The Mantle Transcriptome of Chamelea gallina (Mollusca: Bivalvia) and Shell Biomineralization
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Mantle Transcriptome Sequencing and Assembly
2.2. Transcriptome Refinement
2.3. Gene Expression Analyses
2.4. DEGs Functional Enrichment Analysis
2.5. X-ray Diffraction (XRD) and Scanning Electron Microscopy (SEM) Measurements
2.6. Assessment of Environmental Parameters
3. Results
3.1. Transcriptome Assembly and Annotation
3.2. Gene Expression Analyses
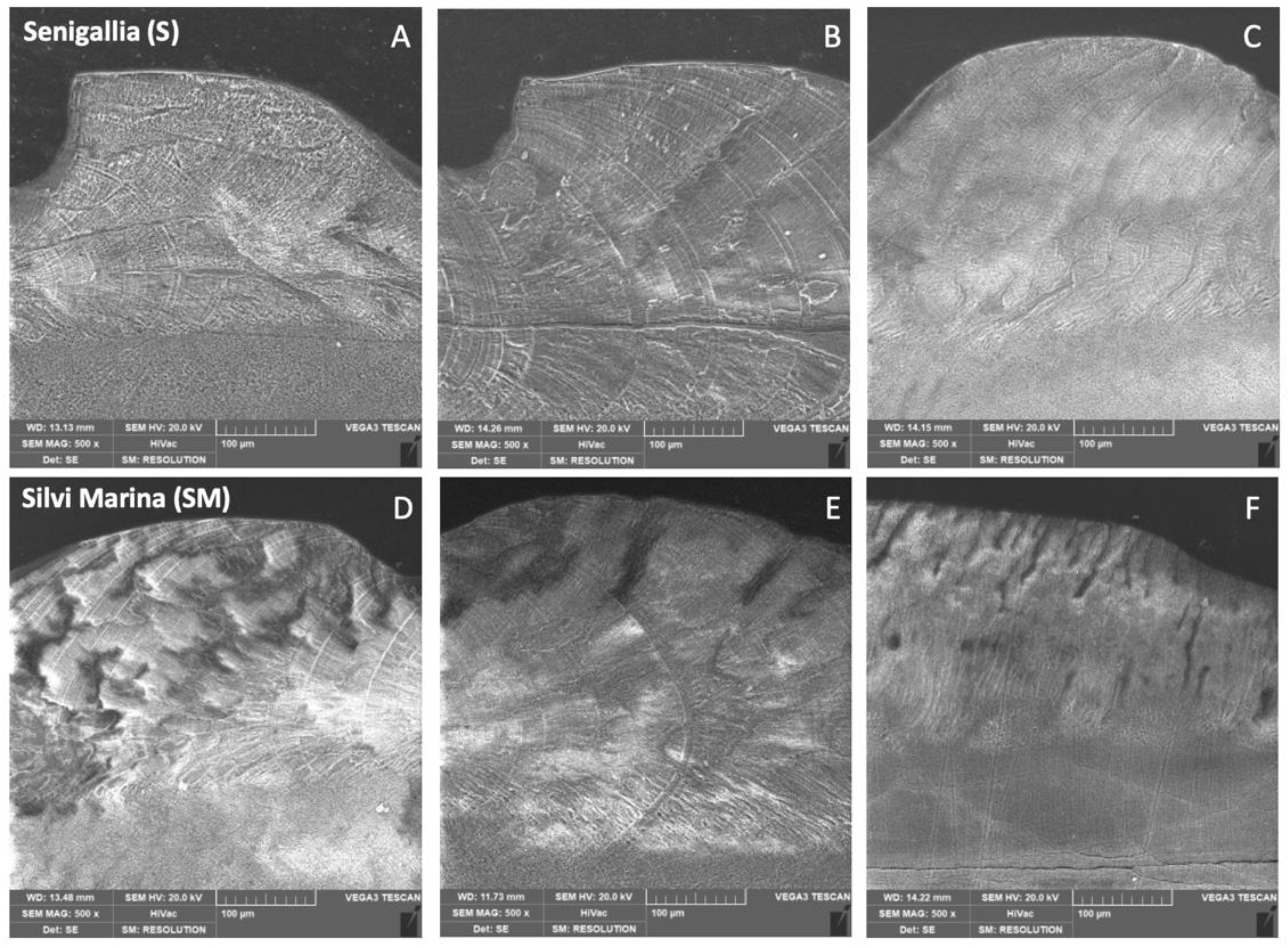
3.3. X-ray Diffraction (XRD) and Scanning Electron Microscopy (SEM) Measurements
3.4. Assessment of Environmental Parameters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bieler, R.; Mikkelsen, P.M.; Collins, T.M.; Glover, E.A.; Gonzalez, V.L.; Graf, D.L.; Harper, E.M.; Healy, J.; Kawauchi, G.Y.; Sharma, P.P.; et al. Investigating the bivalve tree of life-an exemplar-based approach combining molecular and novel morphological characters. Invertebr. Syst. 2014, 28, 32–115. [Google Scholar] [CrossRef] [Green Version]
- Marin, F.; Le Roy, N.; Marie, B. The formation and mineralization of mollusk shell. Front. Biosci. 2012, 4, 1099–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popov, S.V. Composite prismatic structure in bivalve shell. Acta Palaeontol. Pol. 1986, 31, 3–28. [Google Scholar]
- Checa, A. Physical and biological determinants of the fabrication of molluscan shell microstructures. Front. Mar. Sci. 2018, 5, 353. [Google Scholar] [CrossRef] [Green Version]
- Ivanina, A.V.; Falfushynska, H.I.; Beniash, E.; Piontkivska, H.; Sokolova, I.M. Biomineralization-related specialization of hemocytes and mantle tissues of the Pacific oyster Crassostrea gigas. J. Exp. Biol. 2017, 220, 3209–3221. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, J.; Ferreira, V.; Zhang, X.; Anjos, L.; Félix, R.C.; Batista, F.M.; Power, D.M. Evolution and diversity of alpha-carbonic anhydrases in the mantle of the Mediterranean mussel (Mytilus galloprovincialis). Sci. Rep. 2019, 9, 10400. [Google Scholar] [CrossRef]
