Homeostasis of the Intestinal Mucosa in Healthy Horses—Correlation between the Fecal Microbiome, Secretory Immunoglobulin A and Fecal Egg Count
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Methods
2.2.1. Fecal Egg Count (FEC)
2.2.2. Fecal Protein Analysis
2.2.3. Serum IgA and Fecal SIgA Analysis
2.2.4. Fecal Short-Chain Fatty Acid (SCFA) Analysis
2.2.5. Microbiome Analysis
2.2.6. Statistical Analysis
3. Results
3.1. Fecal Egg Count, Serum IgA and sIgA Analysis Results
3.2. SCFA’s Analysis Results
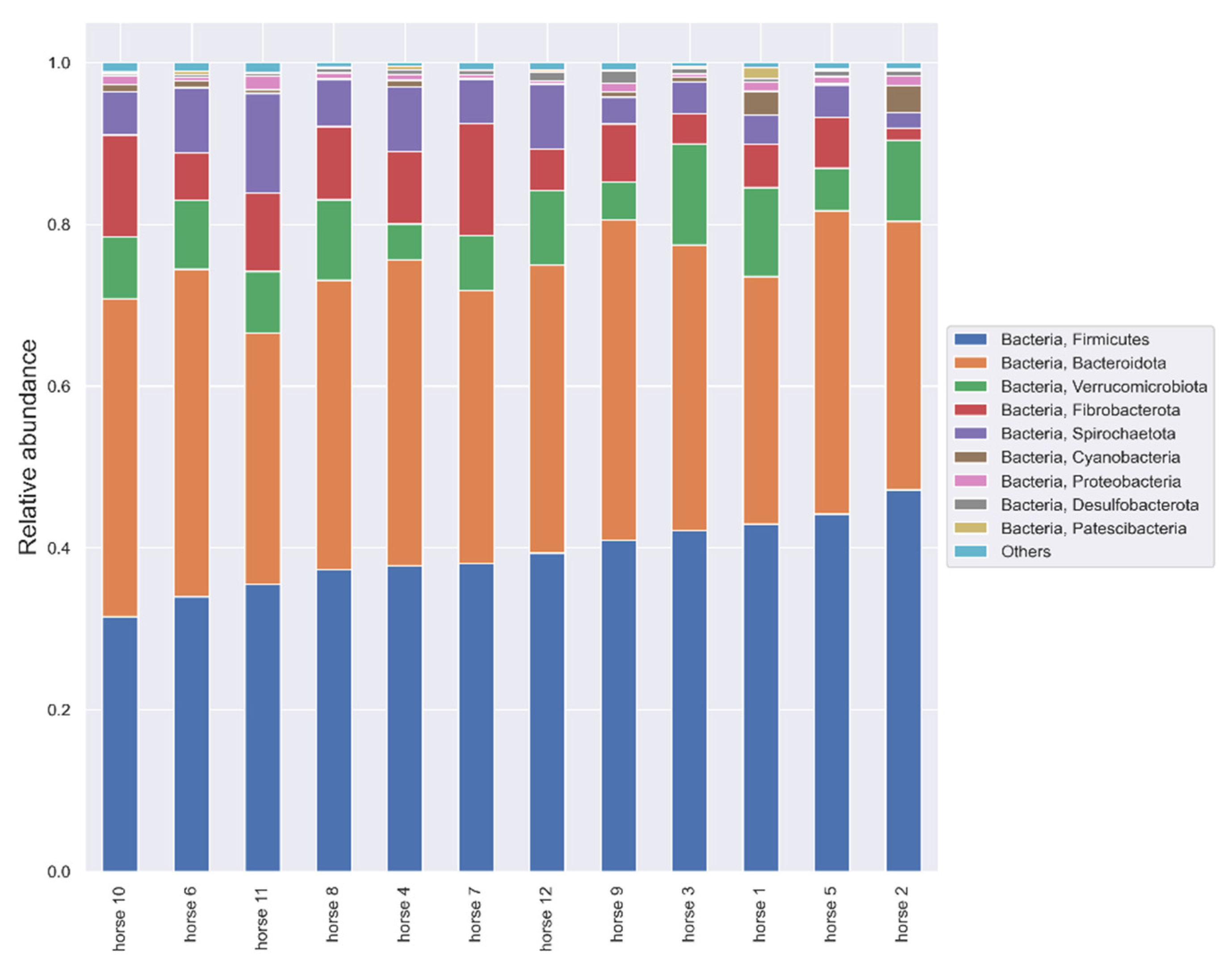
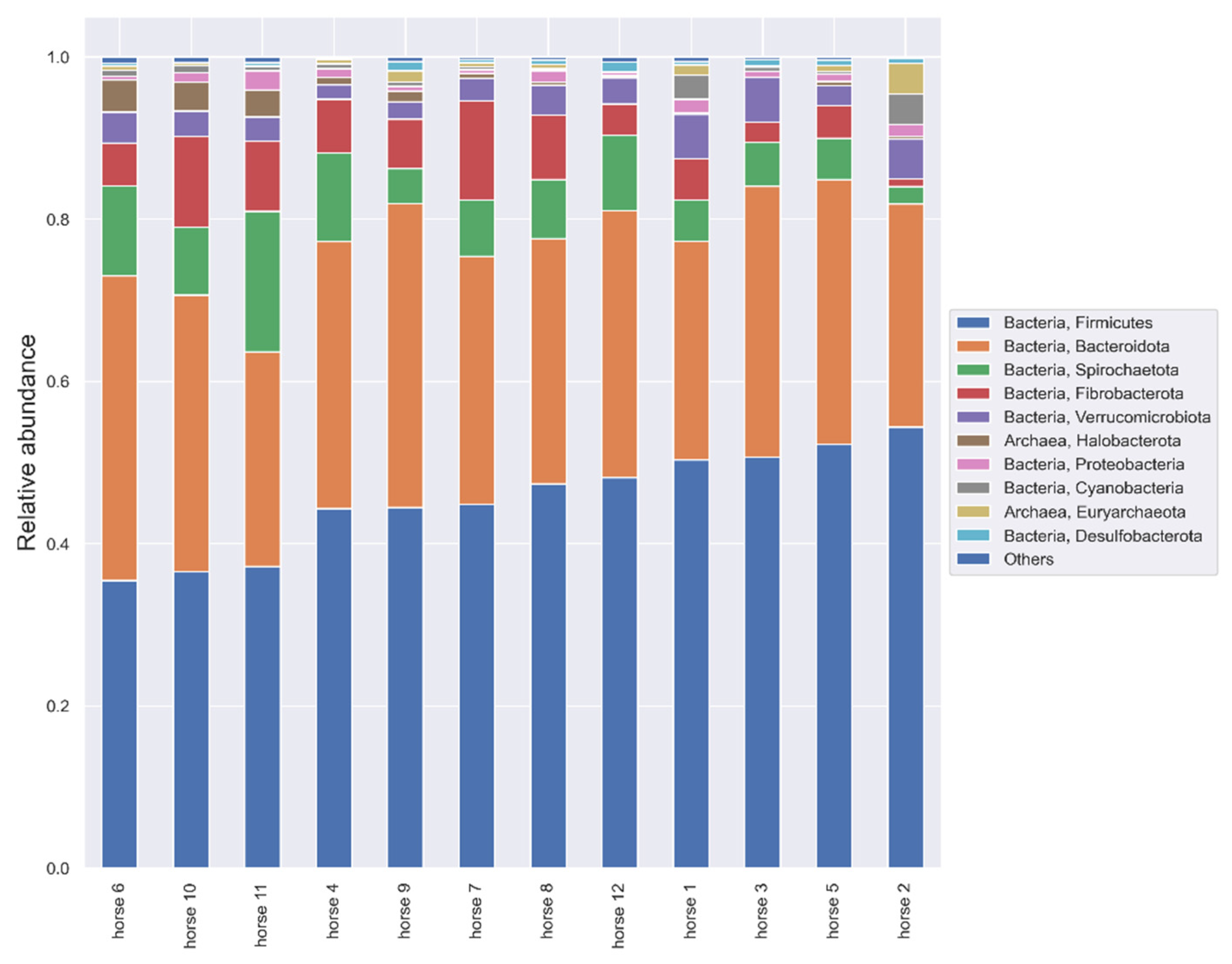
3.3. Microbiome Analysis Results
3.3.1. V3–V4 Hypervariable Regions of the 16S rRNA Gene Analysis
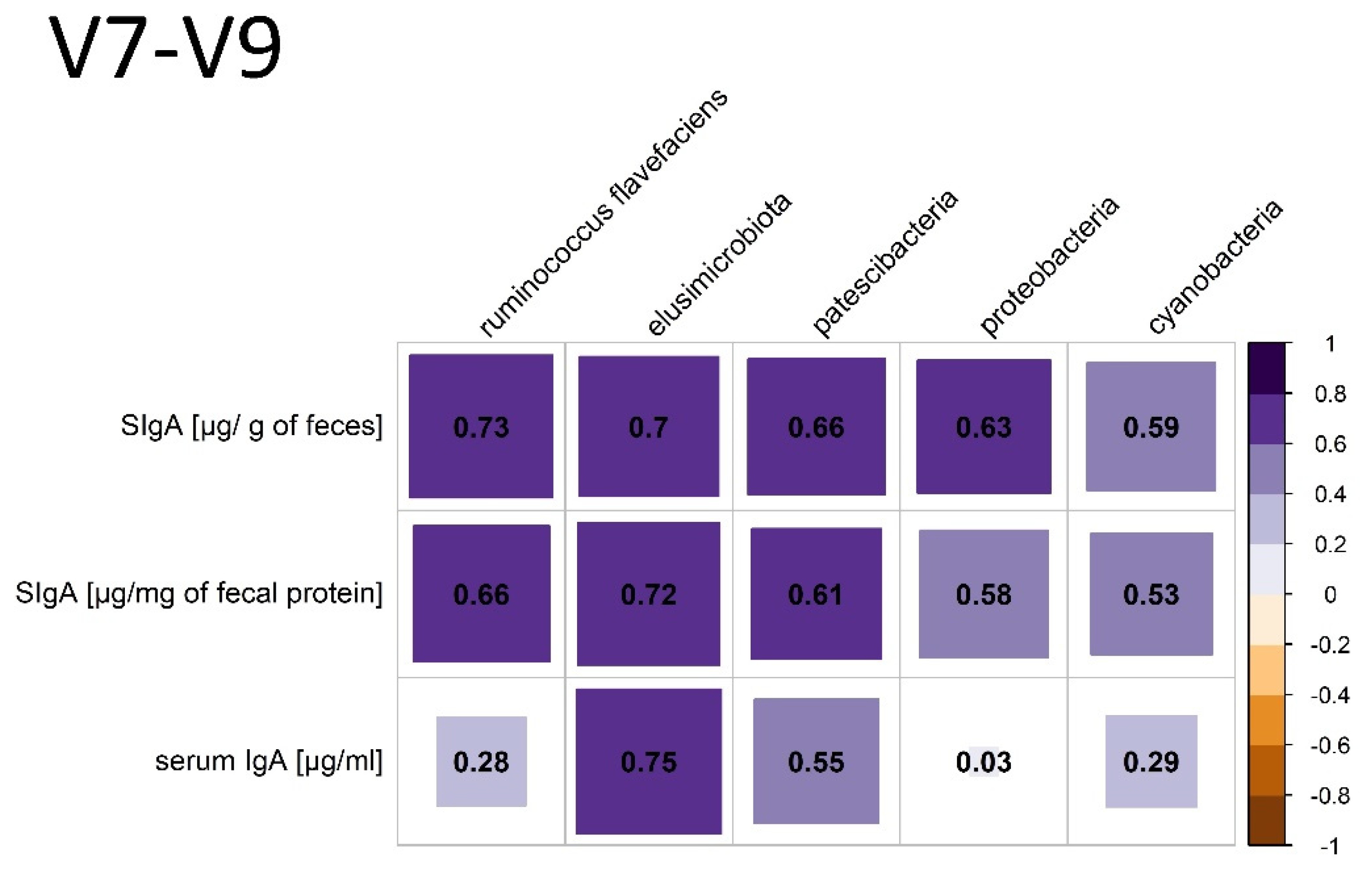
3.3.2. V7-V9 Hypervariable Regions of the 16S rRNA Gene Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gutzeit, C.; Magri, G.; Cerutti, A. Intestinal IgA production and its role in host-microbe interaction. Immunol. Rev. 2014, 260, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.; Elson, C.; Hatton, R.; Weaver, C. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Chase, C.; Kaushik, R.S. Mucosal Immune System of Cattle: All Immune Responses Begin Here. Vet. Clin. N. Am. Food Anim. Pract. 2019, 35, 431–451. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.; Mazmanian, S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 2013, 16, 221–227. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, M.M.; Harris, H.M.; Jeffery, I.B.; Claesson, M.J.; Younge, B.; O’ Toole, P.W.; Ross, R.P. The core faecal bacterial microbiome of Irish Thoroughbred racehorses. Lett. Appl. Microbiol. 2013, 57, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Kauter, A.; Epping, L.; Semmler, T.; Antao, E.; Kannapin, D.; Stoeckle, S.D.; Gehlen, H.; Lübke-Becker, A.; Günther, S.; Wieler, L.H.; et al. The gut microbiome of horses: Current research on equine enteral microbiota and future perspectives. Anim. Microbiome 2019, 1, 14. [Google Scholar] [CrossRef]
