GPX2 Gene Affects Feed Efficiency of Pigs by Inhibiting Fat Deposition and Promoting Muscle Development
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Data Collection
2.2. Genomic DNA and RNA Isolation
2.3. Secondary Structure Predicted by RNAsnp
2.4. 3T3-L1 Cell Culture and Adipogenic Differentiation
2.5. C2C12 Cell Culture and Myogenic Differentiation
2.6. GPX2 Adenovirus Overexpressed
2.7. EdU Proliferation Staining
2.8. Oil Red O Staining and Quantification
2.9. Immunofluorescence Staining
2.10. Quantitative Real-Time PCR (qRT-PCR)
2.11. Western Blotting Analysis
2.12. Statistical Analysis
3. Results
3.1. GPX2 Polymorphisms and Genotype Frequencies and Haplotype Determination
3.2. Prediction of the SNPs’ Effect on the Local RNA Secondary Structure
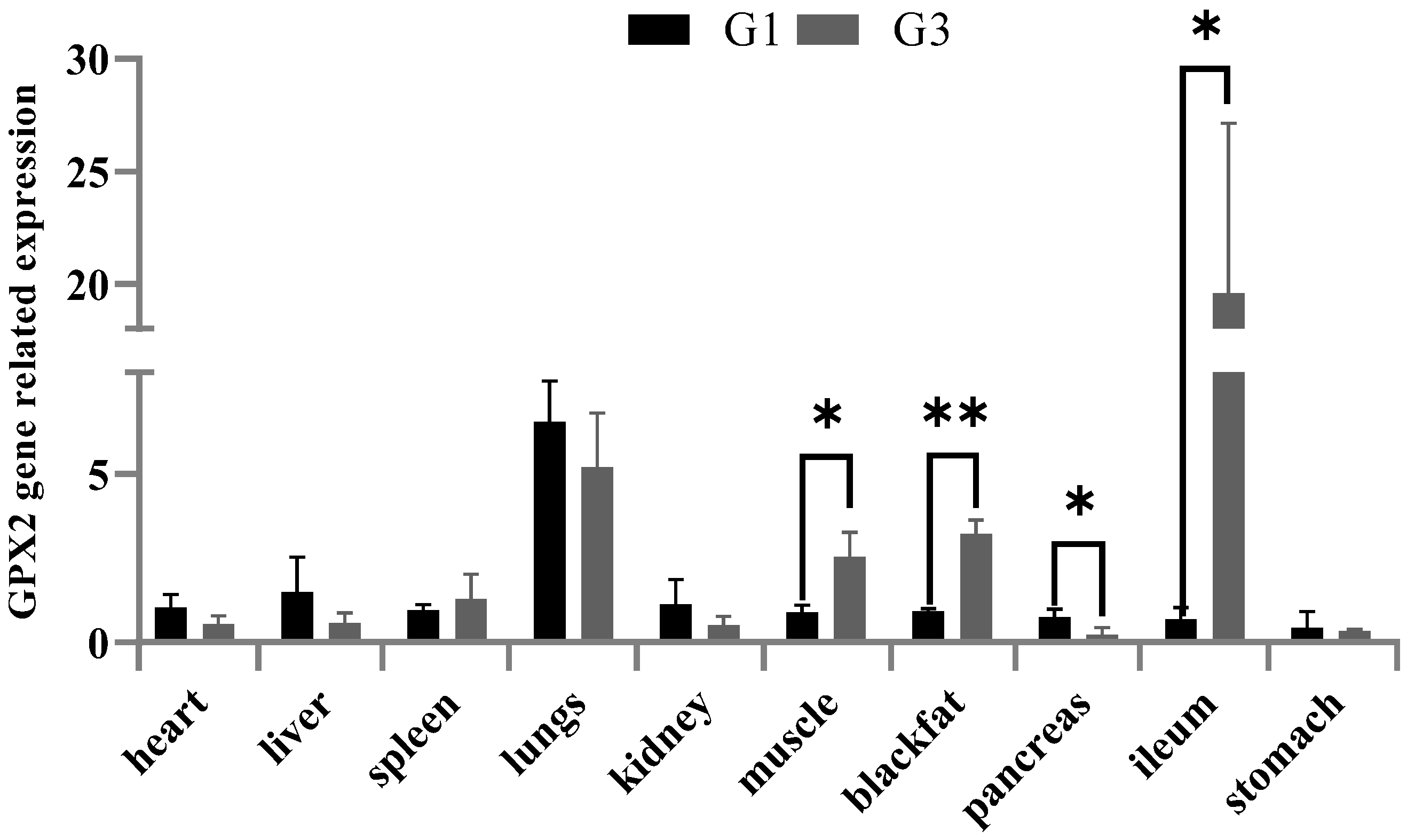
3.3. Genotype and ALGA0043483 Association with Feed Efficiency Traits Expression Analysed in Different Tissues
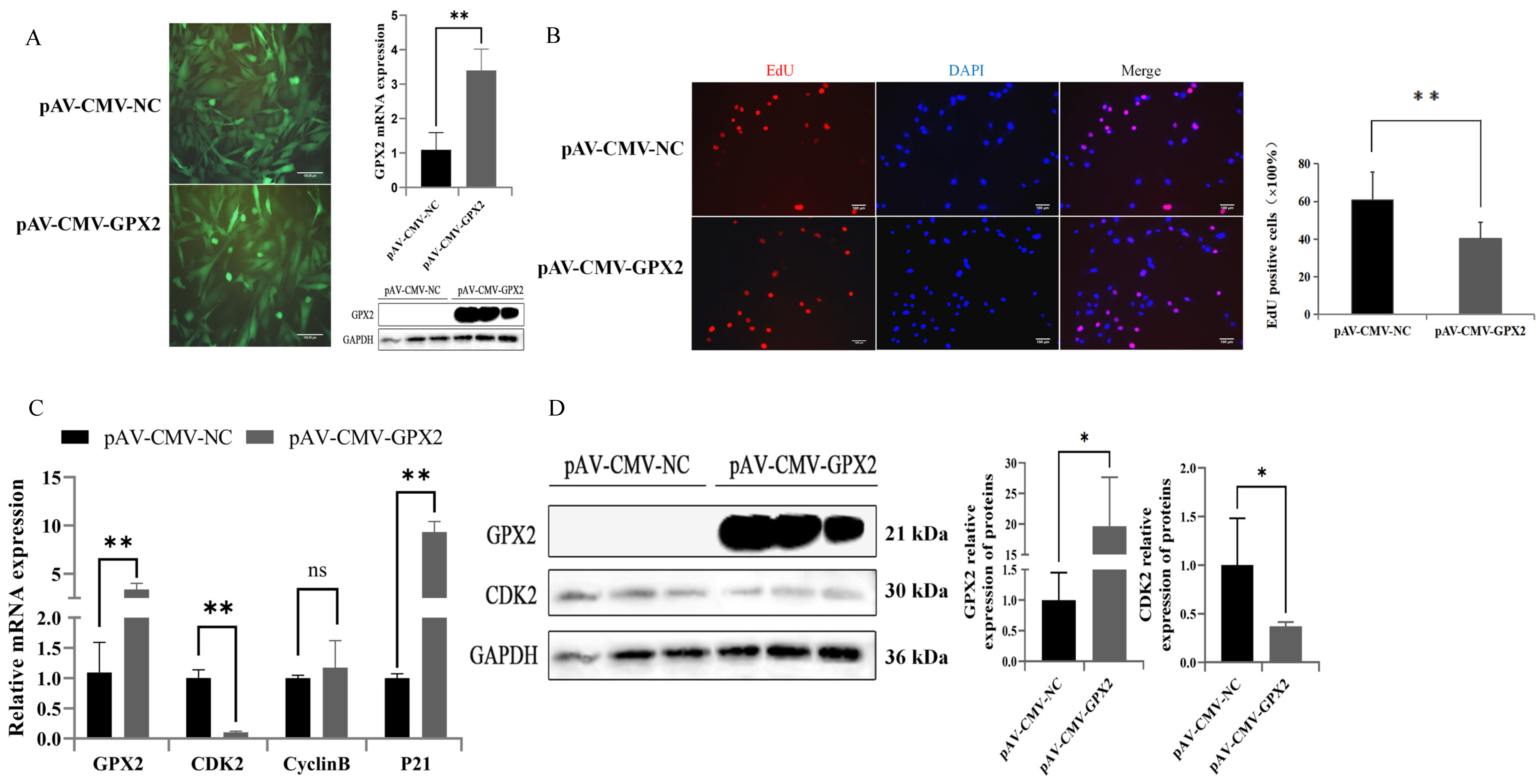
3.4. Overexpression of GPX2 May Inhibit the Proliferation of 3T3-L1
3.5. GPX2 Promoted Lipid Degradation and Inhibit Adipogenic Differentiation of 3T3-L1
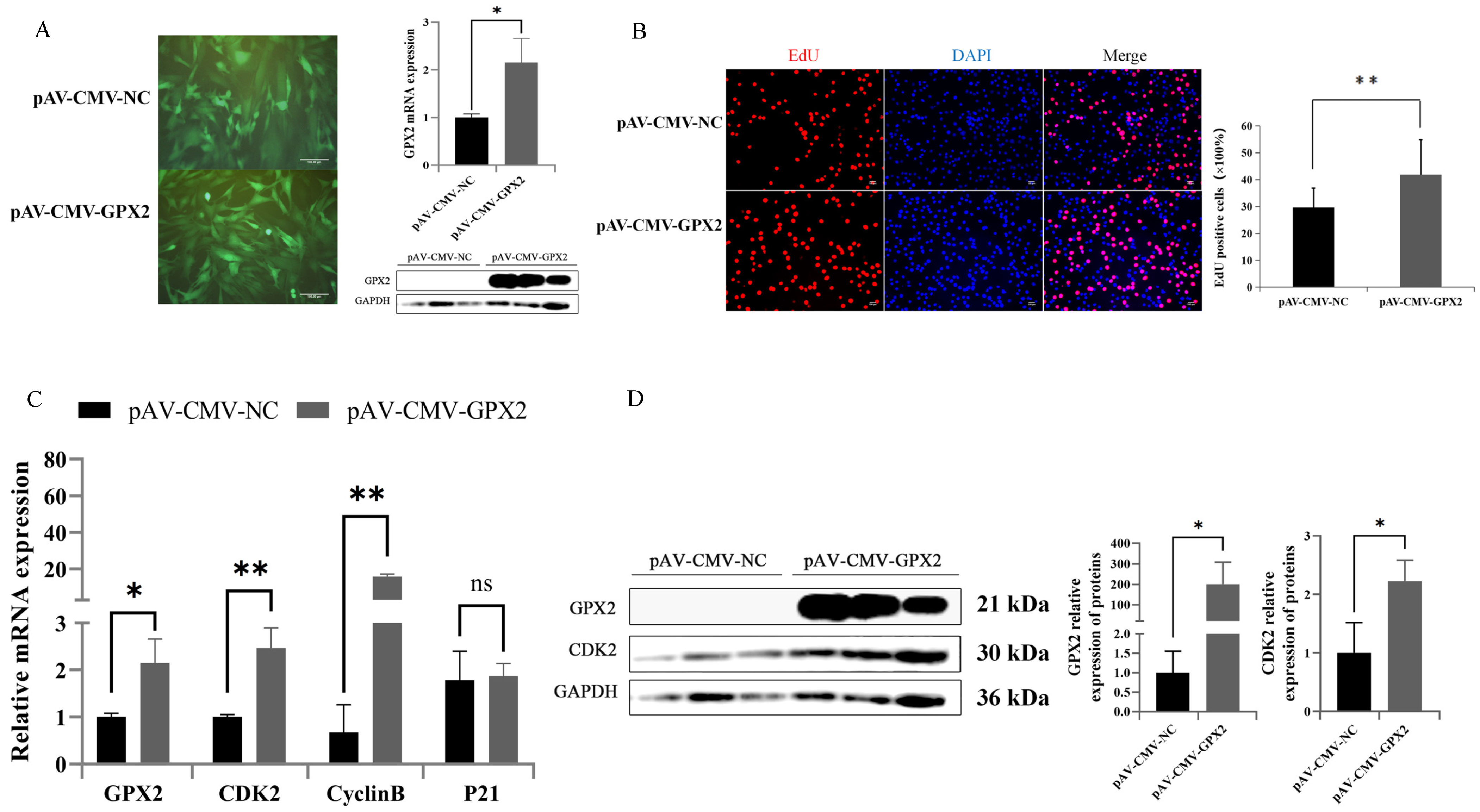
3.6. Overexpression GPX2 Induces the Proliferation of C2C12
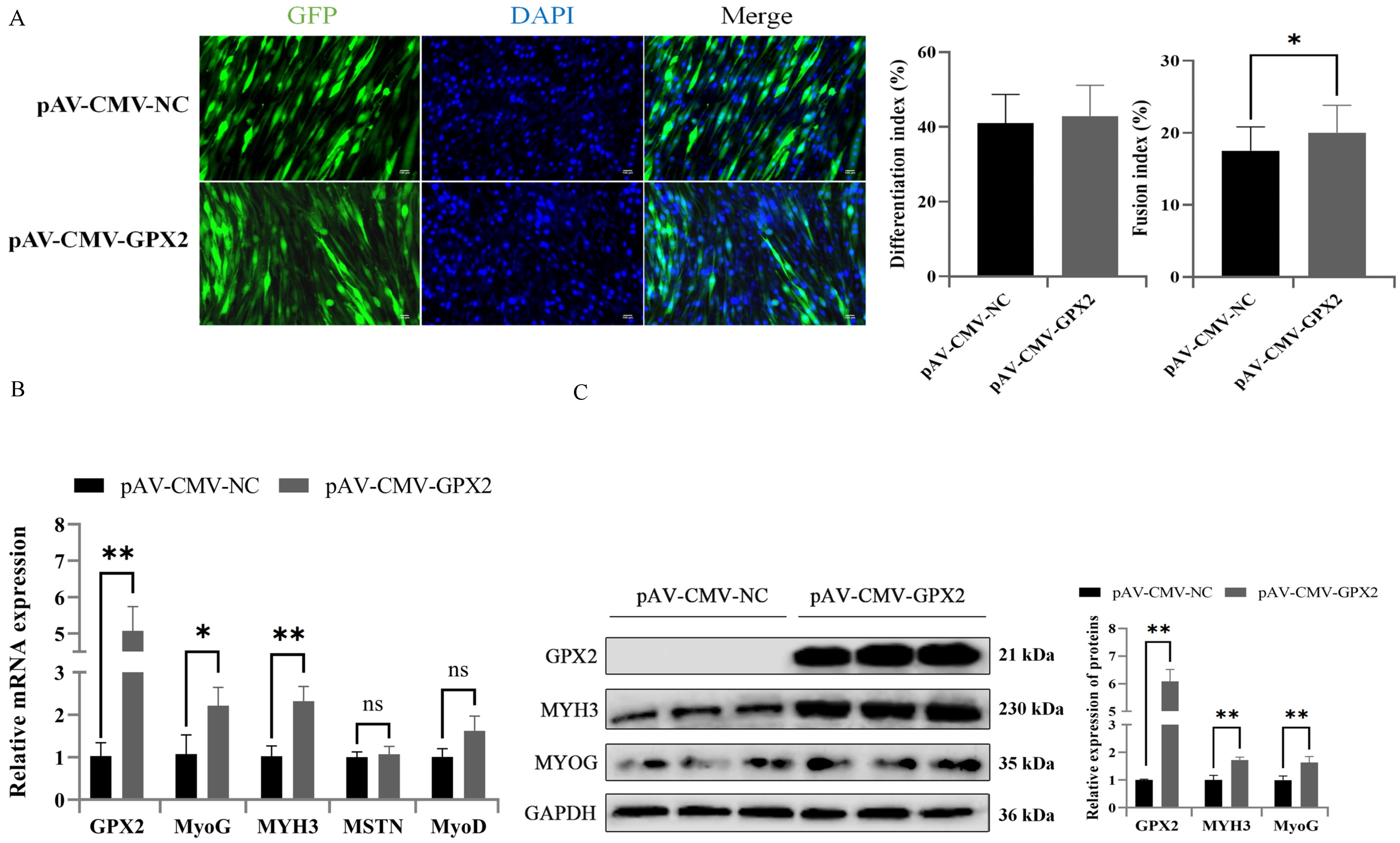
3.7. Overexpression GPX2 Promoted the Myoblastic Differentiation of C2C12
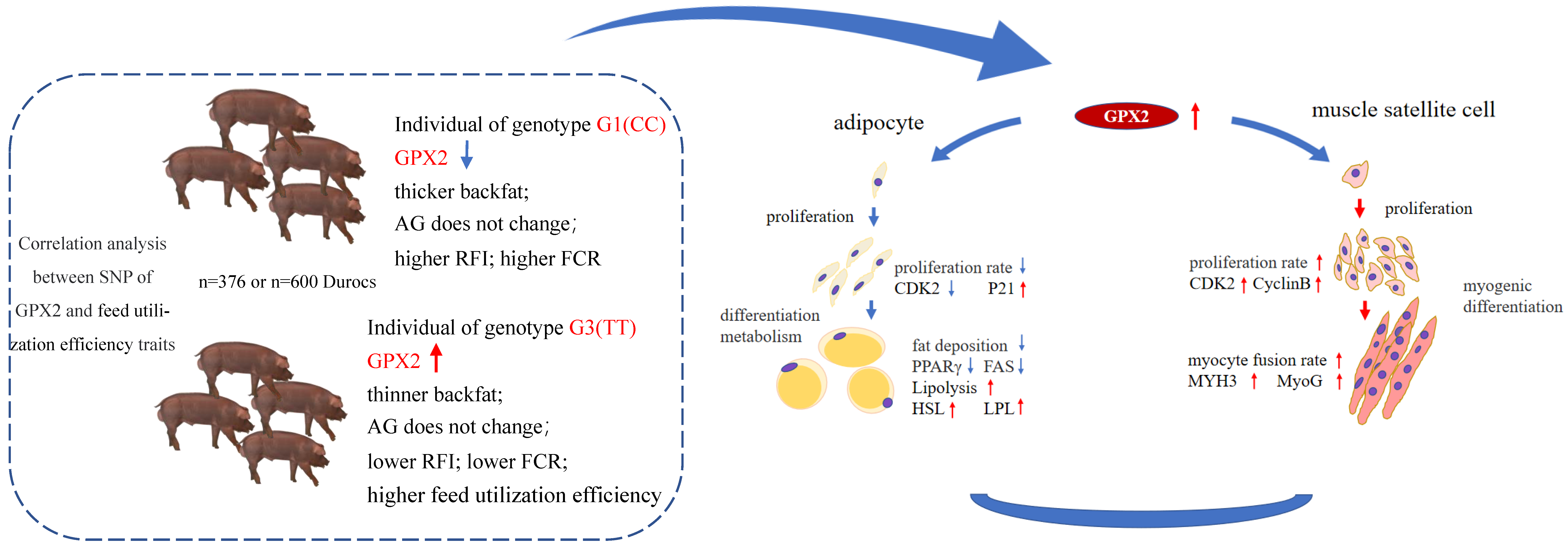
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brigelius-Flohe, R.; Flohe, L. Regulatory Phenomena in the Glutathione Peroxidase Superfamily. Antioxid. Redox Signal. 2020, 33, 498–516. [Google Scholar] [CrossRef] [PubMed]
- Luise, D.; Motta, V.; Bertocchi, M.; Salvarani, C.; Clavenzani, P.; Fanelli, F.; Pagotto, U.; Bosi, P.; Trevisi, P. Effect of Mucine 4 and Fucosyltransferase 1 genetic variants on gut homoeostasis of growing healthy pigs. J. Anim. Physiol. Anim. Nutr. 2019, 103, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Lennicke, C.; Rahn, J.; Wickenhauser, C.; Lichtenfels, R.; Müller, A.S.; Wessjohann, L.A.; Kipp, A.P.; Seliger, B. Loss of epithelium-specific GPx2 results in aberrant cell fate decisions during intestinal differentiation. Oncotarget 2018, 9, 539. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jia, Y.; Li, Q.; Son, K.; Hamilton, C.; Burris, W.R.; Bridges, P.J.; Stromberg, A.J.; Matthews, J.C. Glutathione content and expression of proteins involved with glutathione metabolism differs in longissimus dorsi, subcutaneous adipose, and liver tissues of finished vs. growing beef steers. J. Anim. Sci. 2018, 96, 5152–5165. [Google Scholar] [CrossRef] [PubMed]
- Kipp, A.P.; Müller, M.F.; Göken, E.M.; Deubel, S.; Brigelius-Flohé, R. The selenoproteins GPx2, TrxR2 and TrxR3 are regulated by Wnt signalling in the intestinal epithelium. Biophys. Acta 2012, 1820, 1588–1596. [Google Scholar] [CrossRef]
- Li, Y.-P.; Lin, R.; Chang, M.-Z.; Ai, Y.-J.; Ye, S.-P.; Han, H.-M.; Zhang, Y.-Y.; Mou, H.; Mu, R.-H.; Guo, X. The Effect of GPX2 on the Prognosis of Lung Adenocarcinoma Diagnosis and Proliferation, Migration, and Epithelial Mesenchymal Transition. J. Oncol. 2022, 2022, 7379157. [Google Scholar] [CrossRef]
- Kobayashi, H.; Matsuda, M.; Fukuhara, A.; Komuro, R.; Shimomura, I. Dysregulated glutathione metabolism links to impaired insulin action in adipocytes. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1326–E1334. [Google Scholar] [CrossRef]
- Onteru, S.K.; Gorbach, D.M.; Young, J.M.; Garrick, D.J.; Dekkers, J.C.; Rothschild, M.F. Whole Genome Association Studies of Residual Feed Intake and Related Traits in the Pig. PLoS ONE 2013, 8, e61756. [Google Scholar] [CrossRef]
- Demeure, O.; Sanchez, M.P.; Riquet, J.; Iannuccelli, N.; Demars, J.; Fève, K.; Kernaléguen, L.; Gogué, J.; Billon, Y.; Caritez, J. Exclusion of the swine leukocyte antigens as candidate region and reduction of the position interval for the Sus scrofa chromosome 7 QTL affecting growth and fatness. J. Anim. Sci. 2005, 83, 1979–1987. [Google Scholar] [CrossRef][Green Version]
