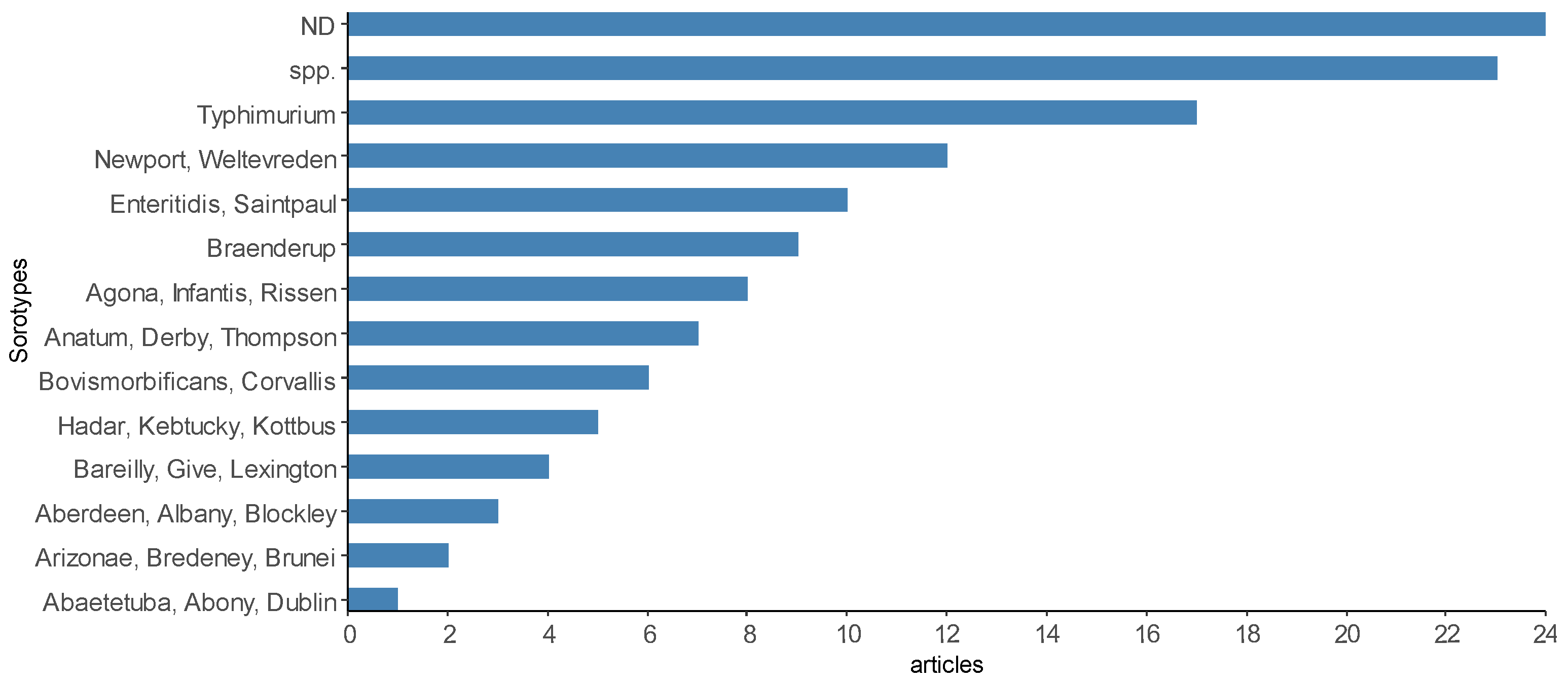Simple Summary
Salmonellosis is characterized by a gastrointestinal infection resulting from the ingestion of water and food contaminated by bacteria of the genus Salmonella, of which causes enterocolitis. Although infections are acute and self-limiting, efforts to prevent this problem of interest to global public health are important. Salmonella spp. is distributed in different environments and animal species; therefore, meat derived from animal production is one of the main routes of human infection, increasing the importance of research focusing on microbiological quality and food safety. Aquaculture is a constantly growing sector in the world, and the monitoring of Salmonella spp. in fish products is important for public health due to the risks of contamination during all stages of production. In this context, the present study carried out a systematic integrative review of the microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020 with the objective of characterizing and contributing to the promotion of measures to control and prevent this pathogen in aquaculture production. A database generated was composed of information that was mined from articles such as the most sampled aquaculture species, the microbiological diagnostic method(s) conducted in the investigation of Salmonella spp., and the main reported serotypes.
Abstract
The present study aimed to characterize, through descriptive statistics, data from scientific articles selected in a systematic integrative review that performed a microbiological diagnosis of Salmonella spp. in aquaculture. Data were obtained from research articles published in the BVS, Scielo, Science Direct, Scopus and Web of Science databases. The selected studies were published between 2000 and 2020 on samples of aquaculture animal production (fish, shrimp, bivalve mollusks, and other crustaceans) and environmental samples of aquaculture activity (farming water, soil, and sediments). After applying the exclusion criteria, 80 articles were selected. Data such as country of origin, categories of fish investigated, methods of microbiological diagnosis of Salmonella spp., sample units analyzed and most reported serovars were mined. A textual analysis of the word cloud and by similarity and descending hierarchical classification with the application of Reinert’s algorithm was performed using R® and Iramuteq® software. The results showed that a higher percentage of the selected articles came from Asian countries (38.75%). Fish was the most sampled category, and the units of analysis of the culture water, muscle and intestine were more positive. The culture isolation method is the most widespread, supported by more accurate techniques such as PCR. The most prevalent Salmonella serovars reported were S. Typhimurium, S. Weltevreden and S. Newport. The textual analysis showed a strong association of the terms “Salmonella”, “fish” and “water”, and the highest hierarchical class grouped 25.4% of the associated text segments, such as “aquaculture”, “food” and “public health”. The information produced characterizes the occurrence of Salmonella spp. in the aquaculture sector, providing an overview of recent years. Future research focusing on strategies for the control and prevention of Salmonella spp. in fish production are necessary and should be encouraged.
Keywords:
Salmonella; fish farming; food safety; public health; word cloud; similarity; Reinert’s algorithm 1. Introduction
The genus Salmonella spp. belongs to the Enterobacteriaceae family and has important metabolic characteristics that not only promote its survival in the gastrointestinal tract environment but also have virulence mechanisms to escape defense cells, reproduce and cause homeostatic disturbance in the host [1,2]. Salmonella spp. causes salmonellosis, a classic infection characterized by enterocolitis, which in most cases is acute and self-limiting, making diagnoses difficult and resulting in underreporting and compromising actions to control and prevent new outbreaks.
Salmonellosis is a foodborne disease (FDA) and represents an important public health problem in several countries around the world [3]. Nontyphoid serovars of the genus Salmonella spp. are responsible for much of the incidence of foodborne outbreaks, with a variety of foods serving as vehicles for the occurrence of salmonellosis in humans [3,4,5]. Such foods include meat (beef, chicken, pork and fish), eggs, milk, cheese, fresh fruits, fruit juices, vegetables and fish [5,6,7].
Salmonella spp. is widely distributed in the environment and in several species of wild animals, production animals and pets. Mammals, fish, birds, reptiles, amphibians and plants can act as reservoirs and disseminators of Salmonella spp. [8,9,10]. However, the adoption of correct and strategic measures in livestock management can help to prevent and reduce the risk of contamination by Salmonella spp., since its spread can occur at several stages along the production chain, including all stages of production, processing, distribution, marketing and handling/preparation [11,12,13,14].
Salmonella spp. has been isolated from points in the fish production chain [1,15]. There is a consensus among researchers that Salmonella spp. does not naturally belong to the aquatic environment, although it is isolated from water, fish and products derived from aquaculture [15,16,17,18]. The contamination of fish by Salmonella spp. is little known, since research has reported that fish act as a host of the bacteria for relatively short periods of time without any description of symptomatic manifestation of the disease [15,16,17,18,19]. Thus, Salmonella spp. has been isolated not only from the viscera but also from the gills and skin of the fish, contributing to an increased risk of cross-contamination during handling, processing, storage and commercialization due to failures in hygienic-sanitary care or using equipment, surfaces and utensils that are inadequately sanitized during the production chain [12,20,21,22].
It is notable that the health of fish depends on the quality of the water. In this way, the chemical, physical and microbiological factors of water are extremely important. Among the possible sources of contamination, water appears to be a probable vehicle, as it is widely exposed to human, agricultural and industrial pollution [23,24,25]. In addition, the water used for aquaculture itself can be a source of contamination. Research has been carried out to assess the presence of Salmonella spp. in surface water and sea water and the subsequent contamination of fish products and aquaculture [11,26]. Fresh water is often contaminated by Salmonella spp. through effluents, and consequently, coastal waters; shellfish and fish farming areas are especially subject to this pathogen [26,27,28]. Even if the presence of Salmonella spp. in aquaculture production species is reported with low prevalence, it is a pathogen of public health interest, and the detection techniques in the prevalence must be observed, tested and improved.
Methods for the rapid and accurate identification of pathogens in the production chain are important for ensuring food quality and for taking control measures through traceability to minimize economic losses and damage to public health. The health risks associated with the consumption of low-quality aquaculture foods make the evaluation and control of food safety topics a global concern [1,7,29]. Microbiological controls must be addressed in designated aquaculture environments for proper production management practices and for consumer education programs.
In this context, an integrative review of investigations of Salmonella spp. in aquaculture between 2000 and 2020 was decided to be performed, with the influence of the systematic review model already observed in other reviews, as it systematizes the research procedures, making the method explicit and a more auditable and reproducible model to facilitate the consolidation of knowledge [30,31]. The selected articles aimed to characterize and produce information through descriptive statistics techniques and text mining [32,33]—this refers to mined data regarding the years of publication, country of origin, area of study, categories of most sampled aquaculture species, sampling origin, methods of microbiological diagnosis of Salmonella spp., most evaluated units of analysis and most identified Salmonella serovars to contribute to the control and prevention of Salmonella spp. in fish production. The textual analysis of the abstracts was performed with word cloud grouping of similarity and descending hierarchical classification with the application of Reinert’s algorithm [34,35]; this was performed to make it possible to identify, organize and classify the most predominant different themes addressed in scientific articles for a better interpretation of the addressed themes in investigations into the microbiological diagnoses of Salmonella spp. in aquaculture.
2. Materials and Methods
2.1. Data Sources and Research Strategy
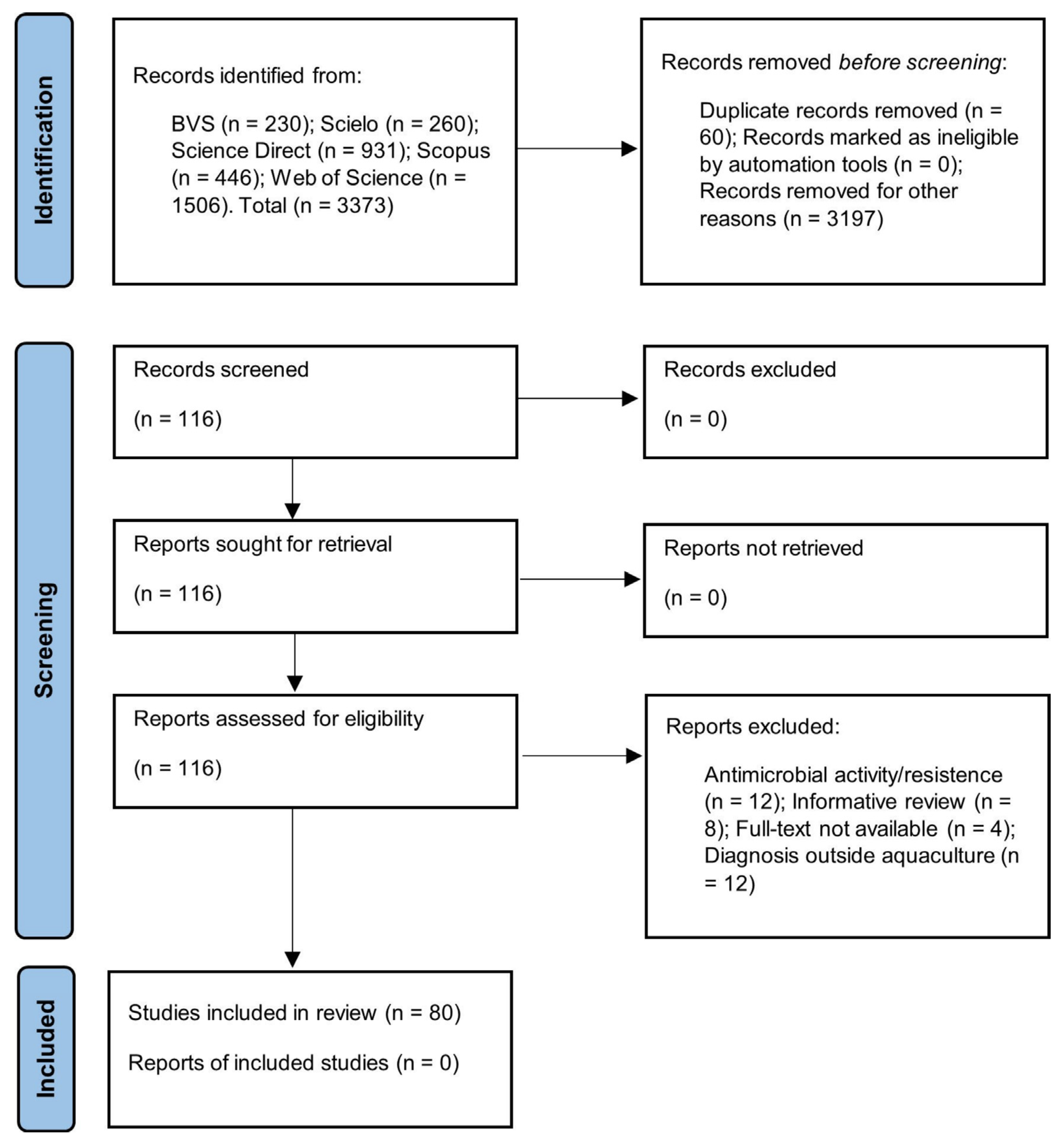
The searches performed in the electronic bibliographic databases BVS, Scielo, Science Direct, Scopus and Web of Science were restricted to the key terms: (fish farming) AND (Salmonella) OR (salmonellosis); the period between 2000 and 2020; English and Portuguese; original article formats. A total of 3373 articles were identified (Figure 1).
