications on mitochondrial genomes of higher anuran groups. Mol. Phylogenetics Evol. 2006, 39, 223–236. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Yoshikawa, N.; Sato, N.; Hayashi, Y.; Oumi, S.; Fujii, T.; Sumida, M. Complete mitochondrial DNA sequence of the endangered frog Odorrana ishikawae (family Ranidae) and unexpected diversity of mt gene arrangements in ranids. Mol. Phylogenetics Evol. 2010, 56, 543–553. [Google Scholar] [CrossRef]
- Wu, Z.; Sainz, A.G.; Shadel, G.S. Mitochondrial DNA: Cellular genotoxic stress sentinel. Trends Biochem Sci. 2021, 46, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M. A Checklist of North American Amphibians and Reptiles: The United States and Canada. Volume 1–Amphibians1. Ichthyol. Herpetol. 2014, 4, 762. [Google Scholar]
- Zhang, J.Y.; Luu, B.E.; Yu, D.N.; Zhang, L.P.; Al-Attar, R.; Storey, K.B. The complete mitochondrial genome of Dryophytes versicolor: Phylogenetic relationship among Hylidae and mitochondrial protein-coding gene expression in response to freezing and anoxia. Int. J. Biol. Macromol. 2019, 132, 461–469. [Google Scholar] [CrossRef]
- Lee, M.Y.; Jeon, H.S.; Min, M.S.; An, J. Sequencing and analysis of the complete mitochondrial genome of Hyla suweonensis (Anura: Hylidae). Mitochondrial DNA Part B 2017, 2, 126–127. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Kreitman, M. Is mitochondrial DNA a strictly neutral marker? Trends Ecol. Evol. 1995, 10, 485–488. [Google Scholar] [CrossRef]
- Wu, L.; Tong, Y.; Ayivi, S.P.G.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. The Complete Mitochondrial Genomes of Three Sphenomorphinae Species (Squamata: Scincidae) and the Selective Pressure Analysis on Mitochondrial Genomes of Limbless Isopachys gyldenstolpei. Animals 2022, 12, 2015. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Insight into the Phylogenetic Relationships among Three Subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with Low-Temperature Selection Pressure Analyses Using Mitogenomes. Insects 2021, 12, 656. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.E.; Lin, Y.F.; Ma, L.; Ding, G.H.; Lin, Z.H. Partial mitochondrial genome of the Sanchiang Tree Toad Hyla sanchiangensis (Anura: Hylidae). Mitochondrial DNA Part B 2020, 5, 2682–2683. [Google Scholar] [CrossRef]
- Zhang, P.; Liang, D.; Mao, R.L.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C. Efficient sequencing of anuran mtDNAs and a mitogenomic exploration of the phylogeny and evolution of frogs. Mol. Biol. Evol. 2013, 30, 1899–1915. [Google Scholar] [CrossRef]
- Cai, Y.Y.; Shen, S.Q.; Lu, L.X.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The complete mitochondrial genome of Pyxicephalus adspersus: High gene rearrangement and phylogenetics of one of the world’s largest frogs. PeerJ 2019, 7, e7532. [Google Scholar] [CrossRef] [PubMed]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Bioinf. Methods Protoc. 2000, 132, 71–91. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Cameron, S. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Fekete, M.; Stadler, P.F. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002, 319, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Bernhart, S.H.; Hofacker, I.L.; Will, S.; Gruber, A.R.; Stadler, P.F. RNAalifold: Improved consensus structure prediction for RNA alignments. BMC Bioinform. 2008, 9, 474. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Borzee, A.; Messenger, K.R.; Chae, S.; Andersen, D.; Groffen, J.; Kim, Y.I.; An, J.; Othman, S.N.; Ri, K.; Nam, T.Y. Yellow sea mediated segregation between North East Asian Dryophytes species. PLoS ONE 2020, 15, e0234299. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, J.Y.; Zheng, R.Q.; Yu, B.G.; Yang, G. Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae). Gene 2009, 447, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.N.; Zhang, J.Y.; Zheng, R.Q. The complete mitochondrial genome of Babina adenopleura (Anura: Ranidae). Mitochondrial DNA 2012, 23, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.N.; Zhang, J.Y.; Zheng, R.Q.; Shao, C. The complete mitochondrial genome of Hoplobatrachus rugulosus (Anura: Dicroglossidae). Mitochondrial DNA 2012, 23, 336–337. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Lima, N.G.d.S.; Carmo, A.O.d.; Martins, A.P.V.; Souza, R.C.C.d.; Kalapothakis, E.; Eterovick, P.C. Complete mitochondrial genome sequence of the high-altitude Brazilian tree frog Bokermannohyla alvarengai (Anura, Hylidae). Mitochondrial DNA Part B 2017, 2, 281–282. [Google Scholar] [CrossRef] [PubMed]
- Fouquet, A.; Marinho, P.; Réjaud, A.; Carvalho, T.R.; Caminer, M.A.; Jansen, M.; Rainha, R.N.; Rodrigues, M.T.; Werneck, F.P.; Lima, A.P. Systematics and biogeography of the Boana albopunctata species group (Anura, Hylidae), with the description of two new species from Amazonia. Syst. Biodivers. Sci. 2021, 19, 375–399. [Google Scholar] [CrossRef]
- Warwick, A.R.; Barrow, L.N.; Smith, M.L.; Means, D.B.; Lemmon, A.R.; Lemmon, E.M. Signatures of north-eastern expansion and multiple refugia: Genomic phylogeography of the Pine Barrens tree frog, Hyla andersonii (Anura: Hylidae). Biol. J. Linn. Soc. 2021, 133, 120–134. [Google Scholar] [CrossRef]
- Barrow, L.N.; Soto-Centeno, J.A.; Warwick, A.R.; Lemmon, A.R.; Moriarty Lemmon, E. Evaluating hypotheses of expansion from refugia through comparative phylogeography of south-eastern Coastal Plain amphibians. J. Biogeogr. 2017, 44, 2692–2705. [Google Scholar] [CrossRef]
- Borzee, A.; Didinger, C.; Jang, Y. Complete mitochondrial genome of Dryophytes suweonensis (Anura Hylidae). Mitochondrial DNA B Resour. 2017, 2, 5–6. [Google Scholar] [CrossRef]
- Ye, L.T.; Zhu, C.C.; Yu, D.N.; Zhang, Y.P.; Zhang, J.Y. The complete mitochondrial genome of Hyla annectans (Anura: Hylidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 1593–1594. [Google Scholar]
- Zhang, P.; Zhou, H.; Chen, Y.Q.; Liu, Y.F.; Qu, L.H. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Syst. Biol. 2005, 54, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.; Kurabayashi, A.; Usuki, C.; Fujii, T.; Sumida, M.J.G. Complete mitochondrial genomes of three neobatrachian anurans: A case study of divergence time estimation using different data and calibration settings. Gene 2008, 407, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Sun, Z.L.; Guo, W.B.; Wu, J.; Qian, L.F.; Pan, T.; Wang, H.; Li, K.; Zhang, B.W. Sequencing of complete mitochondrial genome for Tsinling Tree Toad (Hyla tsinlingensis). Mitochondrial DNA B Resour. 2016, 1, 466–467. [Google Scholar] [CrossRef]