- Clark, M.S.; Peck, L.S.; Arivalagan, J.; Backeljau, T.; Berland, S.; Cardoso, J.C.R.; Caurcel, C.; Chapelle, G.; De Noia, M.; Dupont, S.; et al. Deciphering mollusc shell production: The roles of genetic mechanisms through to ecology, aquaculture and biomimetics. Biol. Rev. 2020, 95, 1812–1837. [Google Scholar] [CrossRef]
- Zhao, R.; Takeuchi, T.; Luo, Y.J.; Ishikawa, A.; Kobayashi, T.; Koyanagi, R.; Villar-Briones, A.; Yamada, L.; Sawada, H.; Iwanaga, S.; et al. Dual gene repertoires for larval and adult shells reveal molecules essential for molluscan shell formation. Mol. Biol. Evol. 2018, 35, 2751–2761. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, Q.; Fan, M.; He, J.; Zhang, X.; Xu, H.; Liao, Z. Recombinant transgelin-like protein 1 from Mytilus shell induces formation of CaCO3 polymorphic crystals in vitro. FEBS Open Bio 2020, 10, 2216–2234. [Google Scholar] [CrossRef]
- Sleight, V.A.; Thorne, M.A.; Peck, L.S.; Arivalagan, J.; Berland, S.; Marie, A.; Clark, M.S. Characterisation of the mantle transcriptome and biomineralisation genes in the blunt-gaper clam, Mya truncata. Mar. Genom. 2016, 27, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Aguilera, F.; McDougall, C.; Degnan, B.M. Evolution of the tyrosinase gene family in bivalve molluscs: Independent expansion of the mantle gene repertoire. Acta Biomater. 2014, 10, 3855–3865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carducci, F.; Biscotti, M.A.; Trucchi, E.; Giuliani, M.E.; Gorbi, S.; Coluccelli, A.; Barucca, M.; Canapa, A. Omics approaches for conservation biology research on the bivalve Chamelea gallina. Sci. Rep. 2020, 10, 19177. [Google Scholar] [CrossRef] [PubMed]
- Gizzi, F.; Caccia, M.G.; Simoncini, G.A.; Mancuso, A.; Reggi, M.; Fermani, S.; Brizi, L.; Fantazzini, P.; Stagioni, M.; Falini, G.; et al. Shell properties of commercial clam Chamelea gallina are influenced by temperature and solar radiation along a wide latitudinal gradient. Sci. Rep. 2016, 6, 36420. [Google Scholar] [CrossRef]