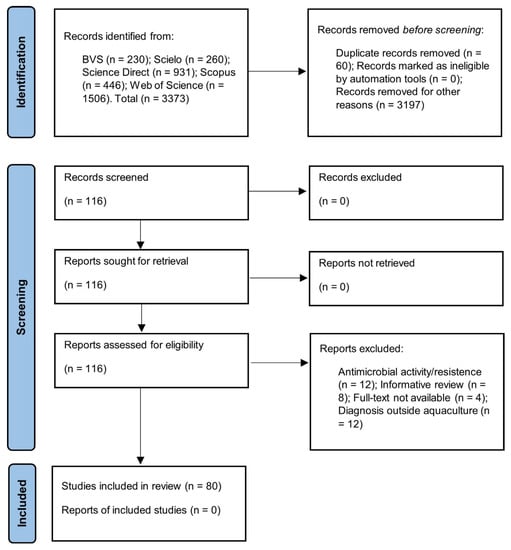
Figure 1.
Strategy for the selection of eligible articles.
2.2. Research Design
The integrative review was based on the steps established by the protocol for Systematic Reviews and Meta-Analyses (PRISMA) [36]. All the steps and strategies for selecting articles are detailed in Figure 1.
2.3. Selection Process
Two reviewers (YDP and AOA) performed the selection of possibly relevant articles by critically reading the titles and abstracts. Thus, 3197 articles were excluded, as they were considered to have no direct relationship with the subject studied. Zotero® Reference Manager version 5.0.96.2. identified 60 duplicate articles that were excluded. A total of 116 articles were retrieved and selected. All disagreements were resolved by consensus.
2.4. Eligibility Criteria
Primary studies tested for Salmonella spp. with reports of positive or negative contamination from samples of species produced in aquaculture, environmental samples (culture water, soil and sediments) from the breeding phase or as a product and derivatives for sale in markets. Therefore, in the screening stage, the studies focused only on the resistance or activity of Salmonella spp. against antimicrobials (12), informative reviews (8), diagnosis of Salmonella spp. in water and other environmental samples not related to aquaculture (12) and studies without access to the full document (4). Thus, 80 productions were chosen for this review.
2.5. Abstract and Data Analysis
Two reviewers (YDP and WST) performed a critical analysis of the contents of the articles that were chosen for data extraction. Two reviewers (WST and AOA) verified the data consistency.
The information extracted was the year of publication, authors, research title, name of the published journal and country. Information such as sample origin, species studied in the investigation, sample rate(s) tested, sample size tested, diagnostic assay(s) performed, number of positive samples and serotypes were also extracted. Some information has been categorized for better interpretation.
All data analysis was performed using the R® statistical package version 4.0.2 (22 June 2020) (R Foundation for Statistical Computing, Vienna, Austria) [37]. The libraries used were “tm” and “SnowballC” for text mining and text stemming [38,39], the “word cloud” generator [40], and the world map generator to analyze the frequency of productions by country [41]. The Iramuteq® program (Interface de R pour les Analyses Multidimensionneles de Textes et de Questionnaires) (LERASS—Laboratoire d’Études et de Recherches Appliquées en Sciences Sociales, Toulouse, France), developed in R®, was used for the analysis of similarity and descending hierarchical classification (DHC) with the application of Reinert’s algorithm of the content of abstracts [34,35].
2.6. Textual Analysis
The use of classical techniques of exploratory data analysis [32] and analytical techniques of text mining [33] for the analysis of unstructured data identified patterns that characterized the production from the perspective of the presence of Salmonella spp. in aquaculture.
A database was generated containing the abstracts of the articles selected in the integrative review, generating a corpus with 17,142 words, in which a word cloud composed of the most frequent words, a similarity graph and a descending hierarchical classification (DHC) graph were produced. From the similarity analysis, it was possible to identify the intensity of the occurrence of the words and the indications of the connectedness between them. The similarity graph presents a tree structure with ramifications; therefore, the content of the selected abstracts can be characterized by the identification of the most used words and the proximity between them. To generate the graphic image that was more visually readable, cleaning was performed by cutting out some words of less or no importance with the context and selecting the terms whose frequencies were equal to or greater than 20. Finally, the descending hierarchical classification technique was applied (DHC) to generate a graph containing the textual corpus of the abstracts divided by groupings of text segments according to the similarity of the terms used. The objective of the entire textual analysis of the abstracts was to enable the identification of the most predominant distinct themes addressed in the scientific articles, organize them, and thus classify them for a better interpretation of the themes addressed in investigations on the microbiological diagnoses of Salmonella spp. in aquaculture.
3. Results
3.1. Number of Articles, Study Themes, Textual Analysis and Countries
The present review identified 3373 scientific articles through the search strategy adopted, where 80 research articles were chosen for samples investigated for the presence or absence of Salmonella spp. in the aquaculture production chain (Figure 1).
The articles were published by 47 different journals to address and disseminate the results obtained with a strong multidisciplinary character on Salmonella spp. as a contaminating pathogen in aquaculture. A comprehensive table summarizing all the essential information on the origin of studies, samples and diagnostic techniques used in each of the 80 included studies is available as Supplementary Material (Table S1).
It was possible to categorize seven main areas of study that the authors used to disseminate their research, emphasizing the importance of surveillance of Salmonella spp. in food produced by aquaculture, and the interest in a microbiological health problem that touches different fields of knowledge. That is, it is considered an essentially multidisciplinary topic that can cover areas related to food science (38 articles), animal production (12 articles), environment (11 articles), microbiology (6 articles), veterinary (5 articles), health (2 articles) and multiple areas (6 articles).
Following the idea of a multidisciplinary approach, the topics of articles involving Salmonella spp. presented proposals that are diverse in objectives and studies. From the abstracts of the articles selected by the integrative review, it was possible to generate the word cloud (Figure 2) and the similarity graph (Figure 3) for textual analysis.
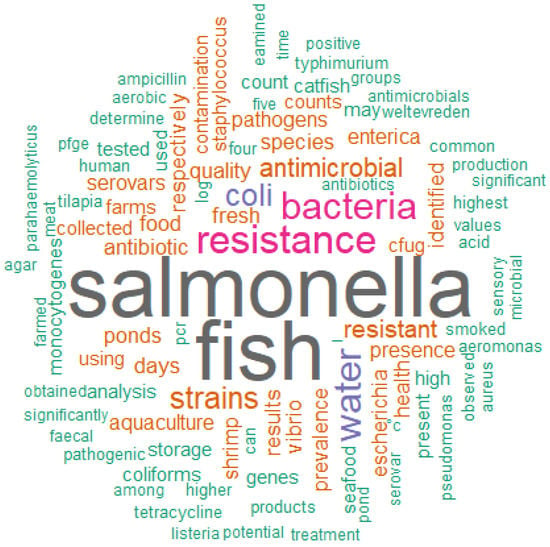
Figure 2.
Word cloud formed from the abstracts of the articles selected by the integrative review on microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020. N total of words = 17142.
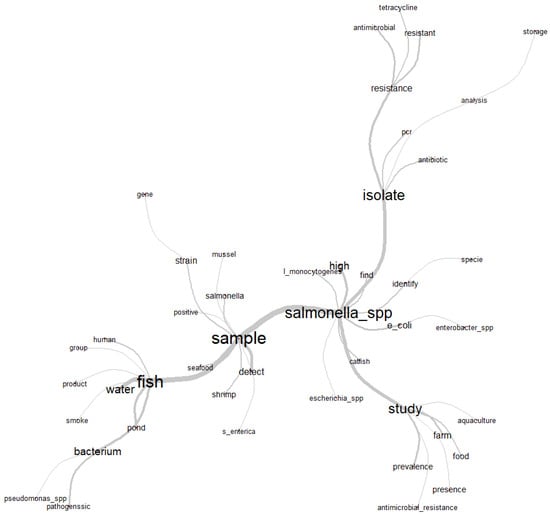
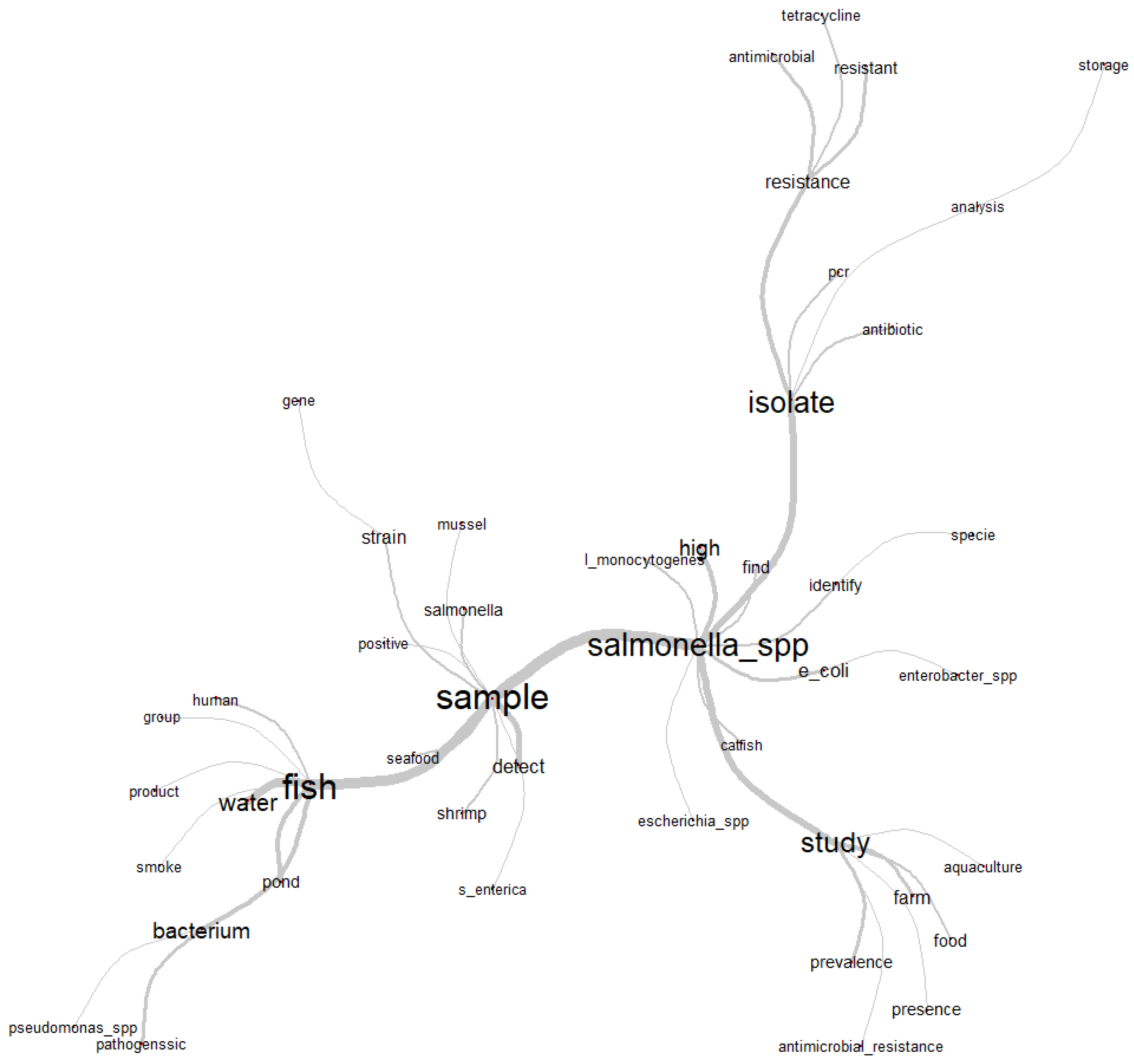
Figure 3.
Similarity chart with the relationship of the most used words in the abstracts of the articles selected by the integrative review on microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020.
The word cloud (Figure 2) was formed by the most frequent words contained in the abstracts. The higher the frequency of the word, the larger the font size of the word represented in the cloud. In this analysis, the five most prominent words in decreasing order of frequency (ni) were “Salmonella” (212), “fish” (207), “resistance” (85), “bacteria” (80) and “water” (79), while the other words contained in the cloud had a frequency (ni) lower than 64. The greater use of these most prominent words indicates that the selected articles emphasized Salmonella spp. (“Salmonella”) as an important pathogen kept under surveillance and microbiological investigation in the aquaculture production chain. In this sense, the word “bacteria” can generally support interest in microbiological safety and safe food for products from aquaculture. The “fish” represented the category of fish most affected or of greatest interest for microbiological surveillance against Salmonella spp. that would pose risks of causing a DTA. The words “water” and “resistance” can represent, respectively, the vehicle that most contributes to the spread and silent contamination of the pathogen in the breeding phase and the concern about the characterization of the isolates regarding the susceptibility or resistance profile to antimicrobials.
For similarity analysis, the composition of the abstracts of the articles generated a graph (Figure 3) with the most frequent central words (ni) “fish” (166), “sample” (151), “isolate” (137), “Salmonella” (126) and “study” (95). The branches make links with other words that were often mentioned simultaneously in the abstracts, and therefore, their relationship with the respective central words is observed. In this way, it was possible to contextualize and understand how the theme of Salmonella spp. in aquaculture was found to be structured in the articles selected by the integrative review.
The co-occurrence of the words “fish”, “water”, “pond” and “bacterium” point to freshwater fish and water, respectively, as the category of fish and the medium that most favors microbiological dispersion; these were most frequently used and analyzed words for the microbiological diagnoses of Salmonella spp. The use of the word “sample” was frequently followed by words that denote the authors’ reports on the investigation, detection and identification of strains (“strain”) or species of Salmonella (“S. enterica”) in several samples of aquaculture species, characterized by the terms “seafood”, “shrimp” and “mussel”.