- Gatto, K.P.; Smith, J.J.; Lourenco, L.B. The mitochondrial genome of the endemic Brazilian paradoxical frog Pseudis tocantins (Hylidae). Mitochondrial DNA B Resour. 2018, 3, 1106–1107. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Martens, P.A.; Clayton, D.A. Mechanism of mitochondrial DNA replication in mouse L-cells: Localization and sequence of the light-strand origin of replication. J. Mol. Biol. 1979, 135, 327–351. [Google Scholar] [CrossRef]
- Yan, P.; Pan, T.; Wu, G.Y.; Kang, X.; Ali, I.; Zhou, W.L.; Li, J.T.; Wu, X.B.; Zhang, B.W. Species Delimitation and Evolutionary History of Tree Frogs in the Hyla chinensis Group (Hylidae, Amphibian). Front. Ecol. Evol. 2020, 8, 234. [Google Scholar] [CrossRef]
- Huang, M.Y.; Duan, R.Y.; Tang, T.; Zhu, C.; Wang, Y. The complete mitochondrial genome of Hyla tsinlingensis (Anura: Hylidae). Mitochondrial DNA Part A 2016, 27, 4130–4131. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Stephens, P.R.; Wiens, J.J. Replicate patterns of species richness, historical biogeography, and phylogeny in Holarctic treefrogs. Evolution 2005, 59, 2433–2450. [Google Scholar] [PubMed]
- McKenzie, M.; Chiotis, M.; Pinkert, C.A.; Trounce, I.A. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol. Biol. Evol. 2003, 20, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Calvaruso, M.A.; Willems, P.; van den Brand, M.; Valsecchi, F.; Kruse, S.; Palmiter, R.; Smeitink, J.; Nijtmans, L. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 2012, 21, 115–120. [Google Scholar] [CrossRef]
- Bernardes, J.S.; Eberle, R.J.; Vieira, F.R.; Coronado, M.A. A comparative pan-genomic analysis of 53 C. Pseudotuberculosis strains based on functional domains. J. Biomol. Struct. Dyn. 2021, 39, 6974–6986. [Google Scholar] [CrossRef]
- Yu, D.N.; Zhang, J.Y.; Li, P.; Zheng, R.Q.; Shao, C. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the Cyt b gene and the complete MT genome. PLoS ONE 2015, 10, e0124825. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O′Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 119. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Vaish, M.; Price-Whelan, A.; Reyes-Robles, T.; Liu, J.; Jereen, A.; Christie, S.; Alonzo III, F.; Benson, M.A.; Torres, V.J.; Krulwich, T.A. Roles of Staphylococcus aureus Mnh1 and Mnh2 antiporters in salt tolerance, alkali tolerance, and pathogenesis. J. Bacteriol. 2018, 200, e00611–e00617. [Google Scholar] [CrossRef]
- Berry, E.A.; Guergova-Kuras, M.; Huang, L.; Crofts, A.R. Structure and function of cytochrome bc complexes. Annu. Rev. Biochem. 2000, 69, 1005–1075. [Google Scholar] [CrossRef]
- Lanciano, P.; Khalfaoui-Hassani, B.; Selamoglu, N.; Ghelli, A.; Rugolo, M.; Daldal, F. Molecular mechanisms of superoxide production by complex III: A bacterial versus human mitochondrial comparative case study. Biochim. Biophys. Acta. 2013, 1827, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Nielsen, R.; Yang, Z.H. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef]