- Mancuso, A.; Stagioni, M.; Prada, F.; Scarponi, D.; Piccinetti, C.; Goffredo, S. Environmental influence on calcification of the bivalve Chamelea gallina along a latitudinal gradient in the Adriatic Sea. Sci. Rep. 2019, 9, 11198. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, 807–811. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- The UniProt Consortiu. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Clementi, E.; Pistoia, J.; Escudier, R.; Delrosso, D.; Drudi, M.; Grandi, A.; Lecci, R.; Cretí, S.; Ciliberti, S.; Coppini, G.; et al. Mediterranean Sea Analysis and Forecast (CMEMS MED-Currents, EAS5 system). Copernicus Monitoring Environment Marine Service (CMEMS). 2019. Available online: https://doi.org/10.25423/CMCC/MEDSEA_ANALYSIS_FORECAST_PHY_006_013_EAS5 (accessed on 15 March 2021).
- Bolzon, G.; Cossarini, G.; Lazzari, P.; Salon, S.; Teruzzi, A.; Feudale, L.; Crise, A.; Solidoro, C. Mediterranean Sea biogeochemical analysis and forecast (CMEMS MED-Biogeochemistry (2018)-Present). Copernicus Monitoring Environment Marine Service (CMEMS). 2020. Available online: https://doi.org/10.25423/CMCC/MEDSEA_ANALYSIS_FORECAST_BIO_006_014_MEDBFM3 (accessed on 15 March 2021).
- Moschino, V.; Deppieri, M.; Marin, M.G. Evaluation of shell damage to the clam Chamelea gallina captured by hydraulic dredging in the Northern Adriatic Sea. ICES J. Mar. Sci. 2003, 60, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Moschino, V.; Chicharo, L.M.Z.; Marin, M.G. Effects of hydraulic dredging on the physiological responses of the target species Chamelea gallina (Mollusca: Bivalvia): Laboratory experiments and field surveys. Sci. Mar. 2008, 72, 493–501. [Google Scholar]
- Knobel, L.; Breusing, C.; Bayer, T.; Sharma, V.; Hiller, M.; Melzner, F.; Stuckas, H. Comparative de novo assembly and annotation of mantle tissue transcriptomes from the Mytilus edulis species complex (M. edulis, M. galloprovincialis, M. trossulus). Mar. Genom. 2020, 51, 100700. [Google Scholar] [CrossRef]
- Yarra, T.; Ramesh, K.; Blaxter, M.; Hüning, A.; Melzner, F.; Clark, M.S. Transcriptomic analysis of shell repair and biomineralization in the blue mussel, Mytilus edulis. BMC Genom. 2021, 22, 437. [Google Scholar] [CrossRef]
- Carter, J.G. Guide to bivalve shell microstructures. In Skeletal Growth of Aquatic Organisms; Rhoads, D.C., Lutz, R.A., Eds.; Plenum Press: New York, NY, USA, 1980. [Google Scholar]