- Lindenberg, F.; Krych, L.; Kot, W.; Fielden, J.; Frøkiær, H.; van Galen, G.; Nielsen, D.S.; Hansen, A.K. Development of the equine gut microbiota. Sci. Rep. 2019, 9, 14427. [Google Scholar] [CrossRef]
- Edwards, J.E.; Shetty, S.A.; van den Berg, P.; Burden, F.; van Doorn, D.A.; Pellikaan, W.F.; Dijkstra, J.; Smidt, H. Multi-kingdom characterization of the core equine fecal microbiota based on multiple equine (sub)species. Anim. Microbiome 2020, 2, 6. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar]
- Parasite Control Guidelines Developed by the AAEP Parasite Control Subcommittee of the AAEP Infectious Disease Committee. 2013. Available online: https://aaep.org/sites/default/files/Guidelines/AAEPParasiteControlGuidelines_0.pdf (accessed on 3 November 2022).
- Maeda, S.; Ohno, K.; Uchida, K.; Nakashima, K.; Fukushima, K.; Tsukamoto, A.; Nakajima, M.; Fujino, Y.; Tsujimoto, H. Decreased immunoglobulin A concentrations in feces, duodenum, and peripheral blood mononuclear cells of dogs with inflammatory bowel disease. J. Vet. Intern. Med. 2013, 27, 47–55. [Google Scholar] [CrossRef]
- Stewart, H.L.; Pitta, D.; Indugu, N.; Vecchiarelli, B.; Engiles, J.B.; Southwood, L.L. Characterization of the fecal microbiota of healthy horses. Am. J. Vet. Res. 2018, 79, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Zak, A.; Siwinska, N.; Slowikowska, M.; Borowicz, H.; Kubiak, K.; Hildebrand, J.; Popiolek, M.; Niedzwiedz, A. Searching for ivermectin resistance in a Strongylidae population of horses stabled in Poland. BMC Vet. Res. 2017, 13, 210. [Google Scholar] [CrossRef]
- Marr, I.; Preisler, V.; Farmer, K.; Stefanski, V.; Krueger, K. Non-invasive stress evaluation in domestic horses (Equus caballus): Impact of housing conditions on sensory laterality and immunoglobulin A. R. Soc. Open Sci. 2020, 7, 191994. [Google Scholar] [CrossRef] [PubMed]