- Shen, Y.Y.; Liang, L.; Zhu, Z.H.; Zhou, W.P.; Irwin, D.M.; Zhang, Y.P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA 2010, 107, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.C.; Shen, X.J.; Irwin, D.M.; Shen, Y.Y.; Zhang, Y.P. Mitogenomic analyses propose positive selection in mitochondrial genes for high-altitude adaptation in galliform birds. Mitochondrion 2014, 18, 70–75. [Google Scholar] [CrossRef]
- Ben Slimen, H.; Awadi, A.; Tolesa, Z.G.; Knauer, F.; Alves, P.C.; Makni, M.; Suchentrunk, F. Positive selection on the mitochondrial ATP synthase 6 and the NADH dehydrogenase 2 genes across 22 hare species (genus Lepus). J. Zool. Syst. Evol. Res. 2018, 56, 428–443. [Google Scholar] [CrossRef]
- Shao′e, S.; Hui, M.; Wang, M.X.; Sha, Z.L. The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): Insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comp. Biochem. Physiol. Part D: Genom. Proteom. 2018, 25, 42–52. [Google Scholar]
- Banguera-Hinestroza, E.; Sawall, Y.; Al-Sofyani, A.; Mardulyn, P.; Fuertes-Aguilar, J.; Cardenas-Henao, H.; Jimenez-Infante, F.; Voolstra, C.R.; Flot, J.F. mtDNA recombination indicative of hybridization suggests a role of the mitogenome in the adaptation of reef-building corals to extreme environments. bioRxiv 2019, 462069. [Google Scholar] [CrossRef]
- Sun, J.T.; Jin, P.Y.; Hoffmann, A.; Duan, X.Z.; Dai, J.; Hu, G.; Xue, X.F.; Hong, X.Y. Evolutionary divergence of mitochondrial genomes in two Tetranychus species distributed across different climates. Insect Mol. Biol. 2018, 27, 698–709. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
| Family | Genus | Species | Length (bp) | GenBank Accession Number | Reference |
|---|---|---|---|---|---|
| Hylidae | Bokermannohyla | Bokermannohyla alvarengai | 17,325 | KY829114 | [57] |
| Boana | Boana aff. courtoisae | 15,840 | MW575899 | [58] | |
| Boana aff. lanciformis | 17,150 | MW575918 | |||
| Boana alfaroi | 17,219 | MW575923 | |||
| Boana almendarizae | 17,439 | MW575920 | |||
| Boana boans | 17,021 | MW575903 | |||
| Boana calcarata | 16,251 | MW575909 | |||
| Boana cinerascens | 17,684 | MW575925 | |||
| Boana dentei | 17,587 | MW575906 | |||
| Boana diabolica | 14,606 | MW575901 | |||
| Boana fasciata | 17,438 | MW575919 | |||
| Boana leucocheila | 18,042 | MW575912 | |||
| Boana maculateralis | 16,301 | MW575922 | |||
| Boana ornatissima | 17,656 | MW575902 | |||
| Boana punctata | 17,899 | MW575907 | |||
| Boana raniceps | 17,180 | MW575911 | |||
| Boana steinbachi | 18,107 | MW575910 | |||
| Boana tetete | 16,107 | MW575921 | |||
| Boana xerophylla | 18,062 | MW575900 | |||
| Dendrobates | Dendrobates auratus | 14,963 | JX564862 | [32] | |
| Dryophytes | Dryophytes andersonii isolate | 15,423 | MW002721 | [59] | |
| Dryophytes cinereus voucher | 15,418 | MF198328 | [60] | ||
| Dryophytes femoralis isolate | 15,415 | MW002723 | [59] | ||
| Dryophytes suweonensis | 17,448 | KX854020 | [61] | ||
| 18,611 | KY419887 | [27] | |||
| 18,288 | KY700829 | Unpublished | |||