- Zhang, C.; Xie, L.P.; Huang, J.; Chen, L.; Zhang, R.Q. A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem. Biophys. Res. Commun. 2006, 342, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Hershey, D.M.; Ren, X.; Melnyk, R.A.; Browne, P.J.; Ozyamak, E.; Jones, S.R.; Chang, M.C.Y.; Hurley, J.H.; Komeili, A. MamO is a repurposed serine protease that promotes magnetite biomineralization through direct transition metal binding in magnetotactic bacteria. PLoS Biol. 2016, 14, e1002402. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Liu, X.J.; Li, J.L. A Kunitz proteinase inhibitor (HcKuPI) participated in antimicrobial process during pearl sac formation and induced the overgrowth of calcium carbonate in Hyriopsis cumingii. Fish Shellfish Immunol. 2019, 89, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, C.; Ma, Z.; Xie, L.; Zhang, R. A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim. Biophys. Acta 2007, 1770, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Jiang, Y.-T.; Sun, Q.; Fan, M.-H.; Wang, J.-X.; Liang, H.-Y. Microstructure and in-depth proteomic analysis of Perna viridis shell. PLoS ONE 2019, 14, e0219699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, K.; Yano, M.; Morimoto, K.; Miyamoto, H. Tyrosinase localization in mollusc shells. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 146, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Arivalagan, J.; Yarra, T.; Marie, B.; Sleight, V.A.; Duvernois-Berthet, E.; Clark, M.S.; Marie, A.; Berland, S. Insights from the shell proteome: Biomineralization to adaptation. Mol. Biol. Evol. 2017, 34, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, C.N.; Morri, C. Marine biodiversity of the Mediterranean Sea: Situation, problems and prospects for future research. Mar. Pollut. Bull. 2000, 40, 367–376. [Google Scholar] [CrossRef]
- Ivanina, A.V.; Jarrett, A.; Bell, T.; Rimkevicius, T.; Beniash, E.; Sokolova, I.M. Effects of seawater salinity and pH on cellular metabolism and enzyme activities in biomineralizing tissues of marine bivalves. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2020, 248, 110748. [Google Scholar] [CrossRef]