Next to the word “Salmonella_spp.” are “Escherichia coli” and “Enterobacter spp.”, representing the authors’ interest and reports in the investigation and microbiological diagnosis of other important bacterial pathogens belonging to the Enterobacteriaceae family in aquaculture species. Associated in this same context is the word “Listeria monocytogenes”, as this is another foodborne bacterial pathogen that causes serious problems of public health importance.
The minor ramifications associated with the term “study” characterize in a more detailed way the directions taken by the authors in relation to the object of investigation (Salmonella), as “aquaculture”, “farm” and “food” are referred to as the primary source of the investigation from livestock and food production, which together with the terms “prevalence” and “presence” reveal worrying results regarding food microbiological safety. This statement can be more evident when associated with the term “antimicrobial_resistance”, as this indicates that research has been carried out regarding the control of a chronic problem of contamination. In this way, the term “isolate” is related to biomolecular techniques based on the polymerase chain reaction (“PCR”) used in diagnostics, as well as bacterial isolates with an antimicrobial resistance profile or those submitted from susceptibility or resistance analysis to groups of antibiotics. “Tetracycline” was the most reported by the authors.
To help identify the themes addressed in the selected articles, and thus verify the privileged lines of studies, a graph was generated using the descending hierarchical classification (DHC) technique. Thus, five different classes or lines of study were obtained by grouping text segments with the greatest similarity in terms of the words used. Figure 4 and Figure 5 present the DHC results of the abstracts included in this review. The most representative class contained 25.4% of the text segments. The least representative class was only 13.9%.
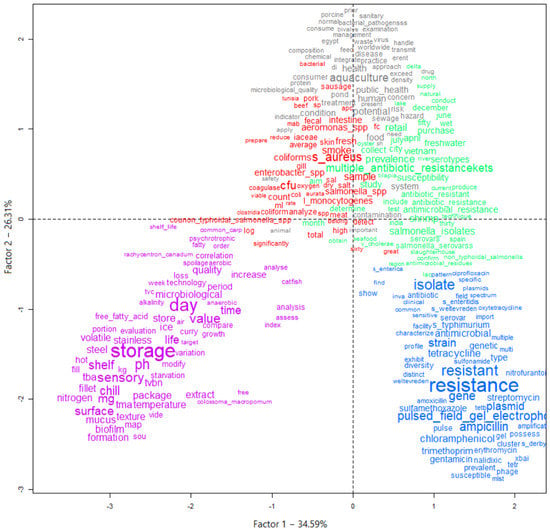
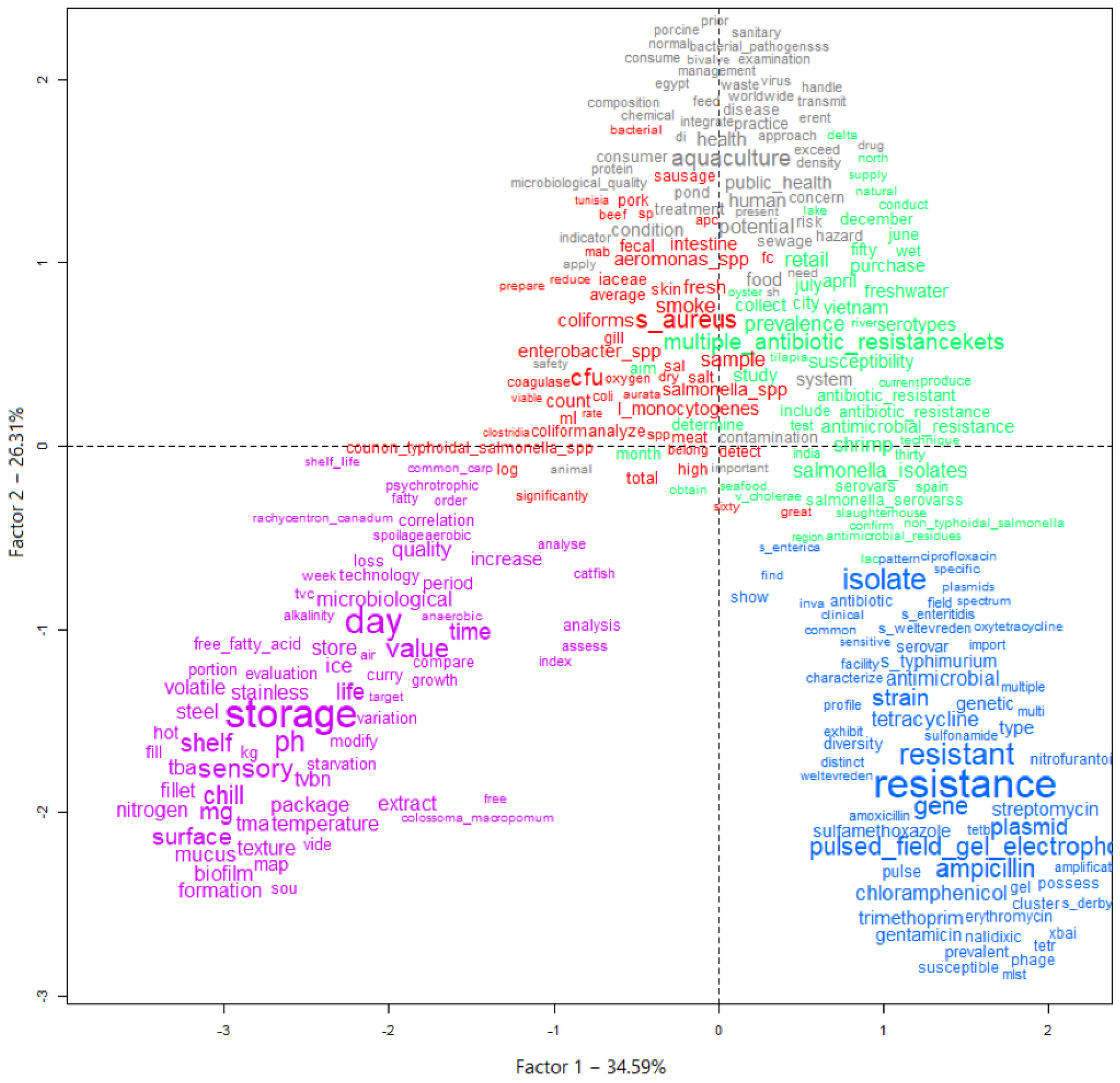
Figure 4.
Graph generated by the descending hierarchical classification technique (DHC) containing the textual corpus of abstracts of articles selected by the integrative review on microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020, divided by groupings of text segments according to the similarity of terms. Legend: Class 1 = Microorganisms (red); Class 2 = One health (gray); Class 3 = Profiles (green); Class 4 = Antimicrobials (blue); Class 5 = Food safety (purple).
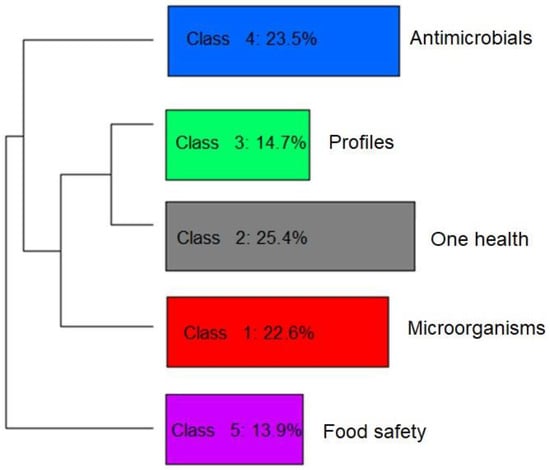
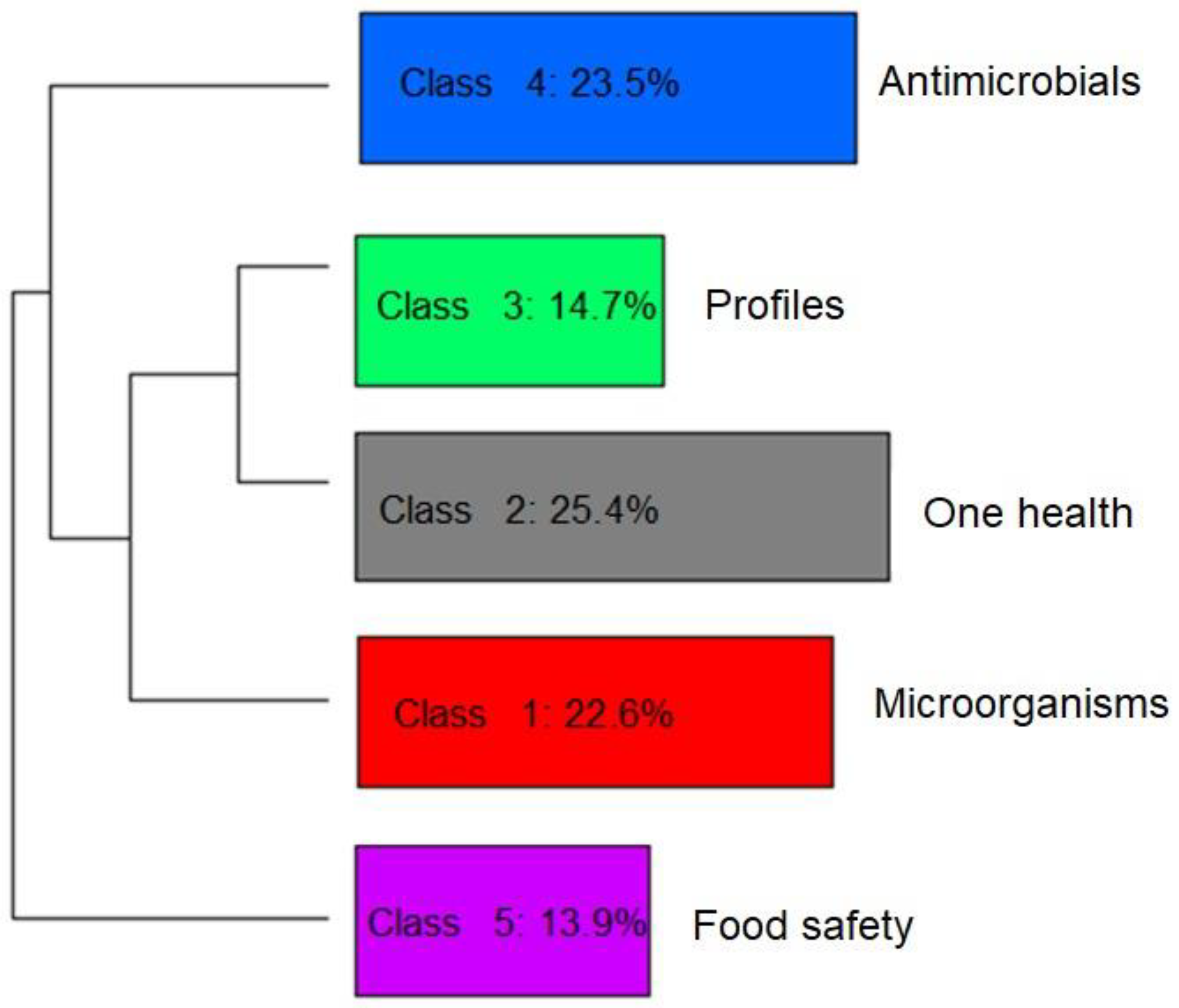
Figure 5.
Hierarchical representation and classification of the five groups formed by dividing the textual corpus of the abstracts of the articles selected by the integrative review on microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020, according to the similarity of terms.
In perspective, Figure 4 and Figure 5 provide a visualization of the terms in each class performed in the two-dimensional space graph so that it is possible to evaluate the composition of each class based on the positioning and intensity and on the size of the words. Note that while classes 4 and 5 are farther apart, each in a different quadrant of the plane, the others show some level of overlap (see Figure 4), which indicates that they share common contents. Based on the frequency of occurrence of terms in each class, it was possible to assign a name that summarizes the general meaning of each grouping of texts (Figure 5).
It was possible to verify that class 5 was separated from the others, which implies a greater differentiation of its content in relation to the other classes (Figure 5). Another division occurred with the separation of class 4, then further division into the three remaining classes occurred based on great similarity with each other.
Class 5, words such as “storage”, “sensory”, “shelf” and “quality” (Figure 4), stood out. This class was named “food safety”, containing terms that mention the period that the food is guaranteed to be free from pathogens or the action of spoiling microorganisms that would modify the quality, making it unfit for consumption. Further and different from class 5 is class 4, which was named “antimicrobials”, where the most frequent terms were “resistance”, “resistant”, “ampicillin” along with several other terms that are names of other antimicrobials. This greater differentiation occurred due to the studies focusing on testing how the isolated pathogens interact with the action of different antimicrobial agents.
Class 1 (“microorganisms”), class 2 (“one health”) and class 3 (“profiles”) have marked similarity to each other. The term “aquaculture” (cluster 2) is found together with the terms “food” and “public health”, indicating the importance of this production activity for human food in a sustainable way through the practice of cultivation, together with collective health policies. Terms referring to other microbiological hazards to be prevented in foods from aquaculture are found in class 1, for example, “Staphylococcus aureus”, “Aeromonas spp.” and “Salmonella spp.”, indicating other pathogens that are potentially investigated and isolated in research in different types of samples. Otherwise, class 3 grouped terms such as “multiple antibiotic resistance”, “prevalence” and “susceptibility”, indicating that it is representative of terms that refer to the occurrence and microbiological profile of the investigated isolates.
