Genetic Diversity, Population Structure and Phylogeny of Indigenous Goats of Mongolia Revealed by SNP Genotyping
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Description
2.2. DNA Extraction and Genotyping
2.3. Construction of the Working Datasets
2.4. Genetic Diversity Estimation
2.4.1. Effective Population Sizes
2.4.2. Runs of Homozygosity (ROH)
2.4.3. Haplotype Sharing
2.5. Genetic Relationships and Population Structure
3. Results
3.1. Dataset Description
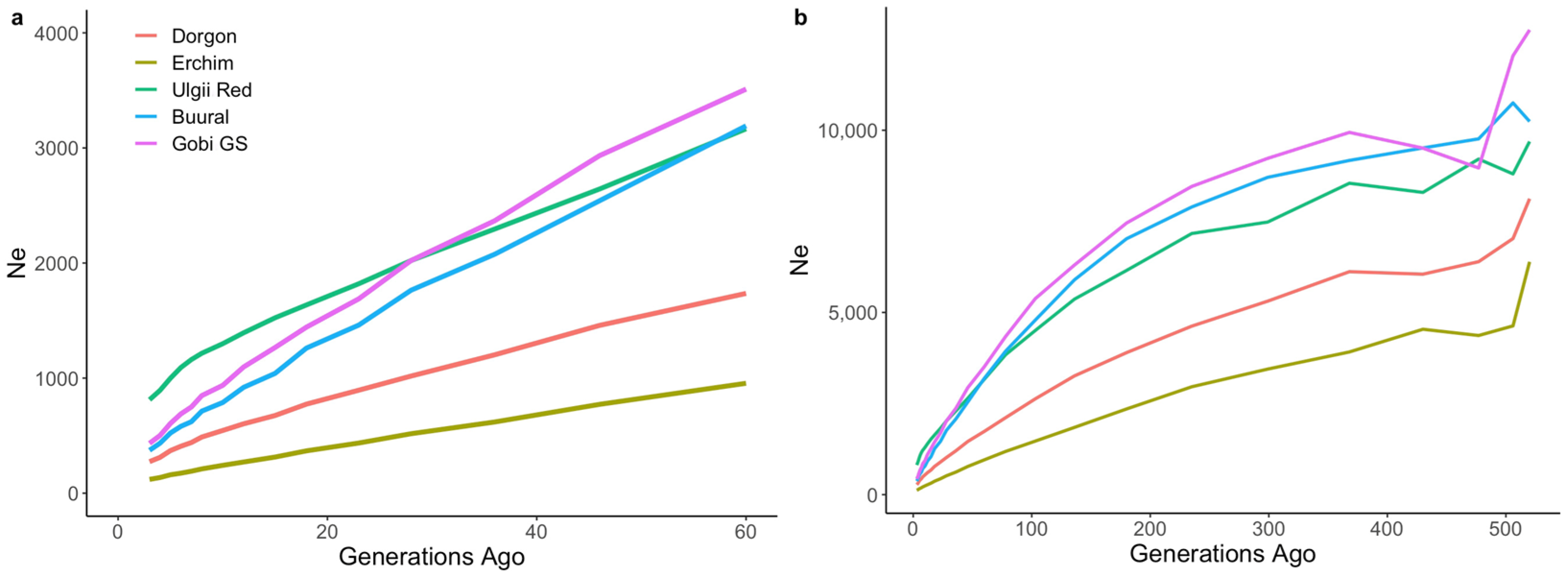
3.2. Genetic Diversity and Effective Population Sizes in Mongolian Populations
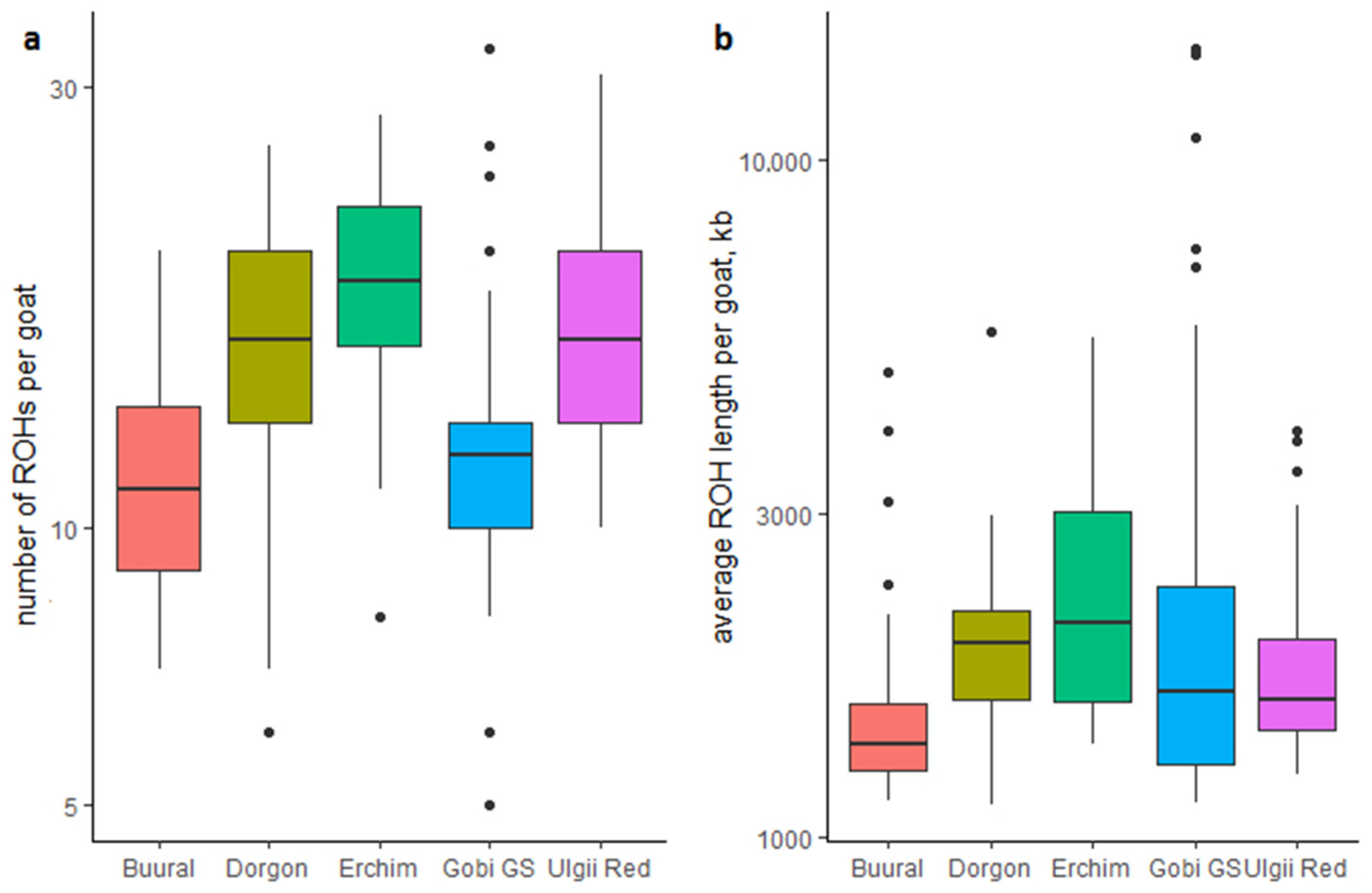
3.3. Runs of Homozygosity (ROH)
3.4. Ancestry of Mongolian Goat Breeds
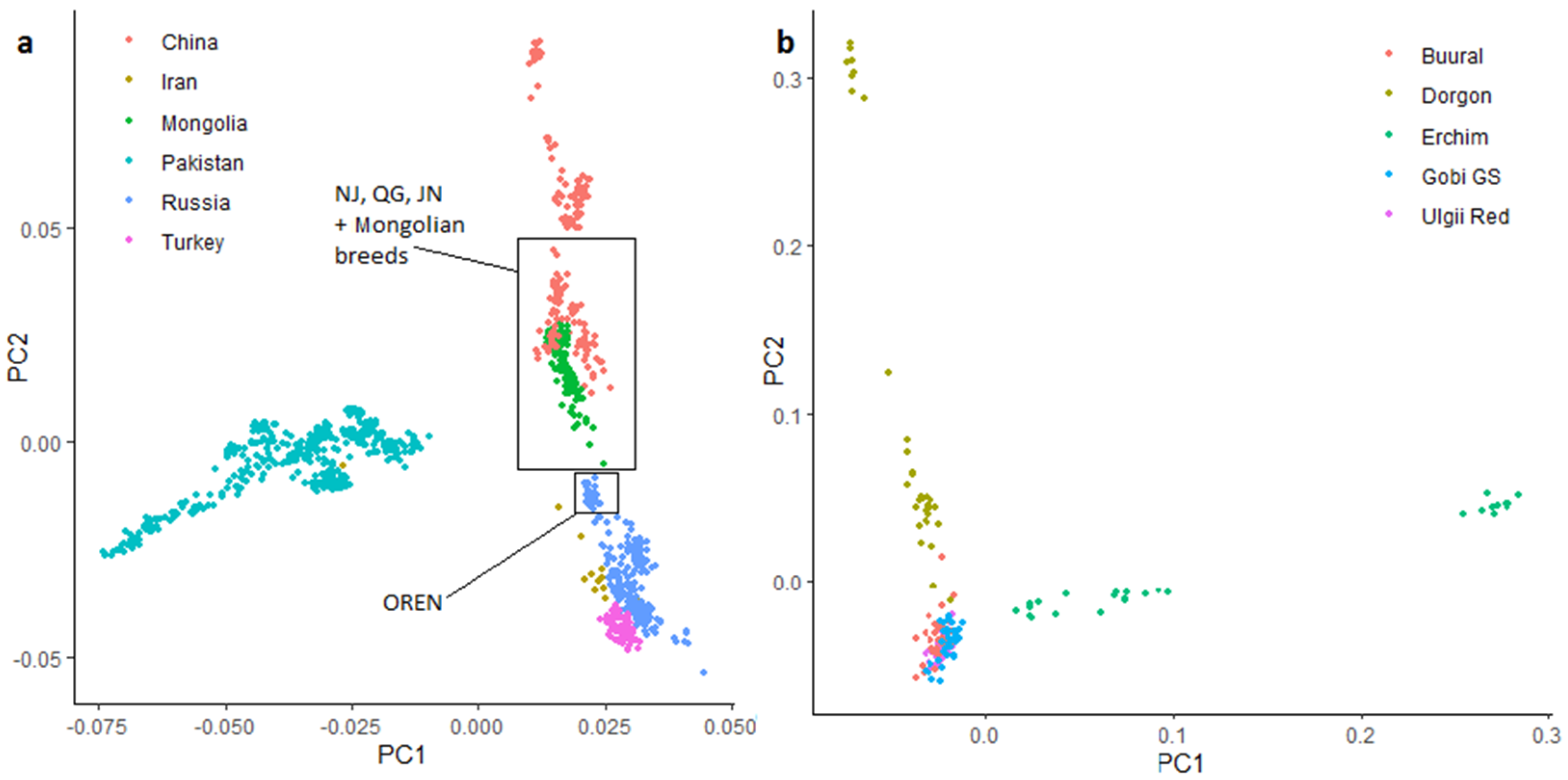
3.4.1. Principal Component Analysis
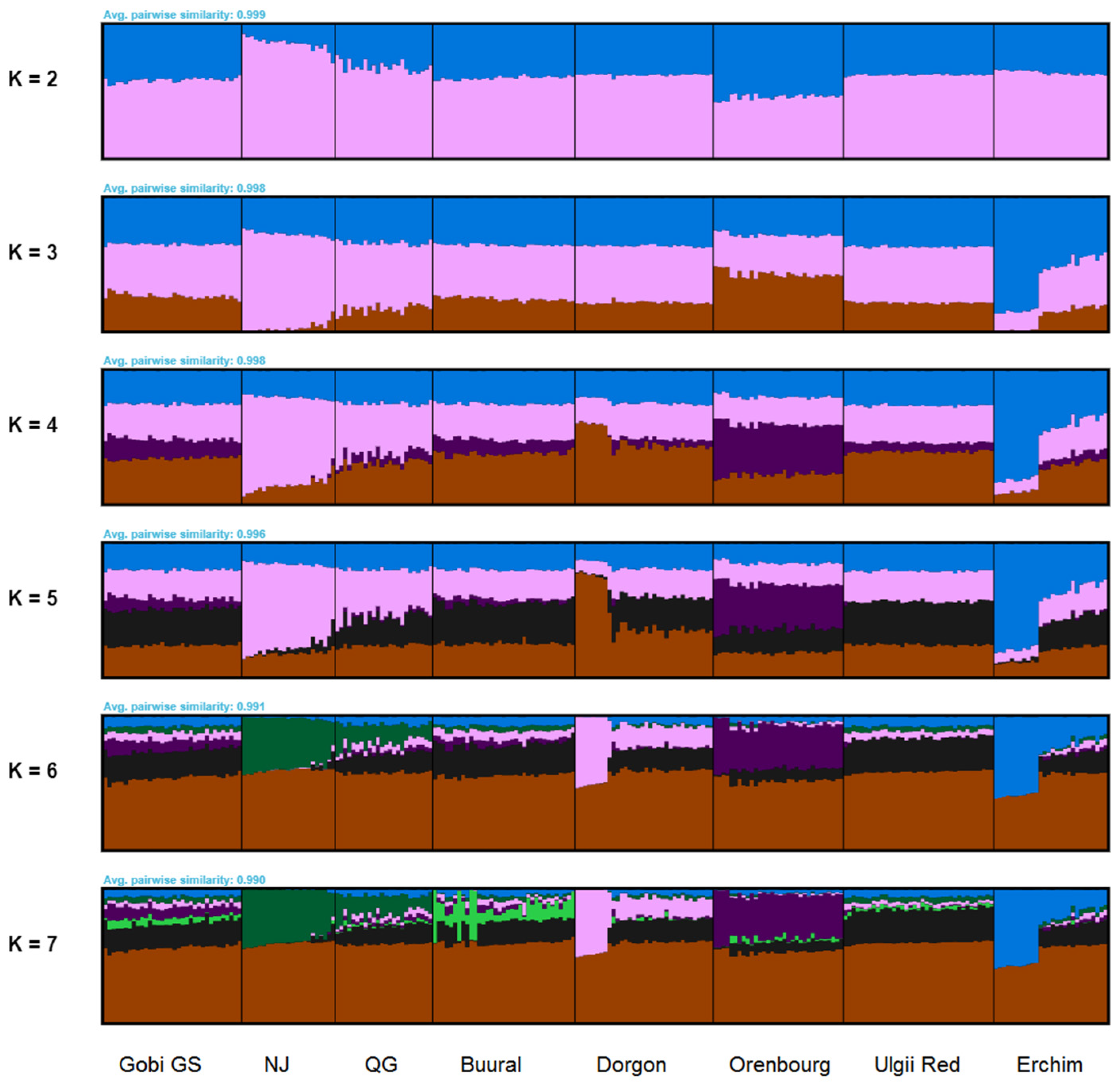
3.4.2. FastSTRUCTURE Analysis
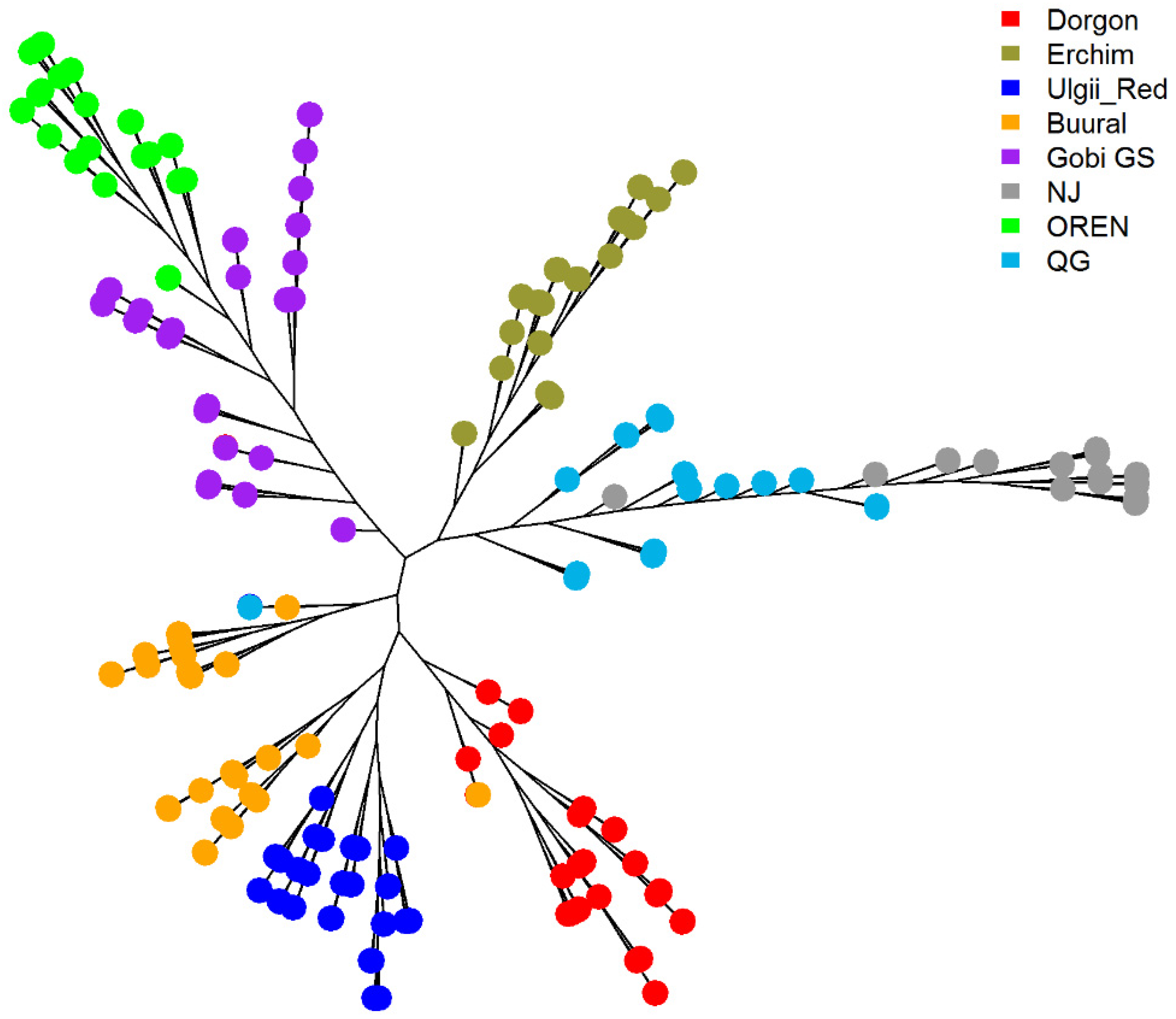