- Ponsuksili, S.; Chomdej, S.; Murani, E.; Bläser, U.; Schreinemachers, H.J.; Schellander, K.; Wimmers, K. SNP detection and genetic mapping of porcine genes encoding enzymes in hepatic metabolic pathways and evaluation of linkage with carcass traits. Anim. Genet. 2005, 36, 477–483. [Google Scholar] [CrossRef]
- Kamendulis, L.M.; Wu, Q.; Sandusky, G.E.; Hocevar, B.A. Perfluorooctanoic acid exposure triggers oxidative stress in the mouse pancreas. Toxicol. Rep. 2014, 1, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. Nutrient Requirements of Swine; National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Li, F.; Sun, J.; Men, J.; Li, H.; Wang, G.; Wang, S.; Wang, J. Selection and validation of reference genes for RT-qPCR analysis in the pericarp of Litchi chinensis. Biol. Plantarum. 2022, 66, 103–111. [Google Scholar] [CrossRef]
- Mathews, D.H.; Turner, D.H.; Zuker, M. RNA secondary structure prediction. Curr. Protoc. Nucleic. Acid. Chem. 2007, 28, 11.2.1–11.2.19. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-C.; Hsu, H.-E.; Lai, J.-C.; Chen, H.-C.; Chuang, S.-W.; Lee, M.-C. Strategy to Estimate Sample Sizes to Justify the Association between MMP1 SNP and Osteoarthritis. Genes 2022, 13, 1084. [Google Scholar] [CrossRef] [PubMed]
- Sabarinathan, R.; Tafer, H.; Seemann, S.E.; Hofacker, I.L.; Stadler, P.F.; Gorodkin, J. RNA snp: Efficient detection of local RNA secondary structure changes induced by SNP s. Hum. Mutat. 2013, 34, 546–556. [Google Scholar] [CrossRef]
- Jurado, J.; Fuentes-Almagro, C.A.; Prieto-Álamo, M.J.; Pueyo, C. Alternative splicing of c-fos pre-mRNA: Contribution of the rates of synthesis and degradation to the copy number of each transcript isoform and detection of a truncated c-Fos immunoreactive species. BMC. Mol. Biol. 2007, 8, 83. [Google Scholar] [CrossRef]