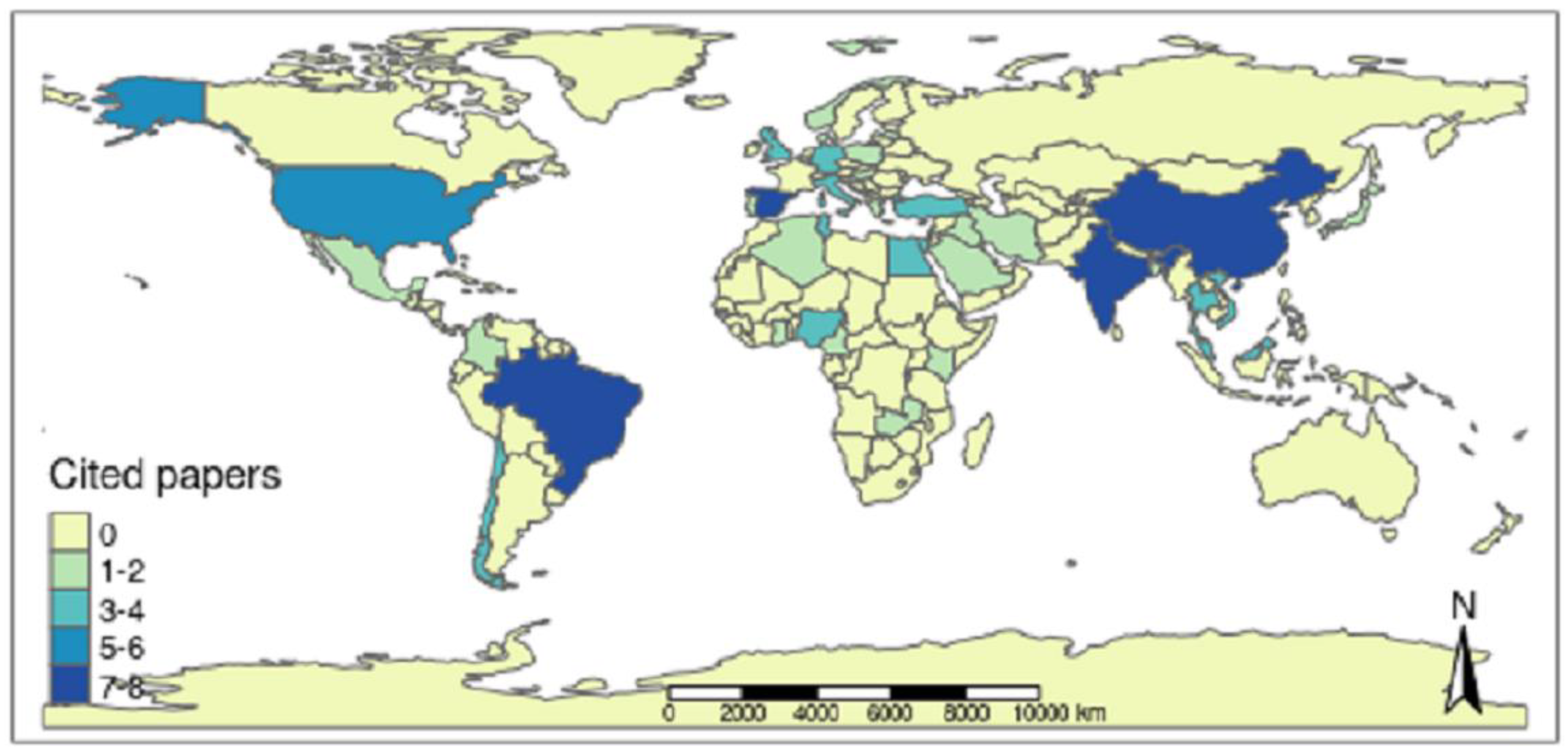
Aquaculture producers from 37 countries located on all continents, except for Oceania, gave rise to the scientific productions chosen by the integrative review (Figure 6). Therefore, it was possible to observe continuous surveillance of Salmonella spp. in the aquaculture sector through the publications analyzed over the last few years.
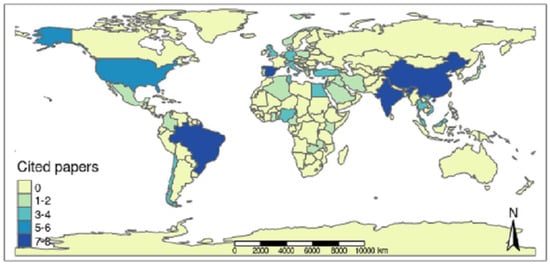
Figure 6.
Frequency of scientific articles selected in the integrative review on microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020 by countries in different continents.
Among all the nations, Brazil was the country with the highest number of articles selected by the integrative review, representing 10% of the selected articles (8 articles). The countries of the Asian continent accounted for 38.75% of the publications (31 articles), and China [17,25,42,43,44,45,46], India [47,48,49,50,51,52,53], Malaysia [54,55,56], Vietnam [57,58,59] and Thailand [60,61] had the highest number of elected publications. The countries of the European continent contributed 25% (20 articles), the second highest percentage of selected publications, with Spain [27,62,63,64,65,66], Italy [67,68,69] and Germany [70,71] having greater prominence. On the American continent, with a percentage of 21.25% (17 articles), the largest number of selected publications is in Brazil [9,18,23,72,73,74,75,76], followed by the USA [14,77,78,79,80] and Chile [81,82], while on the African continent with 15% (12 articles), Egypt [83,84,85], Nigeria [12,86], Tunisia [87,88], Kenya [89,90] and Turkey [91,92] had the highest number of selected works. The nations that had 1 publication elected were Algeria [21], Bangladesh [93], Belgium [94], Cameroon [95], Colombia [96], Denmark [97], England [98], Ghana [28], Greece [99], Hungary [100], Iran [13], Iraq [101], Japan [102], Latvia [103], Lebanon [104], Mexico [105], Norway [106], Poland [107], Portugal [22], Saudi Arabia [108], and Zambia [109].
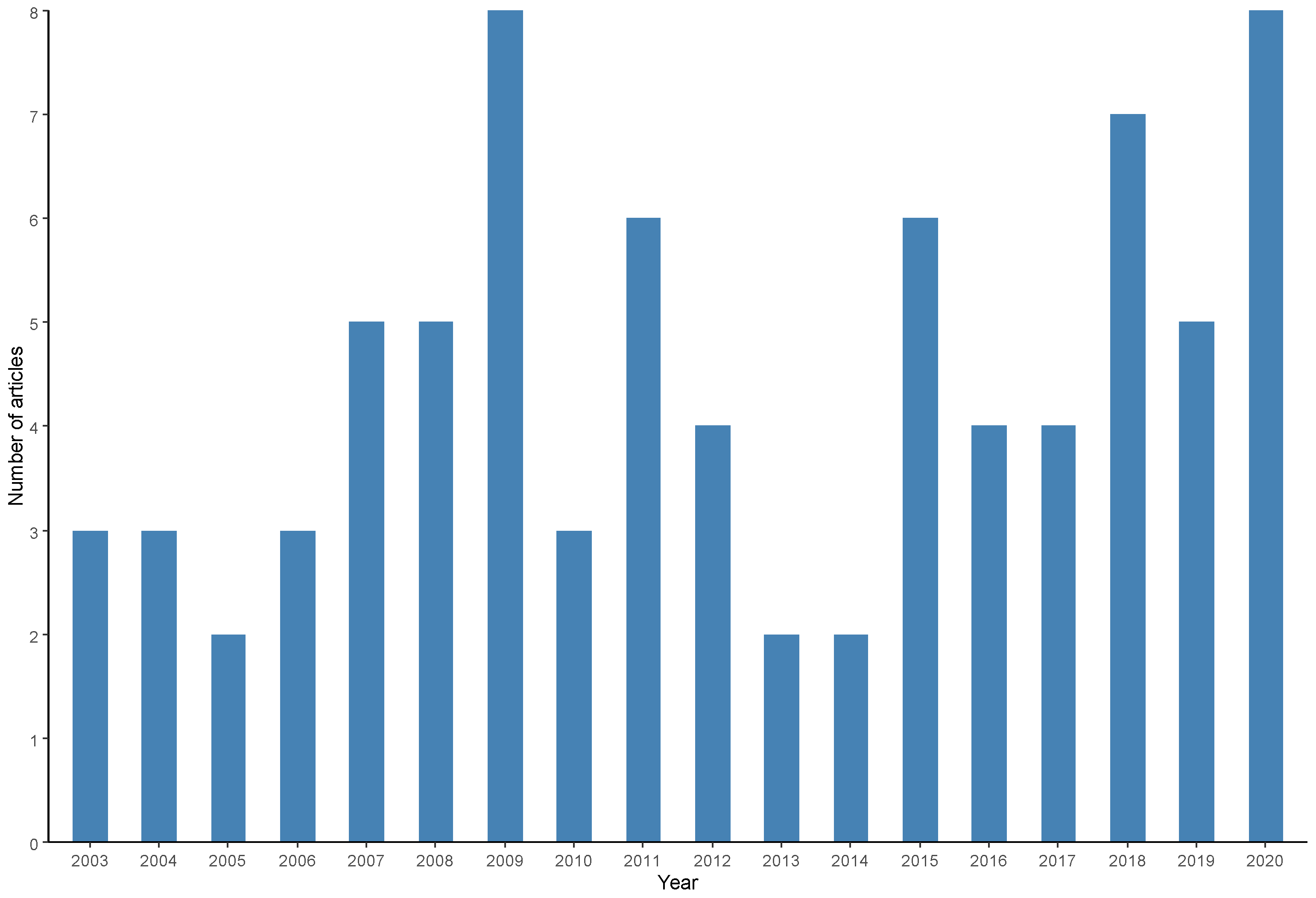
The frequency of scientific publications between 2000 and 2020 was verified (Figure 7). An increase in the number of publications was observed until 2009 and from 2016 onward; however, no general trends of increase or decrease in production were observed during the entire period.
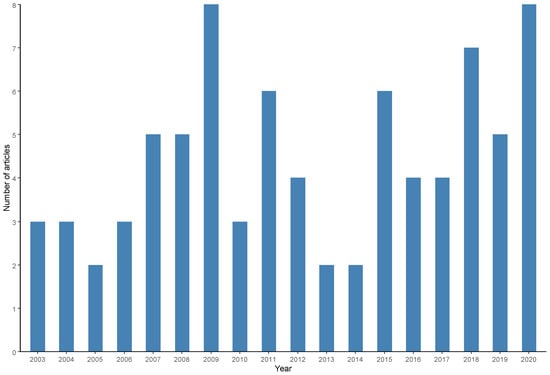
Figure 7.
Frequency of scientific articles on microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020 by year of publication.
Articles published over a longer period of time within the coverage period adopted in the systematic search were published by Ghana [28], Japan [102] and the USA [14] in 2003, while the most recent productions were published by India [50,51], Bangladesh [93], China [44], England [98], Iraq [101], Spain [66] and Vietnam [59]. In 2020, the year with the highest number of publications (8 articles) was 2009, followed by 2018 (7 articles), 2015 and 2011 (6 articles), and 2019, 2008 and 2007 (5 articles). There were no scientific productions selected for this review in the period between 2000 and 2002.
3.2. Sampling and Studied Species
Different frequencies were observed regarding origin, habitat, fish species and environmental samples collected as units of analysis in the articles. In general, these variations occurred due to the different objectives carried out in research on Salmonella spp. regarding microbiological investigation and safety of these foods.
Most of the articles collected samples from aquaculture (54 articles) developed in fresh water (44 articles), indicating greater interest in scientific research in the surveillance of Salmonella spp. about this aquaculture sector.
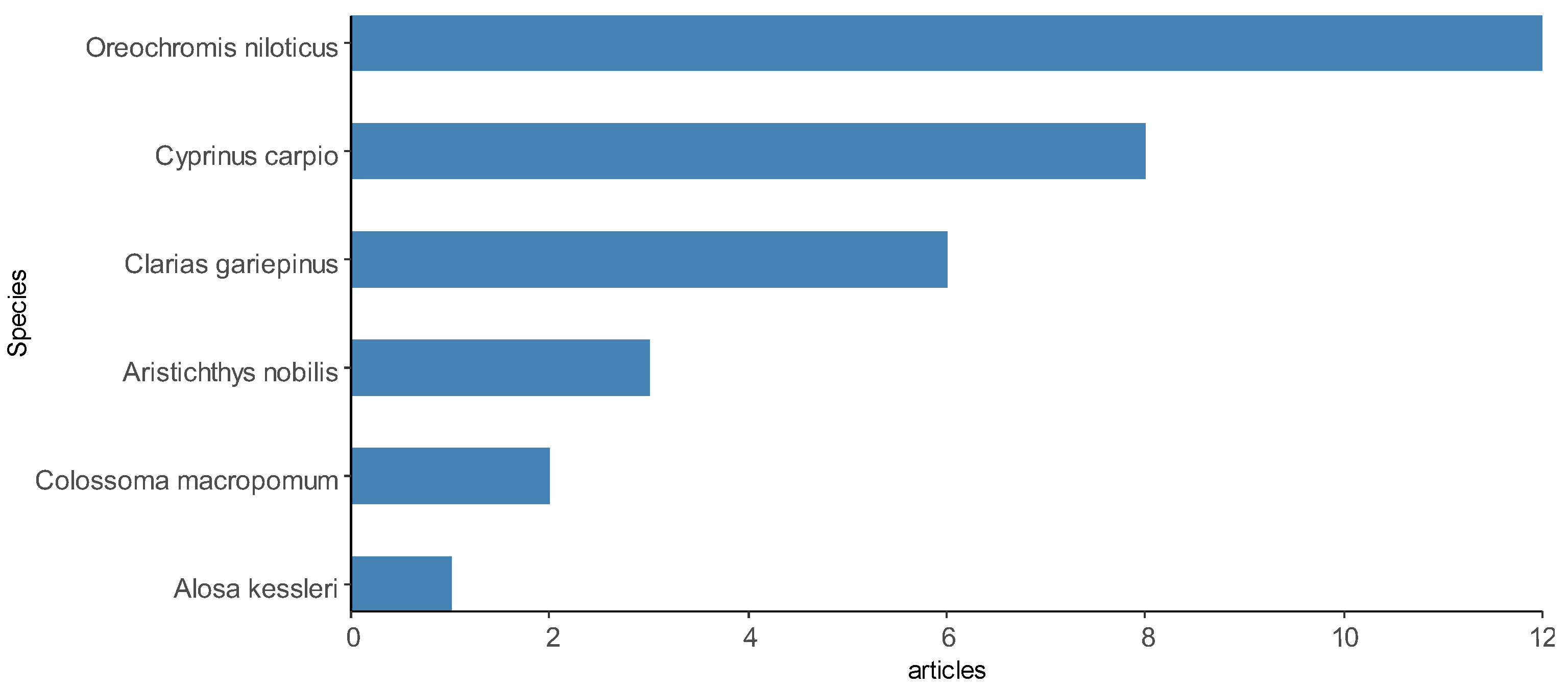
In total, 192 animal species were investigated in the studies included here, including 136 representatives of fish (76 species identified), 20 representatives of shrimp (8 species identified) and 36 representatives of bivalve mollusks, other crustaceans and seafood (18 species identified). Therefore, fish was the category of aquaculture species that was most used for investigations of Salmonella spp. in the selected articles, appearing in at least 58 articles, 31 of which investigated only fish samples. Shrimp was present in 12 articles, while other species of crustaceans and bivalve mollusks were present in 16 articles. Figure 8 demonstrates some of the most reported species in the articles.