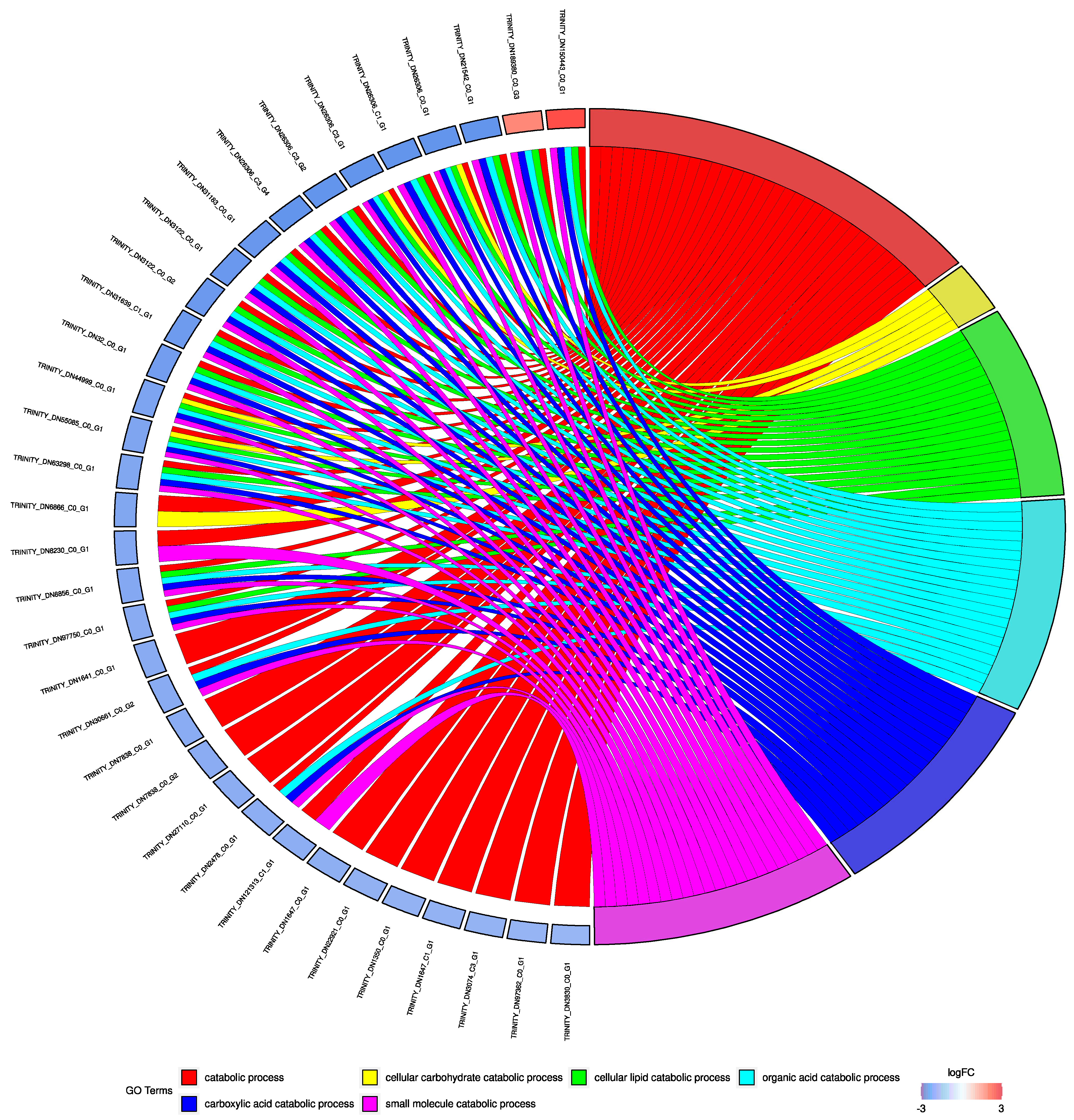

| Upregulated at S Sampling Site | BLAST Results | |
| macrocategories | description_UNIPROT accession | entries found |
| biomineralization related proteins | Putative tyrosinase-like protein tyr-3 (TYR3_CAEEL) | 1 |
| WAP, Kazal, immunoglobulin, Kunitz and NTR domain-containing protein 2 (WFKN2_MOUSE) | 1 | |
| Sarcoplasmic calcium-binding protein (SCP_MIZYE) | 1 | |
| Early growth response 1 (EGR1_XENTR) | 2 | |
| microtubule associated proteins | Janus kinase and microtubule-interacting protein 3 (JKIP3_HUMAN) | 1 |
| Downregulated at S Sampling Site | BLAST Results | |
| macrocategories | description_UNIPROT accession | entries found |
| biomineralization related proteins | insoluble shell matrix protein 6 (IMSP6_RUDPH) | 6 |
| insoluble shell matrix protein 3 (IMSP3_RUDPH) | 3 | |
| insoluble shell matrix protein 2 (IMSP2_RUDPH) | 1 | |
| ubiquitin (UBIQ_LUMTE) | 1 | |
| mucin-like protein (MLP_ACRMI) | 1 | |
| tyrosinase-like protein (TYRO_PINMA) | 1 | |
| perlucin (PLC_HALLA) | 1 | |
| galaxin (GXN_ACRMI) | 1 | |
| carbonic anhydrase 7 (CAH7_HUMAN) | 1 | |
| metal ion binding | protocadherin FAT4 (FAT4_MOUSE) | 1 |
| calcium binding and coiled coil domain-containing protein 2 (CACO2_MACFA) | 1 | |
| CBL-interacting serine-threonine protein kinase 5 (CIPK5_ARATH) | 1 | |
| cAMP and cAMP-inhibited cGMP 3′,5′-cyclic phosphodiesterase 10A (PDE10_HUMAN) | 1 | |
| Zinc finger MYM-type protein 4 (ZMYM4_MOUSE) | 1 | |
| baculoviral IAP repeat containing protein 7 (BIRC7_MOUSE, BIRC_XENTR) | 2 | |
| DNA-directed RNA polymerase II subunit RPB1 (RPB1_PLAFD) | 1 | |
| sodium/potassium-transporting ATPase subunit alpha-B (AT1B_ARTSF) | 1 | |
| thyroid peroxidase (PERT_MOUSE) | 1 | |
| cysteine dioxygenase type 1 (CDO1_BOVIN) | 1 | |
| NADPH oxidase 5 (NOX5_HUMAN) | 1 | |
| metal cation symporter ZIP14 (S39AE_BOVIN) | 1 | |
| sodium/potassium-transporting ATPase subunit alpha (AT1A_TAESO) | 1 | |
| energy metabolism | phosphoenolpyruvate carboxykinase cytoplasmic (PCKCG_BOVIN, PCKG_DROME, PCKGM_MOUSE, PCKCG_CHICK) | 18 |
