RNA Sequencing Reveals circRNA Expression Profiles in Chicken DF1 Cells Infected with H5N1 Influenza Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viral Infection
2.2. RNA Sequencing
2.3. Sequencing Data Analysis
2.4. GO and KEGG Pathway Analysis
3. Results
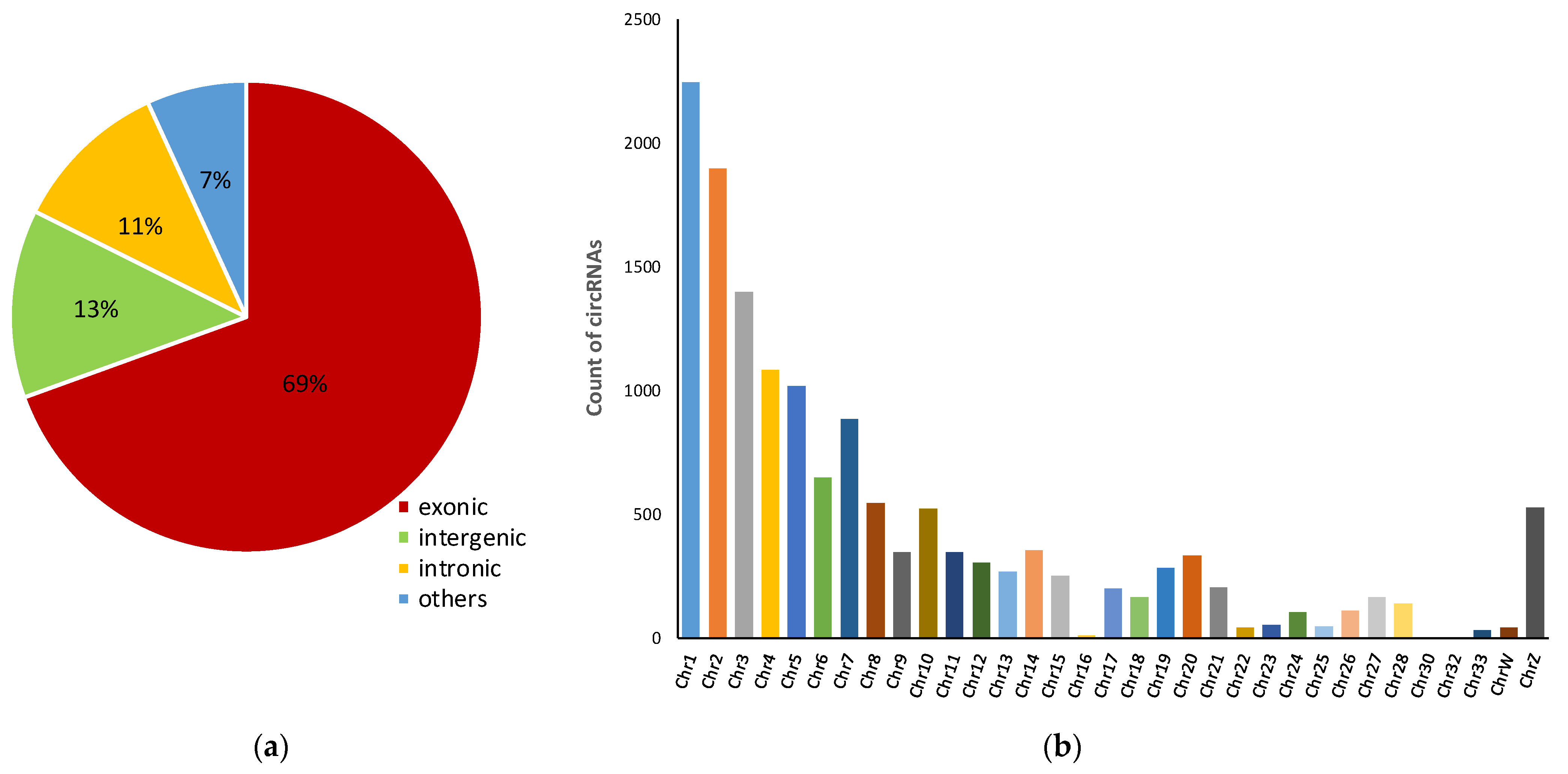
3.1. Characteristics of circRNAs
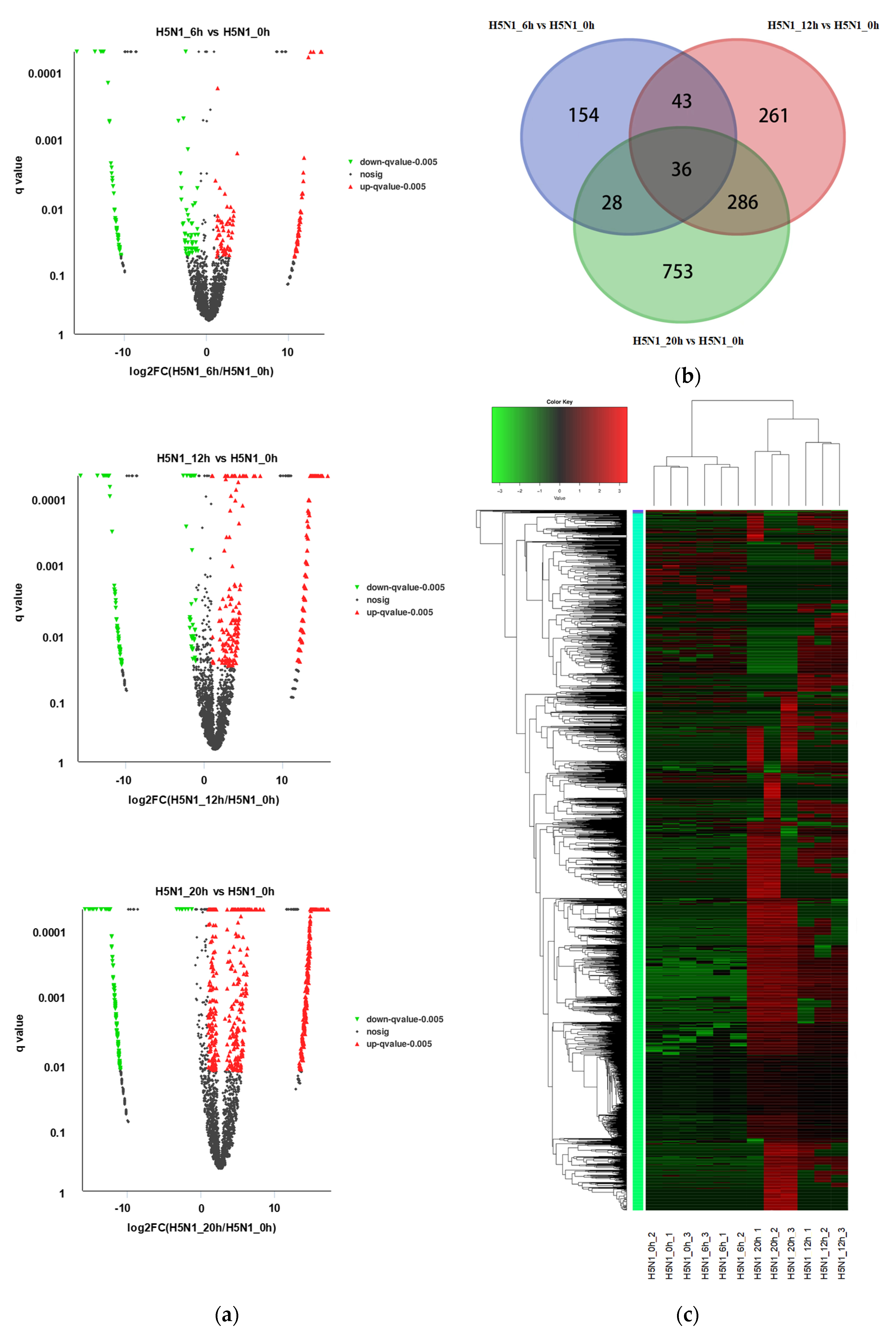
3.2. Identification of Differentially Expressed circRNAs
3.3. Functional Analysis of the Parental Genes of DE circRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Noda, T.; Sagara, H.; Yen, A.; Takada, A.; Kida, H.; Cheng, R.H.; Kawaoka, Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 2006, 439, 490–492. [Google Scholar] [CrossRef]
- MacMahon, K.L.; Delaney, L.J.; Kullman, G.; Gibbins, J.D.; Decker, J.; Kiefer, M.J. Protecting poultry workers from exposure to avian influenza viruses. Public Health Rep. 2008, 123, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Subbarao, K.; Cox, N.J.; Guo, Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: Similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999, 261, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Claas, E.C.; Osterhaus, A.D.; van Beek, R.; De Jong, J.C.; Rimmelzwaan, G.F.; Senne, D.A.; Krauss, S.; Shortridge, K.F.; Webster, R.G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet (Lond. Engl.) 1998, 351, 472–477. [Google Scholar] [CrossRef]
- Subbarao, K.; Klimov, A.; Katz, J.; Regnery, H.; Lim, W.; Hall, H.; Perdue, M.; Swayne, D.; Bender, C.; Huang, J.; et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 1998, 279, 393–396. [Google Scholar] [CrossRef] [Green Version]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Zhang, X.; Odame, E.; Xu, X.; Chen, Y.; Ye, J.; Zhou, H.; Dai, D.; Kyei, B.; Zhan, S.; et al. CircRNA-Protein Interactions in Muscle Development and Diseases. Int. J. Mol. Sci. 2021, 22, 3262. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. Biological Roles and Mechanisms of Circular RNA in Human Cancers. Onco Targets Ther. 2020, 13, 2067–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.G.; Kim, M.V.; Chen, X.; Batista, P.J.; Aoyama, S.; Wilusz, J.E.; Iwasaki, A.; Chang, H.Y. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cell 2017, 67, 228–238.e5. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Zhu, N.; Guo, W.; Wang, X.; Li, K.; Yan, J.; Jiang, C.; Han, S.; Xiang, H.; Wu, X.; et al. RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection. Front. Cell. Infect. Microbiol. 2020, 10, 97C. [Google Scholar] [CrossRef]
- Jost, I.; Shalamova, L.A.; Gerresheim, G.K.; Niepmann, M.; Bindereif, A.; Rossbach, O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018, 15, 1032–1039. [Google Scholar] [CrossRef]
- Cao, L.; Xing, T.; Xu, H.; Ye, M.; Zhang, Q. Differential expression of circular RNA in patients with chronic HBV infection of different stages and related bioinformatic analysis. Chin. J. Clin. Infect. Dis. 2017, 10, 421–427. [Google Scholar]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoint. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 2, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Ranaware, P.B.; Mishra, A.; Vijayakumar, P.; Gandhale, P.N.; Kumar, H.; Kulkarni, D.D.; Raut, A.A. Genome Wide Host Gene Expression Analysis in Chicken Lungs Infected with Avian Influenza Viruses. PLoS ONE 2016, 11, e0153671. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Vijayakumar, P.; Gandhale, P.N.; Ranaware, P.B.; Kumar, H.; Kulkarni, D.D.; Raut, A.A.; Mishra, A. Genome-wide gene expression pattern underlying differential host response to high or low pathogenic H5N1 avian influenza virus in ducks. Acta Virol. 2017, 61, 66–76. [Google Scholar] [CrossRef]