| Dryophytes versicolor | 18,800 | MH087467 | [26] | ||
| Hyla | Hyla annectans | 17,973 | KM271781 | [62] | |
| Hyla chinensis | 18,180 | AY458593 | [63] | ||
| Hyla japonica | 19,519 | AB303949 | [64] | ||
| Hyla sanchiangensis LS | 15,664 | MT561180 | [31] | ||
| Hyla sanchiangensis JX | 15,675 | MK388867 | This study | ||
| Hyla sanchiangensis WC | 15,977 | MK388868 | This study | ||
| Hyla sanchiangensis MX | 15,694 | MZ508281 | Direct Submission | ||
| Hyla tsinlingensis | 18,305 | KU601448 | [65] | ||
| Nyctimystes | Nyctimystes kubori | 14,983 | JX564879 | [32] | |
| Osteocephalus | Osteocephalus taurinus | 14,970 | JX564881 | [32] | |
| Pithecopus | Pithecopus megacephalus | 18,050 | MG772558 | Unpublished | |
| Phyllomedusa | Phyllomedusa tomopterna | 14,926 | JX564887 | [32] | |
| Pseudis | Pseudis tocantins | 15,564 | MH571152 | [66] | |
| Dendrobatidae | Dendrobates | Dendrobates auratus | 14,863 | JX564862 | [32] |
| Mannophryne | Mannophryne trinitatis | 14,939 | JX564878 | ||
| Alsodidae | Odontophrynus | Odontophrynus occidentalis | 14,908 | JX564880 | |
| Rhinodermatidae | Rhinoderma | Rhinoderma darwinii | 14,943 | JX564891 |
| Gene/Region | Position JX/WC | Length (bp) JX/WC | Spacer (+) Overlap (−) JX/WC | Start Codon | Stop Codon | Strand |
|---|---|---|---|---|---|---|
| trnL1 (cta) | 1–72 | 72 | 0 | H | ||
| trnT (aca) | 73–143 | 71 | −1 | H | ||
| trnP (cca) | 143–211 | 69 | −1 | L | ||
| trnF (ttc) | 211–278 | 68 | 0 | H | ||
| 12S rRNA | 279–1211/279–1210 | 933/932 | 0 | H | ||
| trnV (gta) | 1212–1280/1211–1279 | 69 | 0 | H | ||
| 16S rRNA | 1281–2884/1280–2876 | 1604/1597 | 0 | H | ||
| trnL2 (tta) | 2885–2957/2877–2949 | 73 | 0 | H | ||
| ND1 | 2958–3918/2950–3910 | 945 | 0 | TTG | T | H |
| trnI (atc) | 3919–3989/3911–3981 | 71 | −1 | H | ||
| trnQ (caa) | 3989–4059/3981–4051 | 71 | −1 | L | ||
| trnM (atg) | 4059–4127/4051–4119 | 69 | 0 | H | ||
| ND2 | 4128–5162/4120–5154 | 1029 | +7 | ATT | AGA | H |
| trnW (tga) | 5170–5239/5162–5231 | 70 | 0 | H | ||
| trnA (gca) | 5240–5308/5232–5300 | 69 | 0 | L | ||
| trnN (aac) | 5309–5381/5301–5373 | 73 | L | |||
| OL | 5382–5406/5374–5399 | 27/28 | 0 | |||
| trnC (tgc) | 5407–5470/5400–5463 | 64 | 0 | L | ||
| trnY (tac) | 5471–5540/5464–5533 | 70 | +4 | L | ||
| COX1 | 5545–7086/5538–7079 | 1527 | +1 | ATA | AGA | H |
| trnS2 (tca) | 7088–7158/7081–7151 | 71 | +1 | L | ||
| trnD (gac) | 7160–7228/7153–7221 | 69 | +1 | H | ||
| COX2 | 7230–7917/7223–7910 | 672 | 0 | ATG | T | H |
| trnK (aaa) | 7918–7990/7911–7982 | 73/72 | 0 | H | ||
| ATP8 | 7991–8155/7983–8147 | 159 | −25 | ATG | TAA | H |
| ATP6 | 8131–8829/8123–8821 | 699 | −1 | ATC | TAA | H |
| COX3 | 8829–9614/8821–9606 | 783 | −1 | ATG | TAA | H |
| trnG (gga) | 9614–9682/9606–9674 | 69 | 0 | H | ||
| ND3 | 9683–10022/9675–10014 | 327 | 0 | ATG | T | H |
| trnR (cga) | 10023–10091/10015–10083 | 69 | +2 | H | ||
| ND4L | 10094–10396/10086–10388 | 300 | −7 | ATG | TAG | H |
| ND4 | 10390–11754/10382–11746 | 1359 | 0 | ATG | TAA | H |
| trnH (cac) | 11755–11823/11747–11815 | 69 | 0 | H | ||
| trnS1 (agc) | 11824–11890/11816–11882 | 67 | +36 | H | ||
| ND5 | 11927–13729/11919–13721 | 1803 | −17 | ATG | AGA | H |