3.4.3. Phylogenetic Analysis
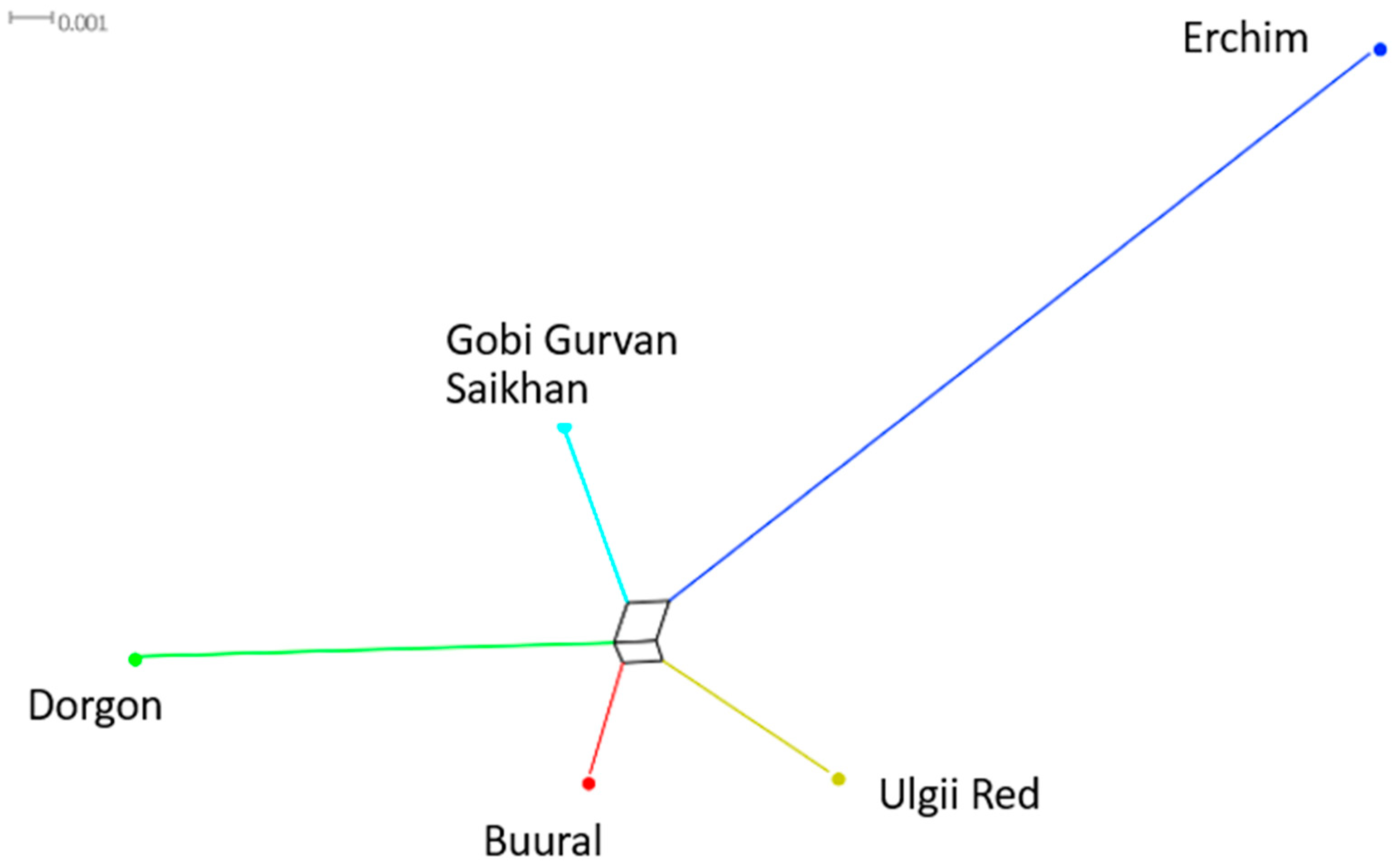
3.4.4. Genetic Distances (FST)
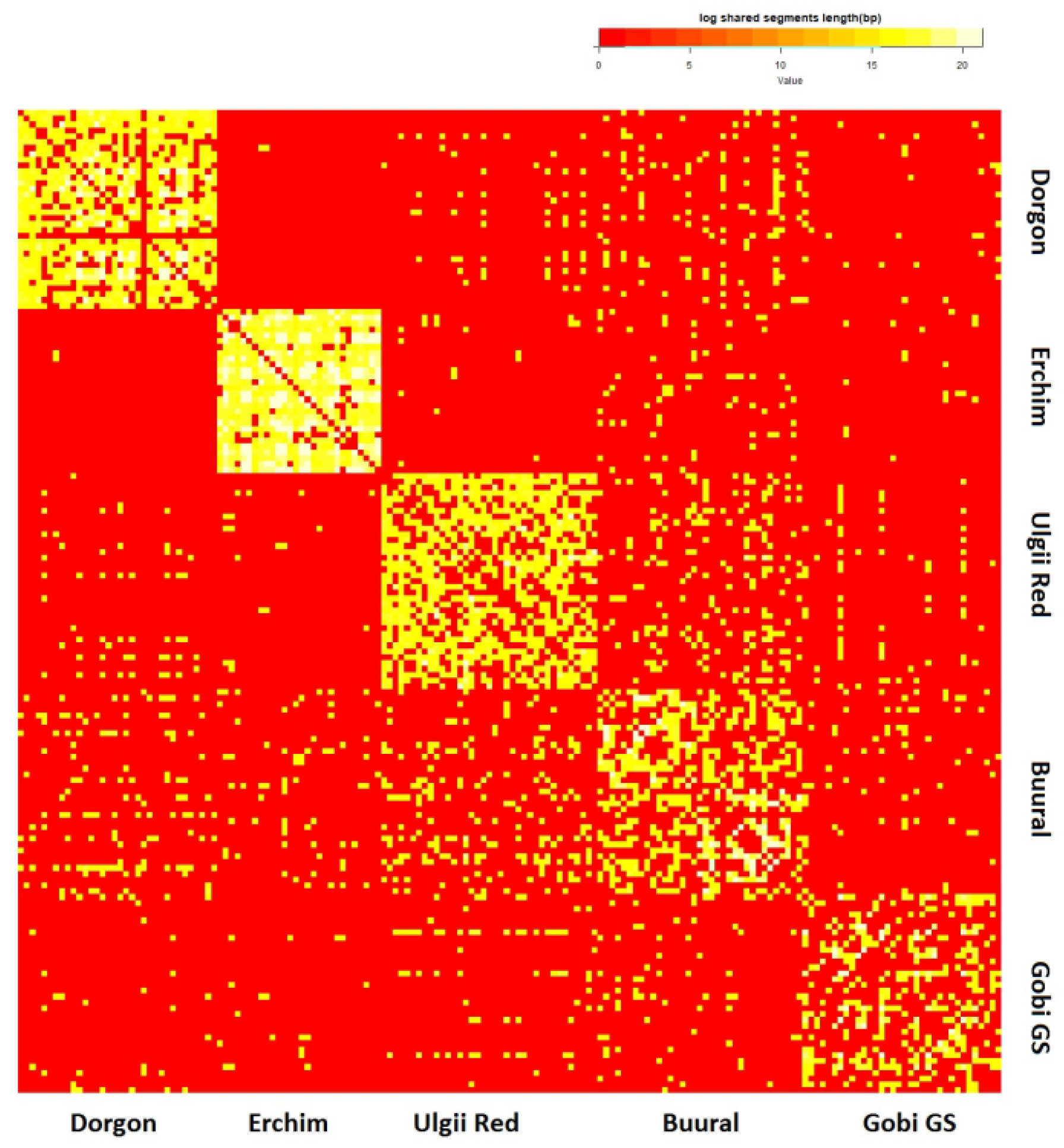
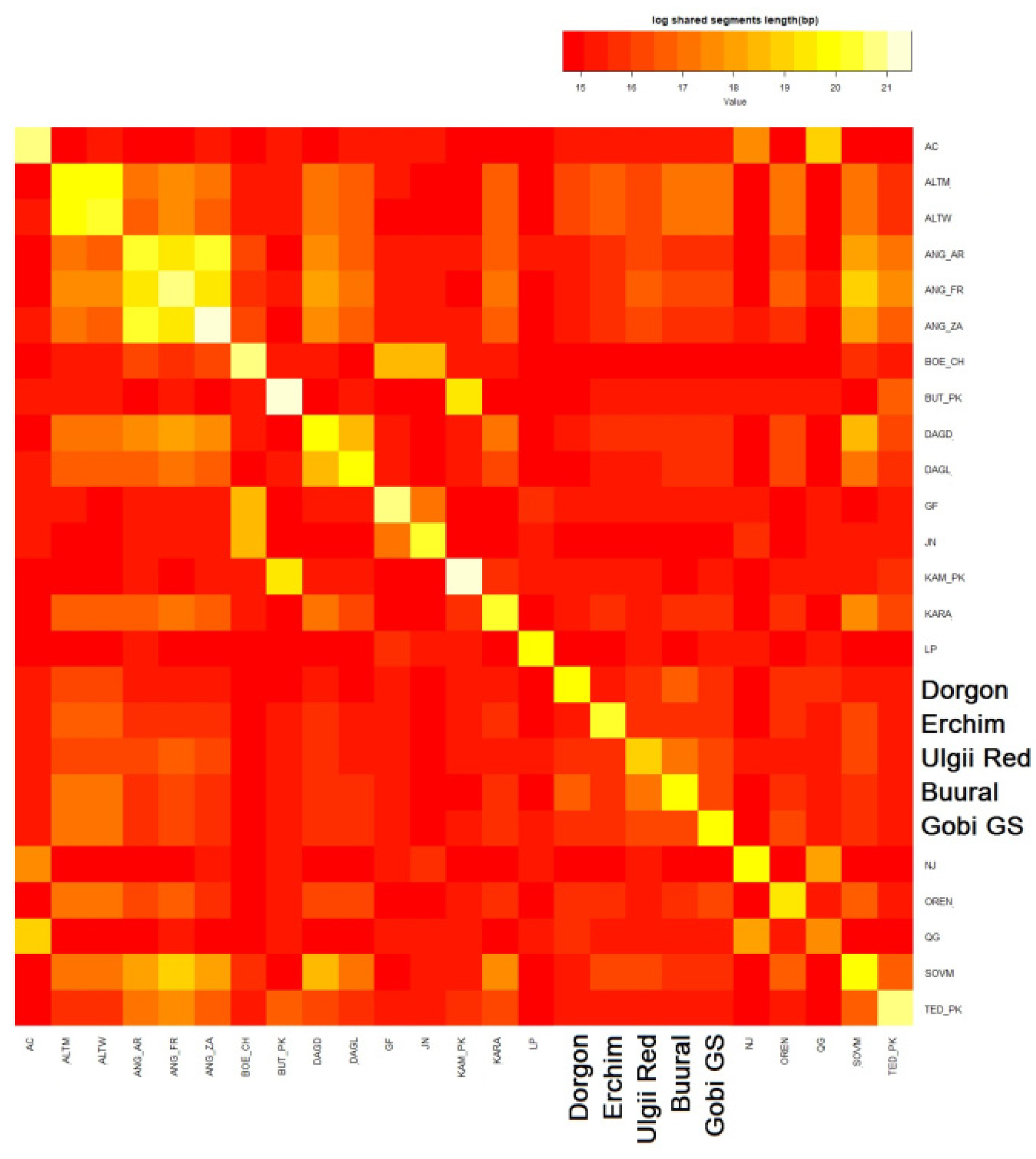
3.4.5. Haplotype Sharing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bélanger, J.; Pilling, D. The State of the World’s Biodiversity for Food and Agriculture; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2019. [Google Scholar]
- Joy, A.; Dunshea, F.R.; Leury, B.J.; Clarke, I.J.; DiGiacomo, K.; Chauhan, S.S. Resilience of small ruminants to climate change and increased environmental temperature: A review. Animals 2020, 10, 867. [Google Scholar] [CrossRef]
- Galal, S. Biodiversity in goats. Small Rumin. Res. 2005, 60, 75–81. [Google Scholar] [CrossRef]
- Olschewsky, A.; Hinrichs, D. An Overview of the Use of Genotyping Techniques for Assessing Genetic Diversity in Local Farm Animal Breeds. Animals 2021, 11, 2016. [Google Scholar] [CrossRef]