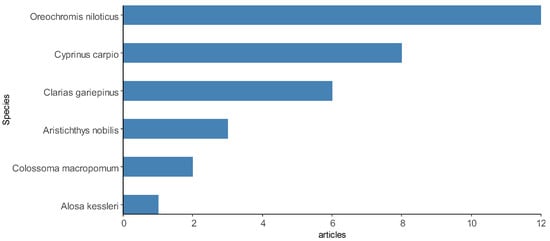
Figure 8.
Some aquaculture species most sampled by the articles selected in the integrative review on the microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020.
Among the fish species are Oreochromis niloticus [23], Cyprinus carpio [101], Salmo salar [82,106], Sparus aurata [62], Catla catla [44], Colossoma macropomum [72], Rachycentron canadum [53], Dicentrarchus labrax [66], Trachurus trachurus [69] and Oncorhynchus mykiss [69]. The shrimp species were Litopenaeus vannamei [58], Paeneus monodon and P. vannamei [61]. Other species of crustaceans and bivalve mollusks reported were Mytilus edulis [71,97], Midye Dolma [92], M. galloprovincialis, Venerupis pullastra, Ruditapes philippinarum, Dosinia exolete and Cerastoderma sp. [27]. Fewer scientific articles have focused on Salmonella spp. only in environmental samples of water [25,74,83,109] and sediments [108]. A description of the animal species investigated in the selected scientific articles can be found in Supplementary Material Table S1.
3.3. Research Methodologies and Analyzed Aliquots
All studies used microbiological culture methodologies for the investigation and isolation of Salmonella spp., of which 21 used the PCR technique and 1 used the qPCR technique [25] concomitantly with culture.
In general, in the diagnostic techniques applied, tests were performed to determine colony characteristics, morphology of the isolates, Gram stain reaction, indole test, methyl red and Voges–Proskauer tests, use of citrate, oxidase test, cell motility, catalase, hydrogen sulfide production, sugar utilization, nitrate reduction, gelatin hydrolysis, starch hydrolysis and test reading.
Some of the studies (17) reported the presence of Salmonella spp. according to standard method ISO 6579 (pre-enrichment in buffered peptone water, incubation at 41.5 °C for 24 h in BOD, enrichment in Rappaport–Vassiliadis (RVS), and with tetrathionate Muller-Kauffmann Novobiocin (MKTTn), incubated at 42 °C and 37 °C for 24 h in BOD, respectively, followed by isolation for typical red colonies with a black center and translucent with a red halo on xylose lysine deoxycholate (XLD) agar incubated at 37 °C for 24 h and later submitted to biochemical confirmations), while 13 studies reported the method recommended by the FDA’s Bacteriological Analytical Manual.
Other methodologies recommended by important bodies, such as the APHA (American Public Health Association) [13,75,85] and Association of Official Analyst’s Chemists (AOAC) [44,73,104], were also used by researchers in at least six studies.
Regarding the PCR techniques used in twenty-one studies, the main target genes were the invA gene [22,27,50,51,52,56,61,75,87] and the 16S rRNA gene [25,66,88,94,95,102]. Other targets, such as the rfb gene [104], hns and invE [52], and the fliB-fliA intergenic region [57], were also used.
The number of samples analyzed was quite heterogeneous. The publication with the lowest number of samples analyzed was 2 portions of fish curry [53], while the study with the highest number was 10,757 samples of water and live bivalve mollusks [68].
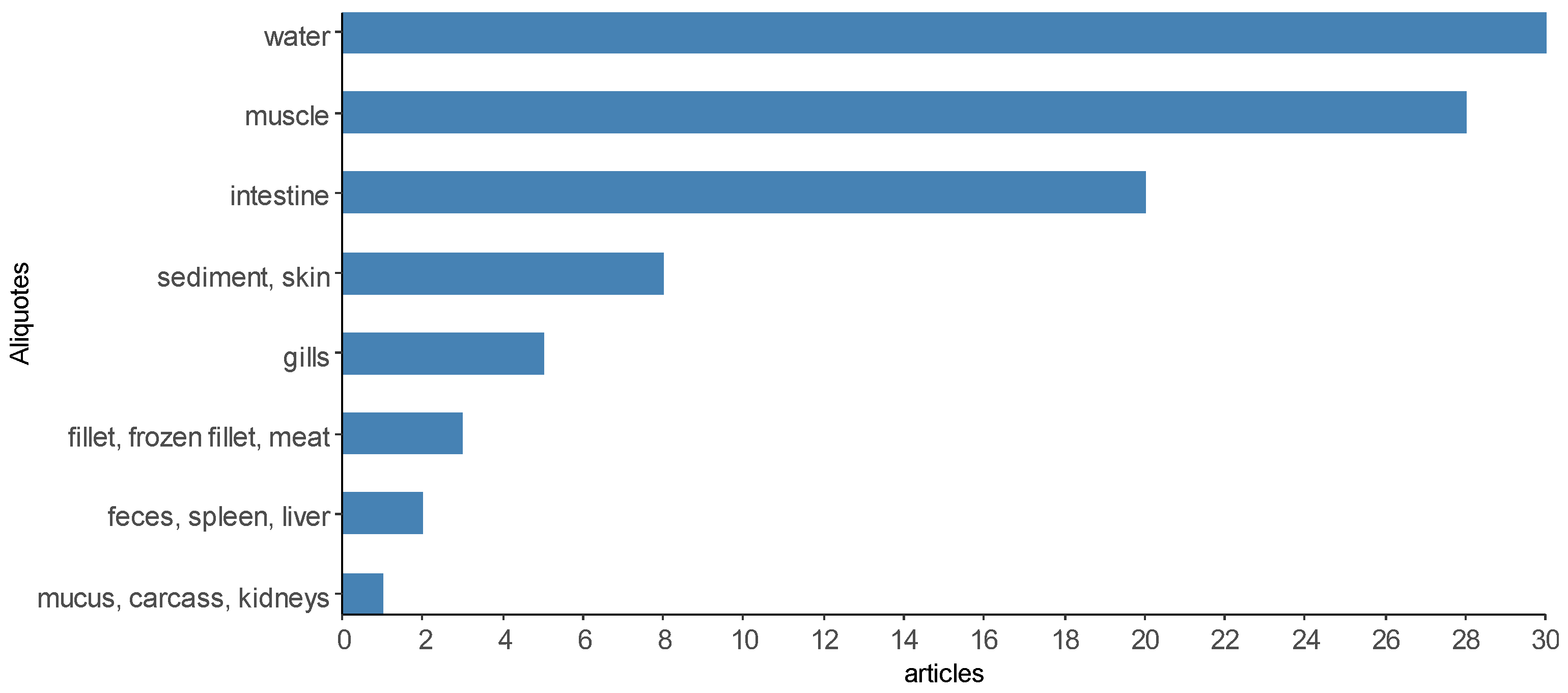
It was possible to verify that the articles used one to seven sample units for the microbiological analysis of Salmonella spp. (Figure 9), only two articles are above that [14,48]. To improve the presentation in Figure 9, the sampling units were categorized into “environment” when the articles performed microbiological analyses for Salmonella spp. In water, ice and sediment samples [9,17,25,28,66,85], “body” for the analysis of body parts such as shell, head, prawns, carapaces, gills, skin, mucus and surface swabs [43,56,59], “viscera” for the analysis of liver, kidneys, spleen, intestine, hepatopancreas, GI tissue [57,93,102,106], “tissue” for analysis of muscle, fillet (fresh, frozen, salted, smoked and vacuum-packed), meat, meat batter, carcass, blood and brain [70,72,73,82], “biofloc” [96] and “feces” [18,51]. A description of the sample rates analyzed in the selected scientific articles can be found in Supplementary Material Table S1.
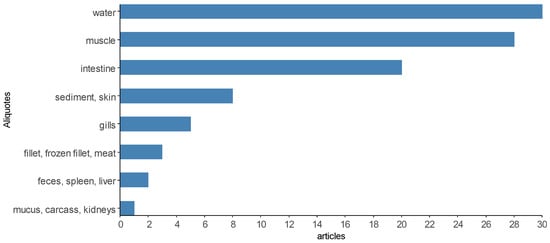
Figure 9.
Most analyzed sample aliquots in the scientific articles selected by the integrative review on the microbiological diagnoses of Salmonella spp. In aquaculture between 2000 and 2020.
3.4. Salmonella spp. Positive Reported
Detection of Salmonella spp. Was positive in 56 (70%) articles, while the remaining 24 (30%) were not detected. Among the positive samples for the presence of Salmonella spp., the lowest percentages of contamination detected were 0.93% and 1.43%, representing positivity for only a single sample [13,99]. The highest number of positives detected was 217 isolates (29.7%) [14]. Most of the “nondetectable” results are in investigations that use only one sample unit (14 articles).
The number of Salmonella serotypes identified in scientific articles was also heterogeneous (Figure 10), but some of the products (21 articles) reported isolation up to the level of gender. Only one Salmonella serotype was identified and reported in some scientific articles, such as S. Dublin [13], S. Enteritidis [89], S. Saintpaul [77], S. Senftenberg [65], S. Typhimurium [80], S. Corvallis [54] and S. Infantis [20], while the largest number of serotypes identified and reported in the same article was sixty-four [79]. The most prevalent serotypes found were S. Typhimurium investigated in fish and shrimp muscle [80,102], fish gills and intestine [56,86] and S. Weltevreden investigated in shrimp muscle and viscera [58], fish muscle [49,79], seafood [14,48], and shrimp and clam muscle [79]. A description of the main serotypes reported in the selected scientific articles can be found in Supplementary Material Table S1.
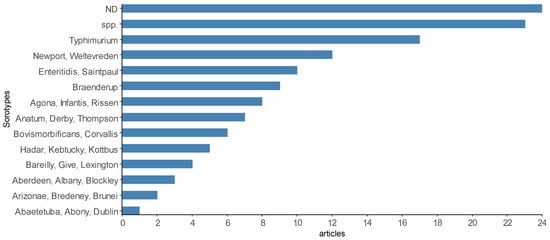
Figure 10.
Some of the most reported Salmonella serotypes by the scientific articles selected in the integrative review on the microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020. Legend: “ND”—undetected Salmonella; “spp.”—Salmonella spp. reported.
4. Discussion
Salmonellosis is a public health concern on a global scale. Through this integrative review, it was possible to demonstrate the continuous surveillance of Salmonella spp. in the aquaculture sector through the analyzed publications from different producing countries located on different continents over the years between 2000 and 2020 (Figure 6 and Figure 7).
In total, 80 articles were included in this study after eliminating those based on eligibility criteria, where the most representative countries in number of productions were Brazil, China, and India. However, not only countries considered to be major world aquaculture producers—such as China [17,43,45], Bangladesh [93], Chile [81], Egypt [83], India [49,51], Norway [106] and Vietnam [57] or even Brazil [18,72,74], which is in production expansion—but also countries with less relevance in activity have carried out surveillance of Salmonella spp. This pathogen has a negative impact not only on public health, since salmonellosis can be transmitted through contaminated fish products, but also on the production rates of flocks, and it acts as an important sanitary barrier to commercial transactions between countries.
Emphasizing the importance and surveillance interest of Salmonella spp., the scientific productions had their results published in several titles of journals covering different areas of knowledge—it is an essentially multidisciplinary theme that can cover different areas related to health [42,61] and the environment [22,25,98], such as microbiology [47], animal production [9,67] and food science [13,72].
From the textual analysis of the most frequent words and analysis of similarity, it can be inferred, in general, that the studies selected in this integrative review present references that are inherent to the diagnosis of pathogens of importance in public health in aquaculture production—this helps to understand how to maintain vigilance when reporting the microbiological diagnosis of Salmonella spp. in aquaculture species, where fish from production stood out. They also reveal other complementary aspects that help to understand the subject more broadly. Among them is the connection that the works make with microbiological diagnosis, encompassing other bacterial pathogens of the Enterobacteriaceae family and other bacterial pathogens such as Listeria monocytogenes, also of public health interest, as they can also be spread through water and cause disease transmitted by contaminated food (DTAs—this shows that different categories of fish can also harbor Salmonella spp. and, therefore, can increase the risk of salmonellosis outbreaks in humans; Salmonella isolates have already been tested for their antimicrobial resistance profile due to the consideration that they are a threat to health.
More than half of the scientific products analyzed reported the presence of Salmonella spp. Over the years (between 2000 and 2020), classic microbiological culture techniques have remained fundamental tools in the microbiological diagnosis of Salmonella spp. in fish [22,58,63]. These data suggest that the isolation method by culture remains widespread, as the analysis steps provide a combination of factors favorable to the isolation of viable cells of Salmonella spp., which can be serotyped, cultured and classified according to the characteristics of virulence, such as biofilm formation capacity and antimicrobial resistance and susceptibility profiles.