| ND6 | 13713–14210/13705–14202 | 495 | 0 | ATG | AGA | L |
| trnE (gaa) | 14211–14278/14203–14270 | 68 | +2 | L | ||
| Cytb | 14281–15429/14273–15421 | 1149 | 0 | ATG | TAG | H |
| Region | H. Sanchiangensis WC | H. sanchiangensis JX | H. sanchiangensis LS (MT561180) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | AT% | AT-Skew | GC-Skew | Length (bp) | AT% | AT-Skew | GC-Skew | Length (bp) | AT% | AT-Skew | GC-Skew | |
| Whole genome * | 15,977 | 60.1 | 0 | −0.275 | 15,675 | 59.9 | 0.002 | −0.278 | 15,664 | 59.8 | 0.001 | −0.275 |
| PCGs | 11,331 | 59.6 | −0.075 | −0.278 | 11,331 | 59.8 | −0.074 | −0.279 | 11,316 | 59.6 | −0.078 | −0.274 |
| tRNA | 1533 | 59.4 | 0.036 | 0.016 | 1533 | 59.9 | 0.053 | −0.012 | 1528 | 59.3 | 0.047 | −0.005 |
| rRNA | 2529 | 59.9 | 0.142 | −0.120 | 2537 | 59.6 | 0.145 | −0.125 | 2533 | 59.8 | 0.144 | −0.119 |
| Model | np | LnL | Estimates of Parameters | Model Compared | LRT p-Value | Positive Sites | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model A | 83 | −169886.61 | Site class | 0 | 1 | 2a | 2b | Model A vs. Model A null | 0.000213429 | 1595 L 0.973 *, 2308 T 0.955 *, 2479 S 0.984 *, 2632 L 0.950 *, 3272 T 0.956 * |
| f | 0.9212 | 0.073 | 0.0056 | 0.00045 | ||||||
| ω0 | 0.0427 | 1 | 0.043 | 1 | ||||||
| Model A null | 82 | −169893.47 | ω1 | 0.0427 | 1 | 149.39 | 149.39 | Not Allowed | ||
| 1 | ||||||||||
| Genes | Postive Selection Sites | Amino Acids | BEB Value | Feature Key | Description | |
|---|---|---|---|---|---|---|
| Foreground | Background | |||||
| Cytb | 316 | T/V/S | L | 0.973 * | Domain | CYTB_CTER |
| ND3 | 85 | V/A/S | T/A/S | 0.955 * | / | / |
| ND4 | 47 | N | S/P/A | 0.984 * | Domain | Oxidored_q5_N |
| 200 | T/M | L/V/M | 0.950 * | Domain | Proton_antipo_M | |
| ND5 | 400 | V | T/A/S/V | 0.956 * | Domain | Proton_antipo_M |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.-H.; Huang, H.-M.; Wu, L.; Storey, K.B.; Zhang, J.-Y.; Zhang, Y.-P.; Yu, D.-N. Characterization of Two Mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with Phylogenetic Relationships and Selection Pressure Analyses of Hylidae. Animals 2023, 13, 1593. https://doi.org/10.3390/ani13101593
Hong Y-H, Huang H-M, Wu L, Storey KB, Zhang J-Y, Zhang Y-P, Yu D-N. Characterization of Two Mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with Phylogenetic Relationships and Selection Pressure Analyses of Hylidae. Animals. 2023; 13(10):1593. https://doi.org/10.3390/ani13101593
Chicago/Turabian StyleHong, Yue-Huan, Hai-Ming Huang, Lian Wu, Kenneth B. Storey, Jia-Yong Zhang, Yong-Pu Zhang, and Dan-Na Yu. 2023. "Characterization of Two Mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with Phylogenetic Relationships and Selection Pressure Analyses of Hylidae" Animals 13, no. 10: 1593. https://doi.org/10.3390/ani13101593
APA StyleHong, Y.-H., Huang, H.-M., Wu, L., Storey, K. B., Zhang, J.-Y., Zhang, Y.-P., & Yu, D.-N. (2023). Characterization of Two Mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with Phylogenetic Relationships and Selection Pressure Analyses of Hylidae. Animals, 13(10), 1593. https://doi.org/10.3390/ani13101593