- Stella, A.; Nicolazzi, E.L.; Van Tassell, C.P.; Rothschild, M.F.; Colli, L.; Rosen, B.D.; Sonstegard, T.S.; Crepaldi, P.; Tosser-Klopp, G.; Joost, S. AdaptMap: Exploring Goat Diversity and Adaptation; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Cortellari, M.; Barbato, M.; Talenti, A.; Bionda, A.; Carta, A.; Ciampolini, R.; Ciani, E.; Crisà, A.; Frattini, S.; Lasagna, E. The climatic and genetic heritage of Italian goat breeds with genomic SNP data. Sci. Rep. 2021, 11, 10986. [Google Scholar] [CrossRef]
- Pogorevc, N.; Simčič, M.; Khayatzadeh, N.; Sölkner, J.; Berger, B.; Bojkovski, D.; Zorc, M.; Dovč, P.; Medugorac, I.; Horvat, S. Post-genotyping optimization of dataset formation could affect genetic diversity parameters: An example of analyses with alpine goat breeds. BMC Genom. 2021, 22, 546. [Google Scholar] [CrossRef]
- Ganbold, O.; Lee, S.-H.; Paek, W.K.; Munkhbayar, M.; Seo, D.; Manjula, P.; Khujuu, T.; Purevee, E.; Lee, J.H. Mitochondrial DNA variation and phylogeography of native Mongolian goats. Asian-Australas. J. Anim. Sci. 2020, 33, 902. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.P.; Davi, N.K.; D’Arrigo, R.D.; Skees, J.; Nachin, B.; Leland, C.; Lyon, B.; Wang, S.-Y.; Byambasuren, O. Dzuds, Droughts, and Livestock Mortality in Mongolia. Environ. Res. Lett. 2015, 10, 074012. [Google Scholar] [CrossRef]
- Colli, L.; Milanesi, M.; Talenti, A.; Bertolini, F.; Chen, M.; Crisà, A.; Daly, K.G.; Del Corvo, M.; Guldbrandtsen, B.; Lenstra, J.A. Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes. Genet. Sel. Evol. 2018, 50, 58. [Google Scholar] [CrossRef] [Green Version]
- Beketov, S.; Piskunov, A.; Voronkova, V.; Petrov, S.; Kharzinova, V.; Dotsev, A.; Zinovieva, N.; Selionova, M.; Stolpovsky, Y.A. Genetic diversity and phylogeny of fleece-bearing goats of Central and Middle Asia. Russ. J. Genet. 