- Das, S.; Coward, P.; Dasgupta, A. A small yeast RNA selectively inhibits internal initiation of translation programmed by poliovirus RNA: Specific interaction with cellular proteins that bind to the viral 5’-untranslated region. J. Virol. 1994, 68, 7200–7211. [Google Scholar] [CrossRef]
- Kipp, A.; Banning, A.; Brigelius-Flohé, R. Activation of the glutathione peroxidase 2 (GPx2) promoter by β-catenin. Biol. Chem. 2007, 388, 1027–1033. [Google Scholar] [CrossRef]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef]
- Esworthy, R.S.; Swiderek, K.M.; Ho, Y.-S.; Chu, F.-F. Selenium-dependent glutathione peroxidase-GI is a major glutathione peroxidase activity in the mucosal epithelium of rodent intestine. Biochim. Biophys. Acta 1998, 1381, 213–226. [Google Scholar] [CrossRef]
- Makki, K.; Froguel, P.; Wolowczuk, I. Adipose tissue in obesity-related inflammation and insulin resistance: Cells, cytokines, and chemokines. ISRN. Inflamm. 2013, 2013, 139239. [Google Scholar] [CrossRef] [PubMed]
- El-Magd, N.F.A.; Barbosa, P.O.; Nick, J.; Covalero, V.; Grignetti, G.; Bermano, G. Selenium, as selenite, prevents adipogenesis by modulating selenoproteins gene expression and oxidative stress–related genes. Nutrition 2022, 93, 111424. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, F.; Farias, C.L.A.; Rea, S.; Capuani, B.; Feraco, A.; Coppola, A.; Mammi, C.; Pastore, D.; Abete, P.; Rovella, V. Tyrosol may prevent obesity by inhibiting adipogenesis in 3T3-L1 preadipocytes. Oxid. Med. Cell. Longev. 2020, 2020, 4794780. [Google Scholar] [CrossRef] [PubMed]
- Su, H.-m.; Feng, L.-n.; Zheng, X.-d.; Chen, W. Myricetin protects against diet-induced obesity and ameliorates oxidative stress in C57BL/6 mice. J. Zhejiang Univ. Sci. B 2016, 17, 437–446. [Google Scholar] [CrossRef]
- Cao, C.; Sun, S.; Li, J.; Song, C.; Meng, Q.; Shi, B.; Shan, A. Lycopene modulates lipid metabolism in rats and their offspring under a high-fat diet. Food Funct. 2021, 12, 8960–8975. [Google Scholar] [CrossRef]