The use of complementary methodologies such as molecular diagnosis by conventional [14] or real-time polymerase chain reaction (PCR) [25] to identify Salmonella spp., techniques such as MALDI-TOF-mass spectrometry [17,21] and MLST (Multilocus Sequence Typing) for serotyping were conducted to a lesser extent in the studies. Although molecular diagnosis by PCR is considered a more sensitive technique with the advantages of not generating false positive or false negative results [110] and having a diagnosis in a shorter time compared to microbiological techniques, there is the limitation of not being able to differentiate the cells in the diagnosis of viable specimens from the killing of a pathogen for further microbial isolation. Shabarinath et al. [52] investigated 100 samples and detected Salmonella spp. in 20% of them using the conventional microbiology technique, while the PCR technique detected positivity in 52%; in contrast, Hollmann et al. [110] performed a microbiological diagnosis technique after a direct PCR examination of the samples, isolating approximately 74% of the positives.
Salmonella is one among many other microbiological risks to be avoided in aquaculture production [76,86]; therefore, as it is an important pathogen in public health, it was possible to observe the inclusion of Salmonella spp. in microbiological investigations in aquaculture [80,104], since this bacterial genus is not part of the natural microbiota of fish, and there is a lack of studies that can conclude that fish are affected by clinical symptoms that characterize infection. Therefore, even being a pathogen of the Enterobacteriaceae family, it has been possible to identify that Salmonella spp. can survive and multiply in places other than the gastrointestinal tract of fish [18,66]. In this sense, Salmonella isolates have been detected in fresh or saltwater collections [22,94] in different external anatomical parts, such as mucus, skin, and gills [17,28,86] of fish and imported frozen food products [80]. In addition to showing that these animals are potential reservoirs of Salmonella, all these findings serve as a basic tool for control or prevention measures to be taken, to avoid dissemination and cross-contamination in breeding and production systems and consequently to prevent the risk of occurrence of salmonellosis outbreaks in humans.
The studies presented several choices for analysis units of samples from different categories of aquaculture species for the microbiological diagnosis of Salmonella spp. This lack of uniformity could be explained by the difference in the different objectives of the production or other contextual factors in the country that could impact the microbial contamination of products from aquaculture to be kept under surveillance. There are also geographic differences in the study sites. Factors related to the study design: type of sample used (muscle or several other parts) and type of tests used to measure bacterial prevalence (microbiology, PCR) or concentration are likely to contribute to the observed differences.
The different ranges of samples and prevalence results may be because most of the fish sampled were collected in a lightly processed form from a farm or market, where the chances of contamination are high and can contribute to many results observed in the studies.
In the 56 studies that reported positive Salmonella, 164 serotypes were identified (Figure 10), with 31 of them cited in at least four articles or more. Extrinsic factors, such as region, environment, species of production and types of samples analyzed in the investigations, are some of the factors that contribute to the diversity of serovars.
S. Typhimurium was the most frequently identified in 17 scientific articles from different countries on the Asian continent [43,49,56,57,61,92,102], North and South American countries [74,80], and African country [86], and has been isolated from freshwater [74], muscle, intestine and gills of freshwater fish [49,86], marine fish [102] and other seafood [14,48]. It is the serotype that most commonly causes salmonellosis among the nontyphoid serotypes [29] and has been reported as the dominant serovar causing human infection in China [111]. It can also be isolated from animals from different livestock sectors, such as swine, cattle and chickens [112]. These results agree with the analyses performed by Ferrari et al. [5], in which the S. Typhimurium serotype presented a cosmopolitan profile, being the most prevalent and disseminated worldwide and being considered an example of a generalist serotype by several food matrices (beef, pork, chicken and fish), but mainly by pork. In recent research, Wang et al. [111] reported that S. Enteritidis, Derby, Typhimurium, Thompsom and Aberdeen were the most common serovars detected in chickens, pigs, ducks, aquatic products and turtles, respectively.
S. Weltevreden was the second most reported serotype in scientific articles from countries in Asia [17,49,57,61] and North America [79], being closely associated with the aquaculture of freshwater fish [17], marine shrimp [58,61] and various seafood imported into the US [14,79]. This serotype displays global importance in seafood and is most prevalent in South and Southeast Asia [5]; however, in a recent study using over 35,000 Salmonella enterica isolates to explore the temporal and spatial dynamics of the dominant serovars in China, the authors found that S. Typhimurium is the dominant serovar [111]. To a lesser extent, this serotype has been the cause of outbreaks in North America [4]. Ferrari and others. [5] suggested that fish imports from Asia may also have imported this pathogen, as in their analysis, there was no evidence that S. Weltevreden was native to North America. In the present review, two articles with samples of seafood imported into the US reporting the presence of S. Weltevreden were selected [14,79]. This is a classic example of the transmission of pathogens from distant regions, forcing the improvement of hygienic-sanitary control measures.
S. Newport was also the second most reported serotype in scientific articles from China [45,46], India [48,52] and the USA [14] in seafood samples. Its highest prevalence was detected in North America, being a pathogen transmissible to humans mainly through the consumption of seafood [5]. However, Zhao et al. [14] reported detecting S. Newport from various seafood imported from 38 countries.
Other reported serotypes, such as S. Stanley (8), S. Kentucky and S. Hadar (5) and S. Heidelberg (2), have also been previously discussed in FSA and NARMS reports as being important in causing salmonellosis in humans [113,114].
All serotypes need to be kept under surveillance, as aquaculture products have become potential sources for the spread of Salmonella spp. During the entire period that comprises production (rearing, capture or removal, processing and retailing phases), fish products are subject to contamination by pathogenic microorganisms naturally present in the aquatic environment and other opportunists introduced through animal and human waste during processing and/or preparation of the production chain [7].
5. Final Considerations
Descriptive analyses on the mined data of scientific articles made it possible to generate information from a qualitative and quantitative perspective on the microbiological diagnoses of Salmonella spp. in aquaculture during the selected years. It was possible to verify a wide variety of journals in which the articles were published, with the area of food science being the topic with the most concentrated articles. The textual analyses of the abstracts organized and classified five main themes that correlate with Salmonella spp. in aquaculture connected to topics such as food safety and public health; however, the absence of words or a set of terms obtained in the word cloud and in the analysis of similarity and descending hierarchical classification (DHC) which refer to measures to prevent or control the contamination of fish was observed. Apparently, the studies included were more focused on characterizing the problem (investigation of the occurrence, aquaculture species sampled, identification and characterization of the profile of the isolates) than on the search for strategies to mitigate the risks of contamination in aquaculture. In general, it was possible to observe from the number of annual publications that there were no increasing or decreasing trends in the surveillance of Salmonella spp. in aquaculture over the years; however, there is a constant number of publications on the subject. In this sense, interest in monitoring Salmonella spp. by classical microbiological diagnosis was shown worldwide, with articles published in several countries in almost all continents (there were no articles selected from Oceania countries). Asian countries are considered the largest aquaculture producers in the world and were the nations that had the most scientific articles selected by this review. As a fish product that is less processed and more subject to microbiological contamination, it was observed that most of the sampling came from aquaculture activities. Several types of fish categories and environmental samples were analyzed by the articles, but fish was the most sampled species in the studies. Salmonella spp. was detected from different units of analysis, and from different anatomical parts of the fish in samples of water from aquaculture cultivation and sediments, suggesting that the pathogen adapts to several favorable environments for multiplication and, consequently, contamination. The conventional microbiological diagnostic techniques used in the articles, sometimes supported by PCR, were unanimous and have shown efficiency in terms of surveillance. Most of the isolates detected were identified up to the genus level; however, it was possible to verify many reported serotypes, which helped to better understand the epidemiological process in aquaculture.
Our research did not include Salmonella genotypic characterization in the eligibility criteria. We encourage new reviews focusing on the molecular characteristics of Salmonella to be carried out.
Finally, the results obtained here can contribute to the promotion of new studies that investigate strategies for the control and prevention of Salmonella spp. in fish production, as the need to increase studies with this focus was observed.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani13010027/s1, Table S1: Characteristics of the articles selected in the integrative review on the microbiological diagnoses of Salmonella spp. in aquaculture between 2000 and 2020.
Author Contributions
Conceptualization, F.H.d.S.F., W.d.S.T. and J.P.L.; methodology, W.d.S.T., A.O.A., E.E.d.S.F. and B.S.V.; formal analysis, A.O.A., W.d.S.T. and Y.D.P.; investigation, Y.D.P., F.H.d.S.F., A.O.A. and W.d.S.T.; data curation, A.O.A., W.d.S.T. and Y.D.P.; writing—original draft preparation, Y.D.P., F.H.d.S.F., A.O.A. and W.d.S.T.; writing—review and editing, E.E.d.S.F., L.K.S.d.S., B.S.V., A.C.N., J.P.L., J.L.d.S. and Y.D.P.; supervision, W.d.S.T., A.O.A., E.E.d.S.F. and L.K.S.d.S.; project administration, F.H.d.S.F., E.E.d.S.F. and W.d.S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful for a D.Sc. (Y.P.) fellowships provided by CNPq (National Council for Scientific and Technological Development), Brazil (Process code, 310181/2021-6).
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Initiative to estimate the global burden of foodborne diseases. In Proceedings of the Fourth Formal Meeting of the Foodborne Disease Burden Epidemiology Reference Group (FERG): Sharing New Results, Making Future Plans and Preparing Ground for the Countries, Geneva, Switzerland, 8–12 November 2010; World Health Organization: Geneva, Switzerland, 2014; p. 108. [Google Scholar]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management, and prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Pinedo, L.C.; Mughini-Gras, L.; Franz, E.; Hald, T.; Pires, S.M. Sources and trends of human salmonellosis in Europe, 2015–2019: An analysis of outbreak data. Int. J. Food Microbiol. 2022, 379, 109850. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Griffin, P.M.; Cole, D.; Walsh, K.A.; Chai, S.J. Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998–2008. Emerg. Infect. Dis. 2013, 8, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Hand, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Ali, A.; Parisi, A.; Conversano, M.C.; Iannacci, A.; D’Emilio, M.C.; Mercurio, V.; Normanno, G. Food-borne bacteria associated with seafoods: A brief review. J. Food Qual. Hazards Control 2020, 7, 4–10. [Google Scholar] [CrossRef]