2021, 57, 816–824. [Google Scholar] [CrossRef]
- Voronkova, V.; Piskunov, A.; Nikolaeva, E.; Semina, M.; Konorov, E.; Stolpovsky, Y.A. Haplotype Diversity of Mongolian and Tuvan Goat Breeds (Capra hircus) Based on mtDNA and Y-Chromosome Polymorphism. Russ. J. Genet. 2021, 57, 1170–1178. [Google Scholar] [CrossRef]
- Porter, V. Mason’s World Dictionary of Livestock Breeds, Types and Varieties, 6th ed.; CABI: West Sussex, UK, 2020; ISBN 978-1-78924-153-2. [Google Scholar]
- Takahashi, H.; Nyamsamba, D.; Mandakh, B.; Zagdsuren, Y.; Amano, T.; Nomura, K.; Yokohama, M.; Ito, S.-I.; Minezawa, M. Genetic structure of Mongolian goat populations using microsatellite loci analysis. Asian-Australas. J. Anim. Sci. 2008, 21, 947–953. [Google Scholar] [CrossRef]
- Chang, C.; Chow, C.; Tellier, L.; Vattikuti, S.; Purcell, S.; Lee, J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Reyer, H.; Sölkner, J.; Fornara, M.S.; Aybazov, A.-M.M.; Wimmers, K.; Brem, G.; Zinovieva, N.A. SNP-based genotyping provides insight into the West Asian origin of Russian local goats. Front. Genet. 2021, 12, 1133. [Google Scholar] [CrossRef]
- Berihulay, H.; Li, Y.; Liu, X.; Gebreselassie, G.; Islam, R.; Liu, W.; Jiang, L.; Ma, Y. Genetic diversity and population structure in multiple Chinese goat populations using a SNP panel. Anim. Genet. 2019, 50, 242–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiveRsity: An R Package for the Estimation and Exploration of Population Genetics Parameters and Their Associated Errors-Keenan-2013-Methods in Ecology and Evolution-Wiley Online Library. Available online: https://besjournals.onlinelibrary.wiley.com/doi/10.1111/2041-210X.12067 (accessed on 10 November 2021).