- Fang, K.; Wu, F.; Chen, G.; Dong, H.; Li, J.; Zhao, Y.; Xu, L.; Zou, X.; Lu, F. Diosgenin ameliorates palmitic acid-induced lipid accumulation via AMPK/ACC/CPT-1A and SREBP-1c/FAS signaling pathways in LO2 cells. Altern. Med. 2019, 19, 255. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, S.; Li, Z.; Zhao, X.; Li, W.; Sun, Y.; Zhang, Z.; Ling, W.; Feng, X. Effects and mechanisms of resveratrol on the amelioration of oxidative stress and hepatic steatosis in KKAy mice. Nutr. Metab. 2014, 11, 35. [Google Scholar] [CrossRef]
- Okuno, Y.; Fukuhara, A.; Hashimoto, E.; Kobayashi, H.; Kobayashi, S.; Otsuki, M.; Shimomura, I. Oxidative stress inhibits healthy adipose expansion through suppression of SREBF1-mediated lipogenic pathway. Diabetes 2018, 67, 1113–1127. [Google Scholar] [CrossRef]
- Kaneto, H.; Kajimoto, Y.; Fujitani, Y.; Matsuoka, T.; Sakamoto, K.; Matsuhisa, M.; Yamasaki, Y.; Hori, M. Oxidative stress induces p21 expression in pancreatic islet cells: Possible implication in beta-cell dysfunction. Diabetologia 1999, 42, 1093–1097. [Google Scholar] [CrossRef]
- Mishra, P.K.; Raghuram, G.V.; Panwar, H.; Jain, D.; Pandey, H.; Maudar, K.K. Mitochondrial oxidative stress elicits chromosomal instability after exposure to isocyanates in human kidney epithelial cells. Free. Radic. Res. 2009, 43, 718–728. [Google Scholar] [CrossRef]
- Drowley, L.; Okada, M.; Beckman, S.; Vella, J.; Keller, B.; Tobita, K.; Huard, J. Cellular antioxidant levels influence muscle stem cell therapy. Mol. Ther. 2010, 18, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Horiike, M.; Ogawa, Y.; Kawada, S. Effects of hyperoxia and hypoxia on the proliferation of C2C12 myoblasts. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021, 321, R572–R587. [Google Scholar] [CrossRef]
- Sivakumar, A.S.; Hwang, I. Effects of Sunphenon and Polyphenon 60 on proteolytic pathways, inflammatory cytokines and myogenic markers in H2O2-treated C2C12 cells. J. Biosci. 2015, 40, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, P.F.; Wu, Y.-C.M.; Staversky, R.J.; O’Reilly, M.A. p21Cip1 protects against oxidative stress by suppressing ER-dependent activation of mitochondrial death pathways. Free. Radic. Biol. Med. 2009, 46, 33–41. [Google Scholar] [CrossRef] [PubMed]