- Torii, T.; Kanemitsu, K.; Wada, T.; Itoh, S.; Kinugawa, K.; Hagiwara, A. Measurement of short-chain fatty acids in human faeces using high-performance liquid chromatography: Specimen stability. Ann. Clin. Biochem. 2010, 47, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Grimm, P.; Philippeau, C.; Julliand, V. Faecal parameters as biomarkers of the equine hindgut microbial ecosystem under dietary change. Animal 2017, 11, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads volume. EMBnet. J. 2011, 1, 10. [Google Scholar] [CrossRef]
- Callahan, B.; McMurdie, P.; Rosen, M.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Robeson, M.S., II; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management for the masse. bioRxiv 2020. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Tombak, K.J.; Budischak, S.A.; Hauck, S.; Martinez, L.A.; Rubenstein, D.I. The non-invasive measurement of faecal immunoglobulin in African equids. Int. J. Parasitol. Parasites Wildl. 2020, 18, 105–112. [Google Scholar] [CrossRef]
- Costa, M.C.; Arroyo, L.G.; Allen-Vercoe, E.; Stämpfli, H.R.; Kim, P.T.; Sturgeon, A.; Weese, J.S. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3–V5 region of the 16S rRNA gene. PLoS ONE 2012, 7, e41484. [Google Scholar] [CrossRef]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; Janečka, J.E. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 27, 231. [Google Scholar] [CrossRef]
- Dougal, K.; Harris, P.A.; Girdwood, S.E.; Creevey, C.J.; Curtis, G.C.; Barfoot, C.F.; Argo, C.M.; Newbold, C.J. Changes in the Total Fecal Bacterial Population in Individual Horses Maintained on a Restricted Diet over 6 Weeks. Front. Microbiol. 2017, 11, 1502. [Google Scholar] [CrossRef] [PubMed]
- Julliand, V.; de Vaux, A.; Millet, L.; Fonty, G. Identification of Ruminococcus flavefaciens as the predominant cellulolytic bacterial species of the equine cecum. Appl. Environ. Microbiol. 1999, 65, 3738–3741. [Google Scholar] [CrossRef] [PubMed]