- Amagliani, G.; Brandi, G.; Schiavano, G.F. Incidence and role of Salmonella in seafood safety. Food Res. Int. 2012, 45, 780–788. [Google Scholar] [CrossRef]
- Dróżdż, M.; Małaszczuk, M.; Paluch, E.; Pawlak, A. Zoonotic potential and prevalence of Salmonella serovars isolated from pets. Infect. Ecol. Epidemiol. 2021, 11, 1975530. [Google Scholar] [CrossRef]
- Pilarski, F.; Júnior, O.T.; Casaca, J.D.M.; Garcia, F.R.M.; Tomazelli, I.B.; Dos Santos, I.R. Integrated fish/pig systems: Environmental feature and fish quality [Consórcio suíno-peixe: Aspectos ambientais e qualidade do pescado]. Rev. Bras. Zootec. 2004, 33, 267–276. [Google Scholar] [CrossRef]
- Meletiadis, A.; Biolatti, C.; Mugetti, D.; Zaccaria, T.; Cipriani, R.; Pitti, M.; Decastelli, L.; Cimino, F.; Dondo, A.; Maurella, C.; et al. Research on exposure to reptile-associated salmonellosis (RAS) in the Piedmont region of Italy. Animals 2022, 12, 906. [Google Scholar] [CrossRef]
- Casanova, L.M.; Sobsey, M.D. Antibiotic-Resistant Enteric Bacteria in Environmental Waters. Water 2016, 8, 561. [Google Scholar] [CrossRef]
- Akinjogunla, O.J.; Inyang, C.U.; Akinjogunla, V.F. Bacterial species associated with anatomical parts of fresh and smoked Bonga fish (Ethmalosa fimbriata): Prevalence and susceptibility to cephalosporins. Res. J. Microbiol. 2011, 6, 87–97. [Google Scholar] [CrossRef]
- Basti, A.A.; Misaghi, A.; Salehi, T.Z.; Kamkar, A. Bacterial pathogens in fresh, smoked and salted Iranian fish. Food Control 2006, 17, 183–188. [Google Scholar] [CrossRef]
- Zhao, S.; Datta, A.R.; Ayers, S.; Friedman, S.; Walker, R.D.; White, D.G. Antimicrobial-resistant Salmonella serovars isolated from imported foods. Int. J. Food Microbiol. 2003, 84, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, D.V.G.S.; Castro, V.S.; Cunha Neto, A.; Figueiredo, E.E.S. Salmonella spp. in the fish production chain: A review. Ciência Rural 2018, 48, e20180141. [Google Scholar] [CrossRef]
- Novotny, L.; Dvorska, L.; Lorencova, A.; Beran, V.; Pavlik, I. Fish: A potential source of bacterial pathogens for human beings. Veterinární Med. 2004, 49, 343–358. [Google Scholar] [CrossRef]
- Li, K.; Petersen, G.; Barco, L.; Hvidtfeldt, K.; Liu, L.; Dalsgaard, A. Salmonella Weltevreden in integrated and non-integrated tilapia aquaculture systems in Guangdong, China. Food Microbiol. 2017, 65, 19–24. [Google Scholar] [CrossRef]
- dos Santos, R.R.; Xavier, R.G.C.; de Oliveira, T.F.; Leite, R.C.; Figueiredo, H.C.P.; Leal, C.A.G. Occurrence, genetic diversity, and control of Salmonella enterica in native Brazilian farmed fish. Aquaculture 2019, 501, 304–312. [Google Scholar] [CrossRef]
- Kodama, H.; Nakanishi, Y.; Yamamoto, F.; Mikami, T.; Izawa, H.; Imagawa, T.; Hashimoto, Y.; Kudo, N. Salmonella arizonae isolated from a pirarucu, Arapaima gigas Cuvier, with septicaemia. J. Fish Dis. 1987, 10, 509–512. [Google Scholar] [CrossRef]
- Gazal, L.E.S.; Brito, K.C.T.; Cavalli, L.S.; Kobayashi, R.K.T.; Nakazato, G.; Otutumi, L.K.; Cunha, A.C.; Neto, J.A.S.P.; Brito, B.G. Salmonella sp. in fish—What is the importance for health in fish farm? Pesqui. Agropecuária Gaúcha 2018, 24, 55–64, ISSN online: 2595-7686. [Google Scholar] [CrossRef]
- Dib, A.L.; Agabou, A.; Chahed, A.; Kurekci, C.; Moreno, E.; Espigares, M.; Espigares, E. Isolation, molecular characterization and antimicrobial resistance of enterobacteriaceae isolated from fish and seafood. Food Control 2018, 88, 54–60. [Google Scholar] [CrossRef]
- Antunes, P.; Campos, J.; Mourão, J.; Pereira, J.; Novais, C.; Peixe, L. Inflow water is a major source of trout farming contamination with Salmonella and multidrug resistant bacteria. Sci. Total Environ. 2018, 642, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Esposto, E.M.; Silva, W.C.P.; Reis, C.M.F.; Reis, E.M.F.; Ribeiro, R.V.; Rodrigues, D.P.; Lázaro, N.S. Enteropatógenos bacterianos em peixes criados em uma estação de reciclagem de nutrientes e no ecossistema relacionado. Pesqui. Veterinária Bras. 2007, 27, 144–148. [Google Scholar] [CrossRef]
- Saingam, P.; Li, B.; Yan, T. Fecal Indicator Bacteria, Direct Pathogen Detection, and Microbial Community Analysis Provide Different Microbiological Water Quality Assessment of a Tropical Urban Marine Estuary. Water Res. 2020, 185, 116280. [Google Scholar] [CrossRef] [PubMed]
- Klase, G.; Lee, S.; Liang, S.; Kim, J.; Zo, Y.-G.; Lee, J. The microbiome and antibiotic resistance in integrated fishfarm water: Implications of environmental public health. Sci. Total Environ. 2019, 649, 1491–1501. [Google Scholar] [CrossRef]
- Mccoy, E.; Morrison, J.; Cook, V.; Johnston, J.; Eblen, D.; Guo, C. Foodborne agents associated with the consumption of aquaculture catfish. J. Food Prot. 2011, 74, 500–516. [Google Scholar] [CrossRef]
- Martinez, O.; Rodriguez-Calleja, J.M.; Santos, J.A.; Otero, A.; Garcia-Lopez, M.L. Foodborne and Indicator Bacteria in Farmed Molluskan Shellfish before and after Depuration. J. Food Prot. 2009, 72, 1443–1449. [Google Scholar] [CrossRef]
- Ampofo, J.A.; Clerk, G.C. Diversity of bacteria in sewage treatment plant used as fish culture pond in southern Ghana. Aquac. Res. 2003, 34, 667–675. [Google Scholar] [CrossRef]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg. Infect. Dis. 2015, 21, 941–949. [Google Scholar] [CrossRef]
- Sutton, A.; Clowes, M.; Preston, L.; Booth, A. Meeting the review family: Exploring review types and associated information retrieval requirements. Health Inf. Libr. J. 2019, 3, 202–222. [Google Scholar] [CrossRef]
- Booth, A.; Noyes, J.; Flemming, K.; Gerhardus, A.; Wahlster, P.; Van Der Wilt, G.J.; Mozygemba, K.; Refolo, P.; Sacchini, D.; Tummers, M.; et al. Guidance on Choosing Qualitative Evidence Synthesis Methods for Use in Health Technology Assessments of Complex Interventions; Integrate-HTA: Bremen (DE), Germany, 2016. [Google Scholar]
- Mishra, P.; Pandey, C.; Singh, U.; Gupta, A.; Sahu, C.; Keshri, A. Descriptive statistics and normality tests for statistical data Annals of Cardiac Anesthesia. Ann. Card. Anaesth. 2019, 22, 67–72. [Google Scholar] [CrossRef]
- Nahm, U.Y.; Mooney, R.J. A Mutually Beneficial Integration of Data Mining and Information Extraction. In Proceedings of the AAAI/IAAI, Austin, TX, USA, 1–3 August 2000; pp. 627–632. [Google Scholar]
- Reinert, M. Alceste, une méthodologie d’analyze dês données textuelles et une application: Aurelia de Gerard de Nerval. Bull. De Méthodol. Sociol. 1990, 26, 24–54. [Google Scholar] [CrossRef]
- Sousa, Y.S.O. The Use of the Iramuteq Software: Fundamentals of Lexicometry for Qualitative Research. Estud. E Pesqui. Em Psicol. 2021, 21, 1541–1560. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 19 April 2020).
- Feinerer, I.; Hornik, K. tm: Text Mining Package. R Package Version 0.7-8. 2020. Available online: https://CRAN.R-project.org/package=tm (accessed on 18 October 2020).
- Bouchet-Valat, M. SnowballC: Snowball Stemmers Based on the C ‘libstemmer’ UTF-8 Library. R Package Version 0.7.0. 2020. Available online: https://CRAN.R-project.org/package=SnowballC (accessed on 18 October 2020).
- Fellows, I. Wordcloud: Word Clouds. R Package Version 2.6. 2018. Available online: https://CRAN.R-project.org/package=wordcloud (accessed on 18 October 2020).
- Tennekes, M. tmap: Thematic Maps in R. J. Stat. Softw. 2018, 84, 1–39. [Google Scholar] [CrossRef]
- Broughton, E.I.; Walker, D.G. Prevalence of Antibiotic-Resistant Salmonella in Fish in Guangdong, China. Foodborne Pathog. Dis. 2009, 6, 519–521. [Google Scholar] [CrossRef]
- Li, Y.; Pei, X.; Yan, J.; Liu, D.; Zhang, H.; Yu, B.; Li, N.; Yang, D. Prevalence of foodborne pathogens isolated from retail freshwater fish and shellfish in China. Food Control 2019, 99, 131–136. [Google Scholar] [CrossRef]
- Pawar, P.P.; Pagarkar, A.U.; Rathod, N.B. Effect of chilled storage on quality characteristics of battered and breaded snack product from large sized Catla (Catla catla). J. Food Sci. Technol. 2020, 57, 52–59. [Google Scholar] [CrossRef]
- Yang, X.; Wu, Q.; Zhang, J.; Huang, J.; Chen, L.; Liu, S.; Yu, S.; Cai, S. Prevalence, enumeration, and characterization of Salmonella isolated from aquatic food products from retail markets in China. Food Control 2015, 57, 308–313. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, X.; Kuang, D.; Shi, X.; Xiao, W.; Zhang, J.; Gu, Z.; Xu, X.; Meng, J. Prevalence of antimicrobial resistance of nontyphoidal Salmonella serovars in retail aquaculture products. Int. J. Food Microbiol. 2015, 210, 47–52. [Google Scholar] [CrossRef]
- Surendraraj, A.; Sabeena Farvin, K.H.; Yathavamoorthi, R.; Thampuran, N. Enteric bacteria associated with farmed freshwater fish and its culture environment in Kerala, India. Res. J. Microbiol. 2009, 4, 334–344. [Google Scholar]
- Kumar, R.; Surendran, P.K.; Thampuran, N. Detection and characterization of virulence factors in lactose positive and lactose negative Salmonella serovars isolated from seafood. Food Control 2009, 20, 376–380. [Google Scholar] [CrossRef]
- Kakatkar, A.S.; Pansare, L.S.; Gautam, R.K.; Shashidhar, R.; Karani, M.; Bandekar, J.R. Molecular characterization of antibiotic resistant Salmonella isolates from Indian foods. Food Res. Int. 2011, 44, 3272–3275. [Google Scholar] [CrossRef]
- Patel, A.; Jeyasekaran, G.; Jeyashakila, R.; Anand, T.; Wilwet, L.; Pathak, N.; Malini, A.H.; Neethiselvan, N. Prevalence of antibiotic resistant Salmonella spp. strains in shrimp farm source waters of Nagapattinam region in South India. Mar. Pollut. Bull. 2020, 155, 111171. [Google Scholar] [CrossRef]
- Saharan, V.V.; Verma, P.; Singh, A.P. High prevalence of antimicrobial resistance in Escherichia coli, Salmonella spp. and Staphylococcus aureus isolated from fish samples in India. Aquac. Res. 2020, 51, 1200–1210. [Google Scholar] [CrossRef]
- Shabarinath, S.; Sanath Kumar, H.; Khushiramani, R.; Karunasagar, I.; Karunasagar, I. Detection and characterization of Salmonella associated with tropical seafood. Int. J. Food Microbiol. 2007, 114, 227–233. [Google Scholar] [CrossRef]
- Shakila, R.J.; Raj, B.E.; Felix, N. Quality and safety of fish curry processed by sous vide cook chilled and hot filled technology process during refrigerated storage. Food Sci. Technol. Int. 2012, 18, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Ooi, M.C.; Shariff, M.; Khatoon, H. Antibiotic resistant Salmonella and Vibrio associated with farmed Litopenaeus vannamei. Sci. World J. 2012, 2012, 130136. [Google Scholar] [CrossRef]
- Budiati, T.; Rusul, G.; Wan-Abdullah, W.N.; Arip, Y.M.; Ahmad, R.; Thong, K.L. Prevalence, antibiotic resistance and plasmid profiling of Salmonella in catfish (Clarias gariepinus) and tilapia (Tilapia mossambica) obtained from wet markets and ponds in Malaysia. Aquaculture 2013, 372–375, 127–132. [Google Scholar] [CrossRef]
- Sing, C.K.; Khan, M.Z.I.; Daud, H.H.M.; Aziz, A.R. Prevalence of Salmonella sp. in African Catfish (Clarias gariepinus) Obtained from Farms and Wet Markets in Kelantan, Malaysia and Their Antibiotic Resistance. Sains Malays. 2016, 45, 1597–1602. [Google Scholar]
- Nguyen, D.T.A.; Kanki, M.; Nguyen, P.D.; Le, H.T.; Ngo, P.T.; Tran, D.N.M.; Le, N.H.; Dang, C.V.; Kawai, T.; Kawahara, R.; et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC beta-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016, 236, 115–122. [Google Scholar] [CrossRef]
- Noor Uddin, G.M.; Larsen, M.H.; Barco, L.; Minh Phu, T.; Dalsgaard, A. Clonal Occurrence of Salmonella Weltevreden in Cultured Shrimp in the Mekong Delta, Vietnam. PLoS ONE 2015, 10, e0134252. [Google Scholar] [CrossRef] [PubMed]
- Yen, N.T.P.; Nhung, N.T.; Van, N.T.B.; Cuong, N.V.; Tien Chau, L.T.; Trinh, H.N.; Tuat, C.V.; Tu, N.D.; Phu Huong Lan, N.; Campbell, J.; et al. Antimicrobial residues, nontyphoidal Salmonella, Vibrio spp. and associated microbiological hazards in retail shrimps purchased in Ho Chi Minh city (Vietnam). Food Control 2020, 107, 106756. [Google Scholar] [CrossRef]
- Dhowlaghar, N.; Abeysundara, P.D.A.; Nannapaneni, R.; Schilling, M.W.; Chang, S.; Cheng, W.H.; Sharma, C.S. Biofilm formation by Salmonella spp. in catfish mucus extract under industrial conditions. Food Microbiol. 2018, 70, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, B.P.; Utrarachkij, F.; Thongshoob, J.; Mahakunkijcharoen, Y.; Wongchinda, N.; Suthienkul, O.; Khusmith, S. Detection of Salmonella inva gene in shrimp enrichment culture by polymerase chain reaction. Southeast Asian J. Trop. Med. Public Health 2010, 41, 426–445. [Google Scholar] [PubMed]
- Álvarez, A.; García García, B.; Garrido, M.D.; Hernández, M.D. The influence of starvation time prior to slaughter on the quality of commercial-sized gilthead seabream (Sparus aurata) during ice storage. Aquaculture 2008, 284, 106–114. [Google Scholar] [CrossRef]