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbin, L.J.; Liu, A.Y.H.; Bishop, S.C.; Woolliams, J.A. Estimation of historical effective population size using linkage disequilibria with marker data. J. Anim. Breed. Genet. 2012, 129, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Sved, J.A.; Feldman, M.W. Correlation and probability methods for one and two loci. Theor. Popul. Biol. 1973, 4, 129–132. [Google Scholar] [CrossRef]
- Meyermans, R.; Gorssen, W.; Buys, N.; Janssens, S. How to study runs of homozygosity using PLINK? A guide for analyzing medium density SNP data in livestock and pet species. BMC Genom. 2020, 21, 94. [Google Scholar] [CrossRef]
- Dunn.Test Function-Rdocumentation. Available online: https://www.rdocumentation.org/packages/dunn.test/versions/1.3.5/topics/dunn.test (accessed on 10 November 2021).
- Delaneau, O.; Marchini, J.; Zagury, J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods 2012, 9, 179–181. [Google Scholar] [CrossRef]
- Browning, B.L.; Browning, S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 2013, 194, 459–471. [Google Scholar] [CrossRef] [Green Version]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Behr, A.A.; Liu, K.Z.; Liu-Fang, G.; Nakka, P.; Ramachandran, S. Pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 2016, 32, 2817–2823. [Google Scholar] [CrossRef] [Green Version]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Bolormaa, S.; Ruvinsky, A.; Walkden-Brown, S.; Van der Werf, J. Genetic relationships among Australian and Mongolian fleece-bearing goats. Asian-Australas. J. Anim. Sci. 2008, 21, 1535–1543. [Google Scholar] [CrossRef]
- Meuwissen, T.H. Accuracy of breeding values of’unrelated’individuals predicted by dense SNP genotyping. Genet. Sel. Evol. 2009, 41, 35. [Google Scholar] [CrossRef] [Green Version]
- Ryman, N.; Laikre, L.; Hössjer, O. Do estimates of contemporary effective population size tell us what we want to know? Mol. Ecol. 2019, 28, 1904–1918. [Google Scholar] [CrossRef] [Green Version]
- Green, R.E.; Gilbert, G.; Wilson, J.D.; Jennings, K. Implications of the prevalence and magnitude of sustained declines for determining a minimum threshold for favourable population size. PLoS ONE 2020, 15, e0228742. [Google Scholar] [CrossRef] [Green Version]
- Islam, R.; Li, Y.; Liu, X.; Berihulay, H.; Abied, A.; Gebreselassie, G.; Ma, Q.; Ma, Y. Genome-wide runs of homozygosity, effective population size, and detection of positive selection signatures in six Chinese goat breeds. Genes 2019, 10, 938. [Google Scholar] [CrossRef] [Green Version]
- Visser, C.; Lashmar, S.F.; Van Marle-Köster, E.; Poli, M.A.; Allain, D. Genetic Diversity and Population Structure in South African, French and Argentinian Angora Goats from Genome-Wide SNP Data. PLoS ONE 2016, 11, e0154353. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, X.; Li, M.; Li, Y.; Yang, Z.; Wang, X.; Pan, X.; Gong, M.; Zhang, Y.; Guo, Y.; et al. The Origin of Domestication Genes in Goats. Sci. Adv. 2020. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, N.G.; Ernst, N.K. Animal Genetic Resources of the USSR. Available online: https://www.fao.org/3/ah759e/ah759e00.htm (accessed on 10 November 2021).