- Bunker, J.J.; Bendelac, A. IgA Responses to Microbiota. Immunity 2018, 49, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Wilmore, J.R.; Gaudette, B.T.; Gomez Atria, D.; Hashemi, T.; Jones, D.D.; Gardner, C.A.; Cole, S.D.; Misic, A.M.; Beiting, D.P.; Allman, D. Commensal Microbes Induce Serum IgA Responses that Protect against Polymicrobial Sepsis. Cell Host Microbe 2018, 23, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Mirpuri, J.; Raetz, M.; Sturge, C.R.; Wilhelm, C.L.; Benson, A.; Savani, R.C.; Hooper, L.V.; Yarovinsky, F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014, 5, 28–39. [Google Scholar] [CrossRef]
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Banfield, J.F.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife 2013, 2, e01102. [Google Scholar] [CrossRef]
- Barb, J.J.; Oler, A.J.; Kim, H.S.; Chalmers, N.; Wallen, G.R.; Cashion, A.; Munson, P.J.; Ames, N.J. Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples. PLoS ONE 2016, 11, e0148047. [Google Scholar] [CrossRef]
- Roediger, W.E.; Moore, A. Effect of short-chain fatty acid on sodium absorption in isolated human colon perfused through the vascular bed. Dig. Dis. Sci. 1981, 26, 100–106. [Google Scholar] [CrossRef]
- Paßlack, N.; Vahjen, W.; Zentek, J. Impact of Dietary Cellobiose on the Fecal Microbiota of Horses. J. Equine Vet. Sci. 2020, 91, 103–106. [Google Scholar] [CrossRef]
- Raspa, F.; Vervuert, I.; Capucchio, M.T.; Colombino, E.; Bergero, D.; Forte, C.; Greppi, M.; Cavallarin, L.; Giribaldi, M.; Antoniazzi, S.; et al. A high-starch vs. high-fibre diet: Effects on the gut environment of the different intestinal compartments of the horse digestive tract. BMC Vet. Res. 2020, 18, 187. [Google Scholar] [CrossRef]
- Nadeau, J.A.; Andrews, F.M.; Patton, C.S.; Argenzio, R.A.; Mathew, A.G.; Saxton, A.M. Effects of hydrochloric, valeric, and other volatile fatty acids on pathogenesis of ulcers in the nonglandular portion of the stomach of horses. Am. J. Vet. Res. 2003, 64, 413–417. [Google Scholar] [CrossRef]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol. 2016, 11, 1081. [Google Scholar] [CrossRef]