- El Haddad, M.; Jean, E.; Turki, A.; Hugon, G.; Vernus, B.; Bonnieu, A.; Passerieux, E.; Hamade, A.; Mercier, J.; Laoudj-Chenivesse, D. Glutathione peroxidase 3, a new retinoid target gene, is crucial for human skeletal muscle precursor cell survival. J. Cell. Sci. Pt. 2012, 125, 6147–6156. [Google Scholar] [CrossRef]
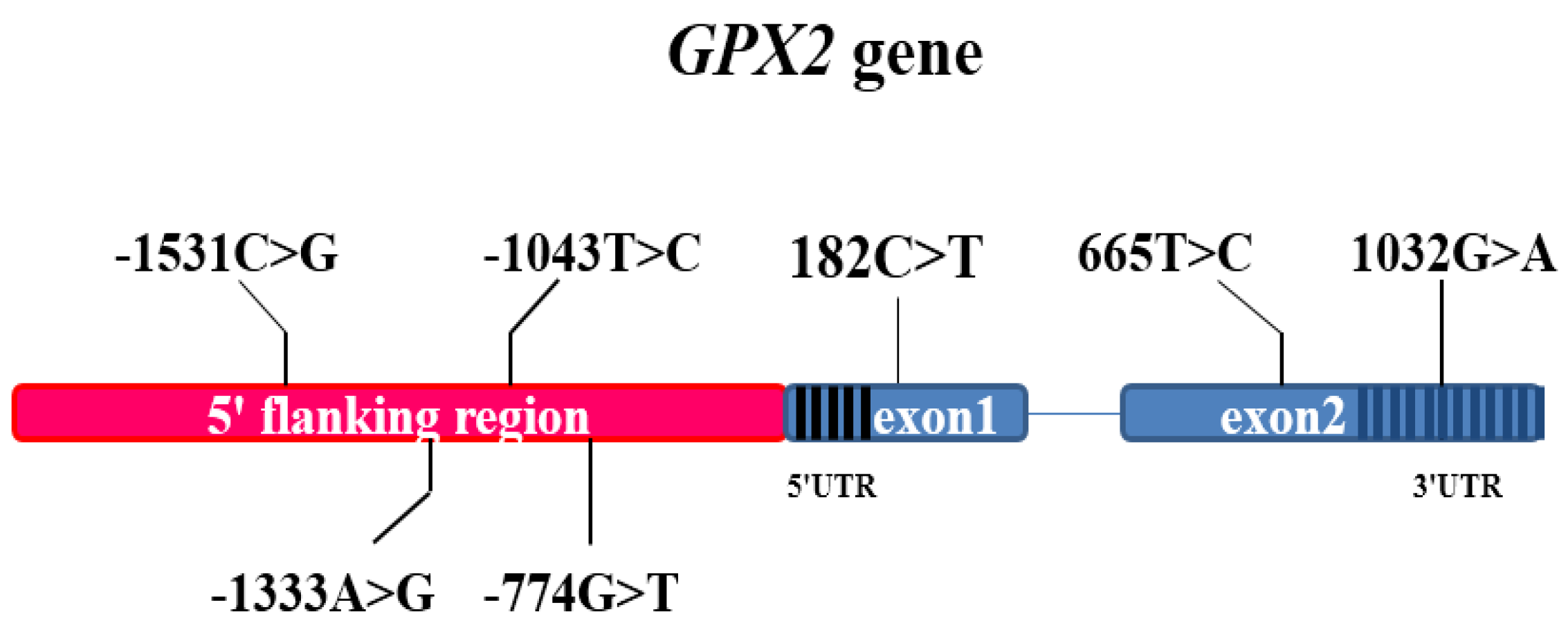

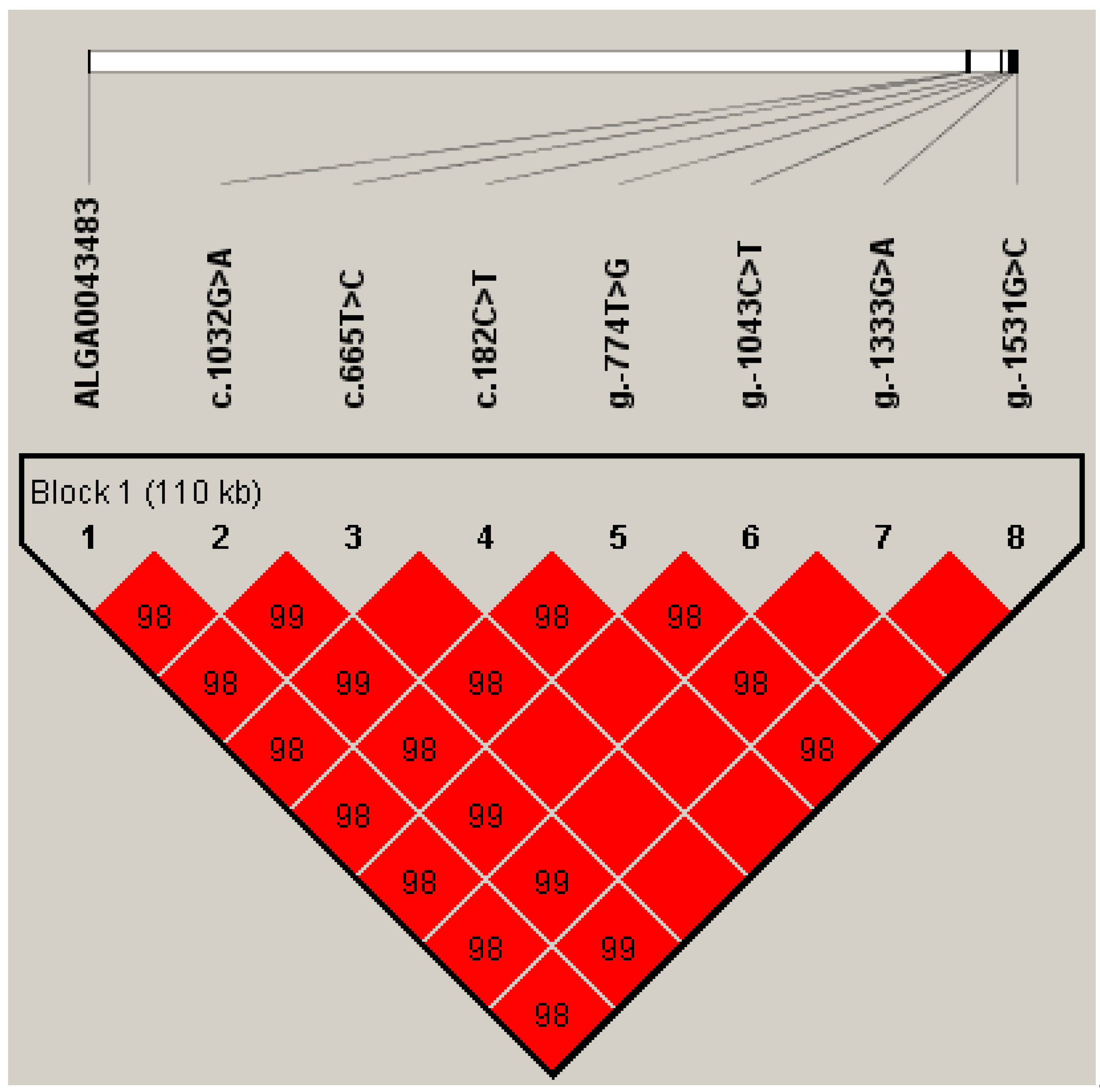

| SNP | Variant Location in Ssc7 | Target Region | Mutation Type | Ensemble Variant ID |
|---|---|---|---|---|
| c.1032 G > A | 89007403 | 3′-untranslated region | 3′-untranslated region | rs328629536 |
| c.665 T > C | 89007770 | Exon2-1 | Synonymous | rs318359750 |
| c.182 C > T | 89011439 | Exon1 | Synonymous | rs81218082 |
| g.-774 T > G | 89012396 | 5′flanking region 2 | 5′flanking region | rs339832589 |
| g.-1043C > T | 89012647 | 5′flanking region 2 | 5′flanking region | rs344114554 |
| g.-1333G > A | 89012937 | 5′flanking region 1 | 5′flanking region | rs318634907 |
| g.-1531G > C | 89013205 | 5′flanking region 1 | 5′flanking region | rs332828021 |
| ALGA0043483 T > C | Near with GPX2 1 | rs80875831 |
| SNP | Num | Genotype Frequency ∗ 100% | Allele Frequency ∗ 100% | He a | Ne b | PIC c | HWE d | |||
|---|---|---|---|---|---|---|---|---|---|---|
| c.1032G > A | 381 | GG | GA | AA | G | A | 0.50 | 1.98 | 0.37 | 1.727 |
| 18.63(71) | 53.28(200) | 28.08(106) | 45.28 | 54.72 | ||||||
| c.665T > C | 383 | TT | TC | CC | T | C | 0.50 | 1.98 | 0.37 | 2.156 |
| 18.54(71) | 53.26(203) | 28.20(107) | 45.17 | 54.83 | ||||||
| c.182C > T | 383 | CC | TC | TT | C | T | 0.50 | 1.98 | 0.37 | 2.013 |
| 18.54(71) | 53.26(203) | 28.20(107) | 45.17 | 54.83 | ||||||
| g.-774T > G | 383 | TT | TG | GG | T | G | 0.50 | 1.99 | 0.37 | 2.171 |