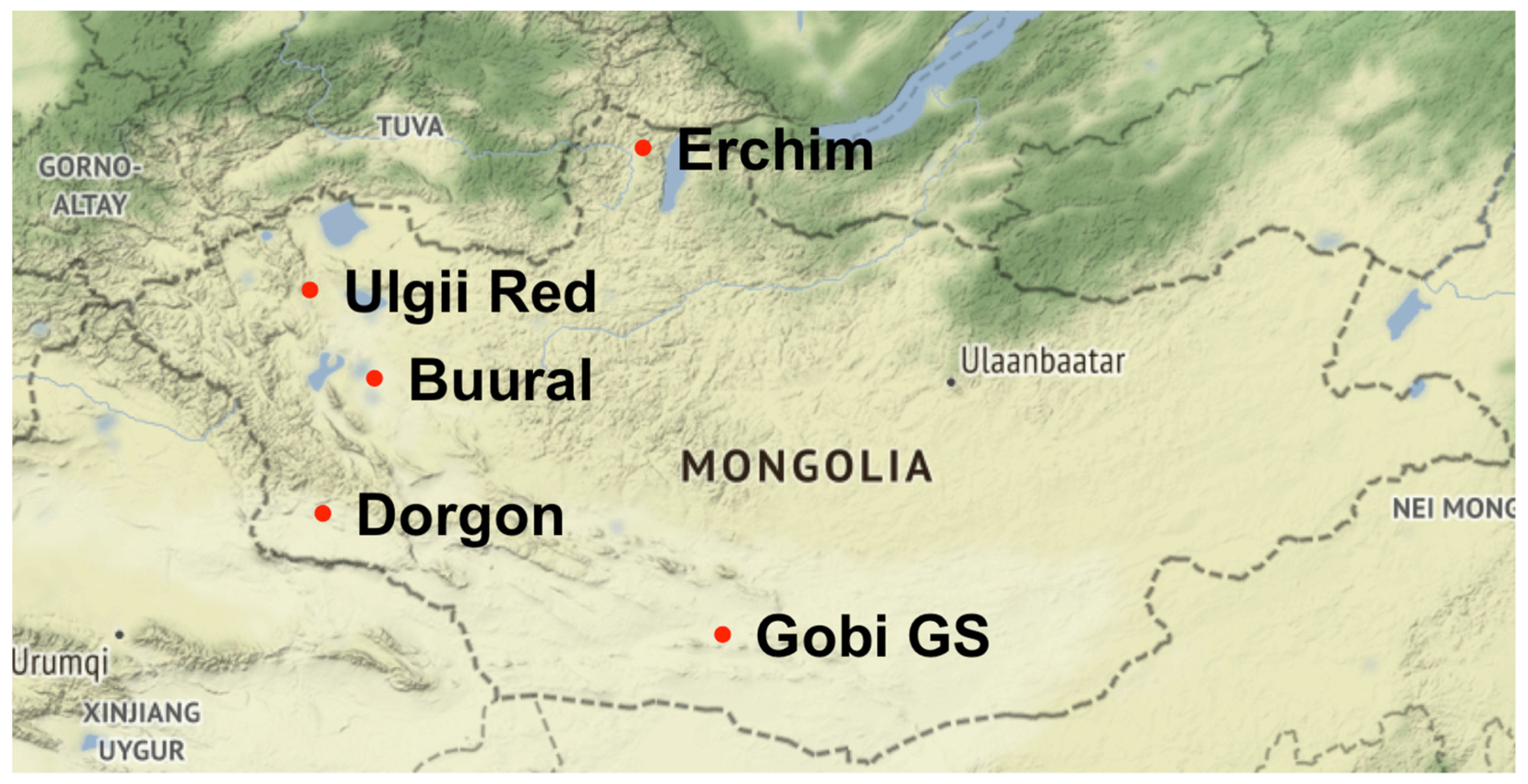
| Breed (Code) | n 1 | Prefecture | Coordinates | Environment | Areas of Benefit | Color |
|---|---|---|---|---|---|---|
| Erchim (EB) | 28 | Huvsgul | 51.42, 99.73 | Mountain areas with extreme continental climate | Milk, meat, and cashmere | Black |
| Dorgon (DG) | 34 | Hovd | 45.80, 92.29 | Western steppes and mountains | Cashmere, meat, and milk | White and red |
| Buural (ZB) | 35 | Zavkhan | 47.95, 93.49 | Gobi-like areas of Great Lake Valleys | Cashmere and milk | Dark brown and black |
| Ulgii Red (UR) | 37 | Uvs | 49.31, 91.99 | Both mountain and steppe areas | Cashmere, meat, and milk | Red and brown |
| Gob Gurvan Saikhan (GGS) | 33 | Gurvan Saikhan | 43.80, 101.58 | South Gobi, Three Beauties’ mountains | Cashmere | Various. Mostly red and white |
| Breed | Ho | He | dH | Fis | FROH |
|---|---|---|---|---|---|
| Dorgon | 0.396 | 0.395 | 0.001 | −0.004 (−0.006; −0.002) | 0.014 |
| Buural | 0.406 | 0.405 | 0.001 | −0.002 (−0.004; 0.000) | 0.007 |
| Erchim | 0.393 | 0.388 | 0.005 | −0.013 (−0.015; −0.011) | 0.019 |
| Gobi GS | 0.400 | 0.410 | −0.010 | 0.023 (0.021; 0.025) | 0.023 |
| Ulgii Red | 0.397 | 0.399 | −0.002 | 0.004 (0.003; 0.005) | 0.013 |
| Breed | Dorgon | Buural | Erchim | Gobi GS | Ulgii Red |
|---|---|---|---|---|---|
| Dorgon | |||||
| Buural | 0.015 | ||||
| Erchim | 0.035 | 0.027 | |||
| Gobi GS | 0.017 | 0.009 | 0.027 | ||
| Ulgi Red | 0.018 | 0.009 | 0.028 | 0.012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukhina, V.; Svishcheva, G.; Voronkova, V.; Stolpovsky, Y.; Piskunov, A. Genetic Diversity, Population Structure and Phylogeny of Indigenous Goats of Mongolia Revealed by SNP Genotyping. Animals 2022, 12, 221. https://doi.org/10.3390/ani12030221
Mukhina V, Svishcheva G, Voronkova V, Stolpovsky Y, Piskunov A. Genetic Diversity, Population Structure and Phylogeny of Indigenous Goats of Mongolia Revealed by SNP Genotyping. Animals. 2022; 12(3):221. https://doi.org/10.3390/ani12030221
Chicago/Turabian StyleMukhina, Vera, Gulnara Svishcheva, Valery Voronkova, Yurii Stolpovsky, and Aleksei Piskunov. 2022. "Genetic Diversity, Population Structure and Phylogeny of Indigenous Goats of Mongolia Revealed by SNP Genotyping" Animals 12, no. 3: 221. https://doi.org/10.3390/ani12030221
APA StyleMukhina, V., Svishcheva, G., Voronkova, V., Stolpovsky, Y., & Piskunov, A. (2022). Genetic Diversity, Population Structure and Phylogeny of Indigenous Goats of Mongolia Revealed by SNP Genotyping. Animals, 12(3), 221. https://doi.org/10.3390/ani12030221