- Mach, N.; Foury, A.; Kittelmann, S.; Reigner, F.; Moroldo, M.; Ballester, M.; Esquerré, D.; Rivière, J.; Sallé, G.; Gérard, P.; et al. The Effects of Weaning Methods on Gut Microbiota Composition and Horse Physiology. Front. Physiol. 2017, 8, 535. [Google Scholar] [CrossRef]
- Lee, S.C.; Tang, M.S.; Lim, Y.A.L.; Choy, S.H.; Kurtz, Z.D.; Cox, L.M.; Gundra, U.M.; Cho, I.; Bonneau, R.; Blaser, M.J.; et al. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl. Trop. Dis. 2014, 8, e288010. [Google Scholar] [CrossRef]
- Broadhurst, M.J.; Ardeshir, A.; Kanwar, B.; Mirpuri, J.; Gundra, U.M.; Leung, J.M.; Wiens, K.E.; Vujkovic-Cvijin, I.; Kim, C.C.; Yarovinsky, F.; et al. Therapeutic Helminth Infection of Macaques with Idiopathic Chronic Diarrhea Alters the Inflammatory Signature and Mucosal Microbiota of the Colon. PLoS Pathog. 2012, 8, e1003000. [Google Scholar] [CrossRef]
- Peachey, L.E.; Molena, R.A.; Jenkins, T.P.; Di Cesare, A.; Traversa, D.; Hodgkinson, J.E.; Cantacessi, C. The relationships between faecal egg counts and gut microbial composition in UK Thoroughbreds infected by cyathostomins. Int. J. Parasitol. 2018, 248, 403–412. [Google Scholar] [CrossRef]
- Walshe, N.; Duggan, V.; Cabrera-Rubio, R.; Crispie, F.; Cotter, P.; Feehan, O.; Mulcahy, G. Removal of adult cyathostomins alters faecal microbiota and promotes an inflammatory phenotype in horses. Int. J. Parasitol. 2019, 49, 489–500. [Google Scholar] [CrossRef]
- Daniels, S.P.; Leng, J.; Swann, J.R.; Proudman, C.J. Bugs and drugs: A systems biology approach to characterising the effect of moxidectin on the horse’s faecal microbiome. Anim. Microb. 2020, 2, 38. [Google Scholar] [CrossRef]
- Clark, A.; Sallé, G.; Ballan, V.; Reigner, F.; Meynadier, A.; Cortet, J.; Koch, C.; Riou, M.; Blanchard, A.; Mach, N. Strongyle Infection and Gut Microbiota: Profiling of Resistant and Susceptible Horses over a Grazing Season. Front. Physiol. 2018, 21, 272. [Google Scholar] [CrossRef]