| 19.06(73) | 53.52(204) | 27.42(104) | 45.82 | 54.18 | ||||||
| g.-1043C > T | 383 | CC | CT | TT | C | T | 0.50 | 1.99 | 0.37 | 1.902 |
| 19.06(73) | 53.00(204) | 27.94(106) | 45.56 | 54.44 | ||||||
| g.-1333G > A | 382 | GG | GA | AA | G | A | 0.50 | 1.98 | 0.37 | 1.997 |
| 18.59(72) | 53.40(204) | 28.01(106) | 45.29 | 54.71 | ||||||
| g.-1531G > C | 382 | GG | GC | CC | G | C | 0.50 | 1.98 | 0.37 | 2.013 |
| 18.59(71) | 53.40(204) | 28.01(107) | 45.29 | 54.71 | ||||||
| ALGA0043483 | 600 | CC | TC | TT | C | T | 0.50 | 2.00 | 0.38 | 2.202 |
| 22.33(134) | 53.00(318) | 24.67(148) | 48.83 | 51.17 | ||||||
| Haplotype | Genotype Sequence | Num | Frequency(%) |
|---|---|---|---|
| G1 | CC-GG-TT-CC-TT-CC-GG-GG | 70 | 18.47 |
| G2 | CT-GA-TC-CT-TG-CT-GA-GC | 200 | 53.03 |
| G3 | TT-AA-CC-TT-GG-TT-AA-CC | 104 | 27.71 |
| G4 | CC-GG-TC-CT-TG-CT-GA-GC | 1 | 0.26 |
| G5 | TT-AA-CC-TT-TG-TT-AA-CC | 2 | 0.53 |
| Trait | Genotype | Value (Mean ± SE) 1 |
|---|---|---|
| Birth weight (kg) | G1 | 1.74 ± 0.04 |
| G2 | 1.70 ± 0.05 | |
| G3 | 1.72 ± 0.04 | |
| Weaning weight (kg) | G1 | 6.94 ± 0.30 |
| G2 | 7.26 ± 0.27 | |
| G3 | 7.48 ± 0.30 | |
| 90d BW (kg) | G1 | 29.70 ± 0.80 |
| G2 | 29.16 ± 0.61 | |
| G3 | 29.27 ± 0.71 | |
| ADFI (kg) | G1 | 1.80 ± 0.03 |
| G2 | 1.83 ± 0.03 | |
| G3 | 1.79 ± 0.03 | |
| AG (kg) | G1 | 0.68 ± 0.02 |
| G2 | 0.71 ± 0.01 | |
| G3 | 0.69 ± 0.02 | |
| 30 kg age (d) | G1 | 90.45 ± 1.24 |
| G2 | 91.30 ± 0.95 | |
| G3 | 91.14 ± 1.11 | |
| 100 kg age (d) | G1 | 193.96 ± 2.33 |
| G2 | 192.82 ± 1.76 | |
| G3 | 193.88 ± 2.05 | |
| 100 kg BF (mm) | G1 | 7.63 ± 0.23 b |
| G2 | 7.27 ± 0.18 ab | |
| G3 | 7.16 ± 0.21 a | |
| FCR | G1 | 2.70 ± 0.06 |
| G2 | 2.62 ± 0.04 | |
| G3 | 2.61 ± 0.05 | |
| RFI (g) | G1 | 3.41 ± 19.33 |
| G2 | 9.37 ± 14.32 | |
| G3 | −10.46 ± 16.72 |
| Trait | Genotypes | Value (Mean ± SE) 1 |
|---|---|---|
| Birth weight (kg) | CC | 1.85 ± 0.043 |
| TC | 1.80 ± 0.038 | |
| TT | 1.83 ± 0.042 | |