| Endoglucanase E-4 (GUN4_THEFU) | 1 | |
| Isocitrate dehydrogenase [NADP] cytoplasmic (IDHC_DICDI) | 1 | |
| cytochrome P450 2D2O (CP2DK_MESAU) | 1 | |
| adenylate kinase isoenzyme 5 (KAD5_HUMAN) | 1 | |
| cytochrome P450 3A18 (CP3AI_RAT) | 1 | |
| ketohexokinase (KHK_HUMAN) | 1 | |
| carboxypeptidase M (CBPM_PONAB) | 1 | |
| alpha-amylase (AMY_PECMA) | 2 | |
| microtubule associated proteins | tubulin alpha 2,4 chain (TBA2_PATVU) | 1 |
| tubulin beta chain (TBB_LYTPI) | 1 | |
| outer dynein arm-docking complex subunit 3(CC151_BOVIN) | 1 | |
| tubulin beta-6 chain (TBB6_ECTVR) | 1 | |
| tubulin beta chain (TBB_PARLI) | 1 | |
| outer dynein arm-docking complex subunit 4 (TTC25_DANRE) | 1 | |
| dynein heavy chain, cytoplasmic (DYHC_EMENI) | 1 | |
| actin, muscle (ACTM_LYTPI) | 1 | |
| alpha-actinin (ACTN_DERFA) | 1 | |
| tubulin beta chain (TBB_STRPU) | 1 | |
| protein trafficking | ADP-ribosylation factor (ARF_ASHGO) | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carducci, F.; Biscotti, M.A.; Mosca, A.; Greco, S.; Gerdol, M.; Memmola, F.; Barucca, M.; Canapa, A. The Mantle Transcriptome of Chamelea gallina (Mollusca: Bivalvia) and Shell Biomineralization. Animals 2022, 12, 1196. https://doi.org/10.3390/ani12091196
Carducci F, Biscotti MA, Mosca A, Greco S, Gerdol M, Memmola F, Barucca M, Canapa A. The Mantle Transcriptome of Chamelea gallina (Mollusca: Bivalvia) and Shell Biomineralization. Animals. 2022; 12(9):1196. https://doi.org/10.3390/ani12091196
Chicago/Turabian StyleCarducci, Federica, Maria Assunta Biscotti, Alessandro Mosca, Samuele Greco, Marco Gerdol, Francesco Memmola, Marco Barucca, and Adriana Canapa. 2022. "The Mantle Transcriptome of Chamelea gallina (Mollusca: Bivalvia) and Shell Biomineralization" Animals 12, no. 9: 1196. https://doi.org/10.3390/ani12091196
APA StyleCarducci, F., Biscotti, M. A., Mosca, A., Greco, S., Gerdol, M., Memmola, F., Barucca, M., & Canapa, A. (2022). The Mantle Transcriptome of Chamelea gallina (Mollusca: Bivalvia) and Shell Biomineralization. Animals, 12(9), 1196. https://doi.org/10.3390/ani12091196