| Stable No | Number of Horses | Mean Age | Last Deworming | Analysis |
|---|---|---|---|---|
| 1 | 12 | Mean 15.5 (sd = 6.6) | 6 months | Serum IgA, FEC, Fecal protein, Fecal SIgA, SCFAs, fecal microbiota |
| 2 | 6 | Mean 12.3 (sd = 4.4) | 1.5 months | FEC, Fecal protein, Fecal SIgA |
| 3 | 42 | Mean 10.9 (sd = 7.2) | 6 months |
| Stable No. | n | Body Weight [kg] | FEC [EPG] 1 | Serum IgA [µg/mL] 1 | Protein [mg/g of Feces] 1 | SIgA [μg/g of Feces] 1 | SIgA [μg/mg of Fecal Protein] 1 |
|---|---|---|---|---|---|---|---|
| 1 | 12 | 610.9 (107.5) | 80.0 (96.1), 20.0 (20.0, 140.0) | 973.9 (532), 949.3 (646.7, 1267.4) | 15.4 (1.8), 15.8 (15.2, 16.5) | 4.2 (3.1), 3.2 (1.8, 6.0)2 | 0.269 (0.181), 0.24 (0.11, 0.36) |
| 2 | 6 | N/A | 20.0 (0.0), 20.0 (20.0, 20.0) | N/A | 18.0 (2.8), 17.2 (16.0, 19.0) | 1.2 (1.2), 8.8 (0.4, 1.2) | 0.06 (0.06), 0.04 (0.02, 0.07) |
| 3 | 42 | N/A | 375.7 (466.7), 190 (20.0, 650.0) | N/A | 20.0 (5.0), 19.3 (15.6, 23.7) | 4.45 (4.62), 2.91 (1.36, 6.09) | 0.25 (0.26), 0.14 (0.08, 0.31) |
| Total | 60 | N/A | 281.0 (417.9), 60 (20.0, 365.0) | N/A | 18.9 (4.7), 17.9 (15.4, 22.3) | 4.1 (4.2), 2.9 (1.3, 5.1) | 0.23 (0.23), 0.14 (0.07, 0.30) |
| SCFAs [mmol/1 g of Feces] | Lactic Acid | C2 | C3 | iC4 | C4 | iC5 | C5 | VFA Ratio |
|---|---|---|---|---|---|---|---|---|
| Mean (SD), Median (IQR) | 3.332 (1.730), 3.437 (2.144, 4.097) | 17.365 (7.030), 14.735 (13.029, 18.567) | 9.606 (3.984), 8.408 (7.121, 10.501) | 3.726 (3.914), 2.327 (1.724, 3.819) | 16.932 (12.737), 11.002 (8.980, 20.601) | 1.302 (2.208), 0.000 (0.000, 2.242) | 2.330 (1.603), 1.824 (1.209, 2.747) | 3.473 (0.433), 3.446 (3.141, 3.625) |
| % SCFAs (mean) | 33.88 | 18.74 | 7.27 | 33.03 | 2.54 | 4.54 |
| β-Diversity Parameters | SIgA [μg/g of Feces] | SIgA [μg/mg of Fecal Protein] | |
|---|---|---|---|
| V3–V4 | Bray–Curtis dissimilarity | p = 0.002, r = 0.59 | p = 0.004, r= 0.49 |
| Jaccard similarity index | p = 0.002, r = 0.55 | p = 0.004, r = 0.44 | |