- Doménech, E.; Jimenez-Belenguer, A.; Amoros, J.A.; Ferrus, M.A.; Escriche, I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in Eastern Spain. Food Control 2015, 47, 120–125. [Google Scholar] [CrossRef]
- Hernández, M.D.; López, M.B.; Álvarez, A.; Ferrandini, E.; García García, B.; Garrido, M.D. Sensory, physical, chemical and microbiological changes in aquacultured meagre (Argyrosomus regius) fillets during ice storage. Food Chem. 2009, 114, 237–245. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Liebana, E. Use of pulsed-field gel electrophoresis to characterize the genetic diversity and clonal persistence of Salmonella senftenberg in mussel processing facilities. Int. J. Food Microbiol. 2005, 105, 153–163. [Google Scholar] [CrossRef]
- Costa, J.C.C.P.; Floriano, B.; Villegas, I.M.B.; Rodríguez-Ruiz, J.P.; Posada-Izquierdo, G.D.; Zurera, G.; Pérez-Rodríguez, F. Study of the microbiological quality, prevalence of foodborne pathogens and product shelf-life of Gilthead sea bream (Sparus aurata) and Sea bass (Dicentrarchus labrax) from aquaculture in estuarine ecosystems of Andalusia (Spain). Food Microbiol. 2020, 90, 103498. [Google Scholar] [CrossRef]
- Caruso, G.; Maimone, G.; Mancuso, M.; Modica, A.; Genovese, L. Microbiological controls across the productive cycle of Dicentrarchus labrax L. and Sparus aurata L.: A study from the environment to the final product. Aquac. Res. 2004, 35, 184–193. [Google Scholar] [CrossRef]
- Rubini, S.; Galletti, G.; D’Incau, M.; Govoni, G.; Boschetti, L.; Berardelli, C.; Barbieri, S.; Merialdi, G.; Formaglio, A.; Guidi, E.; et al. Occurrence of Salmonella enterica subsp. enterica in bivalve mollusks and associations with Escherichia coli in mollusks and fecal coliforms in seawater. Food Control 2018, 84, 429–435. [Google Scholar] [CrossRef]
- Smaldone, G.; Marrone, R.; Zottola, T.; Vollano, L.; Grossi, G.; Cortesi, M.L. Formulation and shelf-life of fish burgers served to preschool children. Ital. J. Food Saf. 2017, 6, 6373. [Google Scholar] [CrossRef] [PubMed]
- Atanassova, V.; Reich, F.; Klein, G. Microbiological quality of Sushi from Sushi bars and retailers. J. Food Prot. 2008, 71, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Brenner, M.; Ramdohr, S.; Effkemann, S.; Stede, M. Key parameters for the consumption suitability of offshore cultivated blue mussels (Mytilus edulis L.) in the German Bight. Eur. Food Res. Technol. 2009, 230, 255–267. [Google Scholar] [CrossRef]
- Araújo, W.S.C.; De Lima, C.L.S.; Peixoto Joele, M.R.S.; Lourenço, L.D.F.H. Development and Application of the Quality Index Method (QIM) for Farmed Tambaqui (Colossoma macropomum) Stored Under Refrigeration. J. Food Saf. 2017, 37, e12288. [Google Scholar] [CrossRef]
- Calixto, F.A.A.; Machado, E.D.S.; Franco, R.M.; de Mesquita, E.D.F. Bacteriological evaluation of fresh, salted and smoked Cobia meat from fish culture of Ilha Grande bay, Rio de Janeiro state, Brazil. Bol. Inst. Pesca 2016, 42, 209–215. [Google Scholar] [CrossRef]
- Palhares, J.C.P.; Kich, J.D.; Bessa, M.C.; Biesus, L.L.; Berno, L.G.; Triques, N.J. Salmonella and antimicrobial resistance in an animal-based agriculture river system. Sci. Total Environ. 2014, 472, 654–661. [Google Scholar] [CrossRef]
- Pastro, D.C.; Mariotto, S.; Santos, E.C.; Ferreira, D.C.; Chitarra, G.S. Use of molecular techniques for the analysis of the microbiological quality of fish marketed in the municipality of Cuiaba, Mato Grosso, Brazil. Food Sci. Technol. 2019, 39 (Suppl. S1), 146–151. [Google Scholar] [CrossRef]
- Ribeiro, R.V.; Reis, E.M.F.; Reis, C.M.F.; Freitas-Almeida, A.C.; Rodrigues, D.P. Incidence and antimicrobial resistance of enteropathogens isolated from an integrated aquaculture system. Lett. Appl. Microbiol. 2010, 51, 611–618. [Google Scholar] [CrossRef]
- Akiyama, T.; Khan, A.A.; Cheng, C.-M.; Stefanova, R. Molecular characterization of Salmonella enterica serovar Saintpaul isolated from imported seafood, pepper, environmental and clinical samples. Food Microbiol. 2011, 28, 1124–1128. [Google Scholar] [CrossRef]
- Pal, A.; Marshall, D.L. Comparison of culture media for enrichment and isolation of Salmonella spp. from frozen Channel catfish and Vietnamese basa fillets. Food Microbiol. 2009, 26, 317–319. [Google Scholar] [CrossRef]
- Ponce, E.; Khan, A.A.; Cheng, C.-M.; Summage-West, C.; Cerniglia, C.E. Prevalence and characterization of Salmonella enterica serovar Weltevreden from imported seafood. Food Microbiol. 2008, 25, 29–35. [Google Scholar] [CrossRef]
- Wang, F.; Jiang, L.; Yang, Q.; Han, F.; Chen, S.; Pu, S.; Vance, A.; Ge, B. Prevalence and Antimicrobial Susceptibility of Major Foodborne Pathogens in Imported Seafood. J. Food Prot. 2011, 74, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Aubourg, S.P.; Quitral, V.; Larraín, M.A.; Rodríguez, A.; Gómez, J.; Maier, L.; Vinagre, J. Autolytic degradation and microbiological activity in farmed Coho salmon (Oncorhynchus kisutch) during chilled storage. Food Chem. 2007, 104, 369–375. [Google Scholar] [CrossRef]
- Dondero, M.; Cisternas, F.; Carvajal, L.; Simpson, R. Changes in quality of vacuum-packed cold-smoked salmon (Salmo salar) as a function of storage temperature. Food Chem. 2004, 87, 543–550. [Google Scholar] [CrossRef]
- Abou-Elela, G.M.; El-Sersy, N.A.; Abd-Elnaby, H.; Wefky, S.H. Distribution and biodiversity of fecal indicators and potentially harmful pathogens in North Delta (Egypt). Aust. J. Basic Appl. Sci. 2009, 3, 3374–3385. [Google Scholar]
- Elsaidy, N.; Abouelenien, F.; Kirrella, G.A.K. Impact of using raw or fermented manure as fish feed on microbial quality of water and fish. Egypt. J. Aquat. Res. 2015, 41, 93–100. [Google Scholar] [CrossRef]
- Mahmoud, M.A.; Abdelsalam, M.; Mahdy, O.A.; El Miniawy, H.M.F.; Ahmed, Z.A.M.; Osman, A.H.; Mohamed, H.M.H.; Khattab, A.M.; Zaki Ewiss, M.A. Infectious bacterial pathogens, parasites and pathological correlations of sewage pollution as an important threat to farmed fishes in Egypt. Environ. Pollut. 2016, 219, 939–948. [Google Scholar] [CrossRef]
- Efuntoye, M.O.; Olurin, K.B.; Jegede, G.C. Bacterial flora from healthy clarias gariepinus and their antimicrobial resistance pattern. Adv. J. Food Sci. Technol. 2012, 4, 121–128. [Google Scholar]
- Abbassi-Ghozzi, I.; Jaouani, A.; Hammami, S.; Martinez-Urtaza, J.; Boudabous, A.; Gtari, M. Molecular analysis and antimicrobial resistance of Salmonella isolates recovered from raw meat marketed in the area of “Grand Tunis”, Tunisia. Pathol. Biol. 2012, 60, e49–e54. [Google Scholar] [CrossRef]
- Boulares, M.; Mejri, L.; Hassouna, M. Study of the Microbial Ecology of Wild and Aquacultured Tunisian Fresh Fish. J. Food Prot. 2011, 74, 762–1768. [Google Scholar] [CrossRef] [PubMed]
- Wanja, D.W.; Mbuthia, P.G.; Waruiru, R.M.; Bebora, L.C.; Ngowi, H.A.; Nyaga, P.N. Antibiotic and Disinfectant Susceptibility Patterns of Bacteria Isolated from Farmed Fish in Kirinyaga County, Kenya. Int. J. Microbiol. 2020, 2020, 8897338. [Google Scholar] [CrossRef] [PubMed]
- Miruka, D.O.; Ochieng, J.O.; Were, J.W.; Waindi, E.N. Microbial assessment of selected earthen fish ponds in western Kenya. Ecohydrol. Hydrobiol. 2013, 13, 261–266. [Google Scholar] [CrossRef]
- Bingol, E.B.; Colak, H.; Hampikyan, H.; Muratoglu, K. The microbiological quality of stuffed mussels (Midye Dolma) sold in Istanbul. Br. Food J. 2008, 110, 1079–1087. [Google Scholar] [CrossRef]
- Yildirim, Z.; Sakin, T.; Çoban, F. Isolation of lytic bacteriophages infecting Salmonella typhimurium and Salmonella enteritidis. Acta Biol. Hung. 2018, 69, 350–369. [Google Scholar] [CrossRef] [PubMed]
- Mannan, M.; Islam, S.R.; Osman, M.H.; Rahman, M.K.; Uddin, M.N.; Kamal, M.; Reza, M.S. Antibacterial activity of oxytetracycline on microbial ecology of Nile tilapia (Oreochromis niloticus) gastrointestinal tract under laboratory condition. Aquac. Res. 2020, 51, 2125–2133. [Google Scholar] [CrossRef]
- Huys, G.; Bartie, K.; Cnockaert, M.; Hoang Oanh, D.T.; Phuong, N.T.; Somsiri, T.; Chinabut, S.; Yusoff, F.M.; Shariff, M.; Giacomini, M.; et al. Biodiversity of chloramphenicol-resistant mesophilic heterotrophs from Southeast Asian aquaculture environments. Res. Microbiol. 2007, 158, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Kaktcham, P.M.; Temgoua, J.-B.; Ngoufack Zambou, F.; Diaz-Ruiz, G.; Wacher, C.; Pérez-Chabela, M.L. Quantitative analyses of the bacterial microbiota of rearing environment, tilapia and common carp cultured in earthen ponds and inhibitory activity of its lactic acid bacteria on fish spoilage and pathogenic bacteria. World J. Microbiol. Biotechnol. 2017, 33, 32. [Google Scholar] [CrossRef] [PubMed]
- Ayazo-Genes, J.; Pertuz-Buelvas, V.; Jimenez-Velasquez, C.; Espinosa-Araujo, J.; Atencio-Garcia, V.; Prieto-Guevara, M. Describing the planktonic and bacterial communities associated with bocachico Prochilodus magdalenae fish culture with biofloc technology. Rev. Mvz Cordoba 2019, 24, 7209–7217. [Google Scholar] [CrossRef]
- Krog, J.S.; Larsen, L.E.; Schultz, A.C. Enteric porcine viruses in farmed shellfish in Denmark. Int. J. Food Microbiol. 2014, 186, 105–109. [Google Scholar] [CrossRef]
- Sagoo, S.K.; Little, C.L.; Greenwood, M. Microbiological study of cooked crustaceans and molluskan shellfish from UK production and retail establishments. Int. J. Environ. Health Res. 2007, 17, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulos, A.; Plessas, S.; Voidarou, C.; Noussias, H.; Stavropoulou, E.; Mantzourani, I.; Tzora, A.; Skoufos, I.; Bezirtzoglou, E. Microbial ecology of fish species ongrowing in Greek sea farms and their watery environment. Anaerobe 2011, 17, 264–266. [Google Scholar] [CrossRef]
- Hudecova, K.; Buchtova, H.; Steinhauserova, I. The Effects of Modified Atmosphere Packaging on the Microbiological Properties of Fresh Common Carp (Cyprinus carpio L.). Acta Vet. Brno 2010, 79, S93–S100. [Google Scholar] [CrossRef]
- ALameer, A.H.A.; Atshan, O.F.; Mahmood, M.M.; Al-Jewari, W.M.; Mohammed, A.A. Detection of Salmonella species in viscera of Carp fish. Plant Arch. 2020, 20, 2683–2686. [Google Scholar]
- Furushita, M.; Shiba, T.; Maeda, T.; Yahata, M.; Kaneoka, A.; Takahashi, Y.; Torii, K.; Hasegawa, T.; Ohta, M. Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl. Environ. Microbiol. 2003, 69, 5336–5342. [Google Scholar] [CrossRef] [PubMed]
- Terentjeva, M.; Eizenberga, I.; Valciņa, O.; Novoslavskij, A.; Strazdiņa, V.; Berziņš, A. Prevalence of foodborne pathogens in freshwater fish in Latvia. J. Food Prot. 2015, 78, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Harakeh, S.; Yassine, H.; El-Fadel, M. Antimicrobial-resistant patterns of Escherichia coli and Salmonella strains in the aquatic Lebanese environments. Environ. Pollut. 2006, 143, 269–277. [Google Scholar] [CrossRef]
- Valenzuela-Armenta, J.A.; Díaz-Camacho, S.P.; Cabanillas-Ramos, J.A.; de Jesus Uribe-Beltrán, M.; de la Cruz MD, C.; Osuna-Ramírez, I.; Báez-Flores, M.E. Microbiological analysis of tilapia and water in aquaculture farms from Sinaloa. Biotecnia 2018, 20, 20–26. [Google Scholar] [CrossRef]
- Nesse, L.L.; Løvold, T.; Bergsjø, B.; Nordby, K.; Wallace, C.; Holstad, G. Persistence of orally administered Salmonella enterica Serovars Agona and Montevideo in Atlantic salmon (Salmo salar L.). J. Food Prot. 2005, 68, 1336–1339. [Google Scholar] [CrossRef]
- Pyz-Lukasik, R.; Paszkiewicz, W. Microbiological quality of farmed grass carp, bighead carp, Siberian sturgeon, and wels catfish from Eastern Poland. J. Vet. Res. 2018, 62, 145–149. [Google Scholar] [CrossRef]
- Al-Harbi, A.H.; Uddin, M.N. Seasonal changes in bacterial flora of fish pond sediments in Saudi Arabia. J. Appl. Aquac. 2006, 18, 35–45. [Google Scholar] [CrossRef]
- Ntengwe, F.W.; Edema, M.O. Physico-chemical and microbiological characteristics of water for fish production using small ponds. Phys. Chem. Earth Parts A/B/C 2008, 33, 701–707. [Google Scholar] [CrossRef]
- Hollmann, I.; Lingens, J.B.; Wilke, V.; Homann, C.; Teich, K.; Buch, J.; Chuppava, B.; Visscher, C. Epidemiological Study on Salmonella Prevalence in Sow Herds Using Direct and Indirect Detection Methods. Microorganisms 2022, 10, 1532. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Lyu, N.; Li, Z.; Ma, S.; Cao, D.; Pan, Y.; Hu, Y.; Huang, H.; Gao, G.F.; et al. The temporal dynamics of antimicrobial-resistant-Salmonella enterica and predominant serovars in China. Natl. Sci. Rev. 2022, nwac269. [Google Scholar] [CrossRef]
- Wallis, T.S.; Barrow, P.A. Salmonella epidemiology and pathogenesis in food-producing animals. Am. Soc. Microbiol. 2005, 1, 2324–6200. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Surveillance Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).