| Weaning weight (kg) | CC | 8.12 ± 0.24 a |
| TC | 8.18 ± 0.21 a | |
| TT | 8.51 ± 0.23 b | |
| 90 d BW (kg) | CC | 30.19 ± 0.71 |
| TC | 29.89 ± 0.62 | |
| TT | 30.42 ± 0.70 | |
| ADFI (kg) | CC | 1.70 ± 0.04 |
| TC | 1.70 ± 0.03 | |
| TT | 1.69 ± 0.04 | |
| AG (kg) | CC | 0.66 ± 0.02 |
| TC | 0.67 ± 0.01 | |
| TT | 0.66 ± 0.02 | |
| 30 kg age (d) | CC | 89.71 ± 1.09 |
| TC | 90.174 ± 0.96 | |
| TT | 89.35 ± 1.08 | |
| 100 kg age (d) | CC | 192.34 ± 1.86 |
| TC | 193.57 ± 1.57 | |
| TT | 192.80 ± 1.84 | |
| 100 kg BF (mm) | CC | 7.87 ± 0.18 A |
| TC | 7.88 ± 0.16 A | |
| TT | 7.46 ± 0.18 B | |
| FCR | CC | 2.62 ± 0.05 |
| TC | 2.61 ± 0.04 | |
| TT | 2.59 ± 0.05 | |
| RFI (g) | CC | −8.56 ± 21.36 |
| TC | −3.65 ± 18.52 | |
| TT | −18.58 ± 21.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, L.; Luo, Y.; Wen, Z.; Dai, Y.; Zheng, C.; Zhu, X.; Qin, L.; Zhang, C.; Liang, H.; Zhang, J.; et al. GPX2 Gene Affects Feed Efficiency of Pigs by Inhibiting Fat Deposition and Promoting Muscle Development. Animals 2022, 12, 3528. https://doi.org/10.3390/ani12243528
Pu L, Luo Y, Wen Z, Dai Y, Zheng C, Zhu X, Qin L, Zhang C, Liang H, Zhang J, et al. GPX2 Gene Affects Feed Efficiency of Pigs by Inhibiting Fat Deposition and Promoting Muscle Development. Animals. 2022; 12(24):3528. https://doi.org/10.3390/ani12243528
Chicago/Turabian StylePu, Lei, Yunyan Luo, Zuochen Wen, Yuxin Dai, Chunting Zheng, Xueli Zhu, Lei Qin, Chunguang Zhang, Hong Liang, Jianbin Zhang, and et al. 2022. "GPX2 Gene Affects Feed Efficiency of Pigs by Inhibiting Fat Deposition and Promoting Muscle Development" Animals 12, no. 24: 3528. https://doi.org/10.3390/ani12243528
APA StylePu, L., Luo, Y., Wen, Z., Dai, Y., Zheng, C., Zhu, X., Qin, L., Zhang, C., Liang, H., Zhang, J., Guo, L., & Wang, L. (2022). GPX2 Gene Affects Feed Efficiency of Pigs by Inhibiting Fat Deposition and Promoting Muscle Development. Animals, 12(24), 3528. https://doi.org/10.3390/ani12243528