| Weighted UniFrac | p = 0.002, r = 0.33 | x | |
| Unweighted UniFrac | p = 0.007, r = 0.41 | p = 0.048, r = 0.29 | |
| V7–V9 | Bray–Curtis dissimilarity | p = 0.003, r = 0.55 | p = 0.004, r = 0.46 |
| Jaccard similarity index | p = 0.004, r = 0.56 | p = 0.007, r = 0.48 | |
| Weighted UniFrac | p = 0.019, r = 0.37 | p = 0.026, r = 0.31 | |
| Unweighted UniFrac | p = 0.002, r = 0.51 | p = 0.003, r = 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Żak-Bochenek, A.; Bajzert, J.; Sambor, D.; Siwińska, N.; Szponar, B.; Łaczmański, Ł.; Żebrowska, P.; Czajkowska, A.; Karczewski, M.; Chełmońska-Soyta, A. Homeostasis of the Intestinal Mucosa in Healthy Horses—Correlation between the Fecal Microbiome, Secretory Immunoglobulin A and Fecal Egg Count. Animals 2022, 12, 3094. https://doi.org/10.3390/ani12223094
Żak-Bochenek A, Bajzert J, Sambor D, Siwińska N, Szponar B, Łaczmański Ł, Żebrowska P, Czajkowska A, Karczewski M, Chełmońska-Soyta A. Homeostasis of the Intestinal Mucosa in Healthy Horses—Correlation between the Fecal Microbiome, Secretory Immunoglobulin A and Fecal Egg Count. Animals. 2022; 12(22):3094. https://doi.org/10.3390/ani12223094
Chicago/Turabian StyleŻak-Bochenek, Agnieszka, Joanna Bajzert, Dominika Sambor, Natalia Siwińska, Bogumiła Szponar, Łukasz Łaczmański, Paulina Żebrowska, Aleksandra Czajkowska, Maciej Karczewski, and Anna Chełmońska-Soyta. 2022. "Homeostasis of the Intestinal Mucosa in Healthy Horses—Correlation between the Fecal Microbiome, Secretory Immunoglobulin A and Fecal Egg Count" Animals 12, no. 22: 3094. https://doi.org/10.3390/ani12223094
APA StyleŻak-Bochenek, A., Bajzert, J., Sambor, D., Siwińska, N., Szponar, B., Łaczmański, Ł., Żebrowska, P., Czajkowska, A., Karczewski, M., & Chełmońska-Soyta, A. (2022). Homeostasis of the Intestinal Mucosa in Healthy Horses—Correlation between the Fecal Microbiome, Secretory Immunoglobulin A and Fecal Egg Count. Animals, 12(22), 3094. https://doi.org/10.3390/ani12223094