- Watanabe, C.; Uchida, Y.; Ito, H.; Ito, T.; Saito, T. Host immune-related gene responses against highly pathogenic avian influenza virus infection in vitro differ among chicken cell lines established from different organs. Vet. Immunol. Immunopathol. 2011, 144, 187–199. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Tan, S.; Liu, W.R.; Lei, Q.; Qiao, W.; Wu, Y.; Liu, X.; Cheng, W.; Wei, Y.Q.; Peng, Y.; et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol. Cancer 2019, 18, 134. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Y.; Zhao, X.; Liu, J.; Yang, W. Epstein-Barr virus circRNAome as host miRNA sponge regulates virus infection, cell cycle, and oncogenesis. Bioengineered 2019, 10, 593–603. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Ding, Y.; Zhang, Y.; Liu, Y.; Li, Y.; Lei, J.; Zhou, J.; Song, S.; Hu, B. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 2019, 231, 238–245. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Guo, Y.; Zhao, L.; Liu, Q.; Tian, M.; Huang, N.; Fan, M.; Yu, M.; Xia, H.; Ping, J. Analysis of the circRNAs expression profile in mouse lung with H7N9 influenza A virus infection. Genomics 2021, 113 Pt 2, 716–727. [Google Scholar] [CrossRef]
- Shi, N.; Zhang, S.; Guo, Y.; Yu, X.; Zhao, W.; Zhang, M.; Guan, Z.; Duan, M. CircRNA_0050463 promotes influenza A virus replication by sponging miR-33b-5p to regulate EEF1A1. Vet. Microbiol. 2021, 254, 108995. [Google Scholar] [CrossRef] [PubMed]
| CircRNA | Regulation | Gene Symbol | Terms/Pathways | Annotated Database |
|---|---|---|---|---|
| CIRI_circ_006230 | down | - | ||
| CIRI_circ_0013435 | down (6:0); up (12:0; 20:0) | ALKBH8 | ||
| CIRI_circ_0011785 | down | - | ||
| CIRI_circ_007242 | down | RPL14 | Influenza virus RNA transcription and replication. | Gene cards |
| CIRI_circ_002201 | down | - | ||
| CIRI_circ_008390 | down | - | ||
| CIRI_circ_006022 | down | - | ||
| CIRI_circ_008353 | down | CUX1 | Component of nf-munr repressor; binds to the matrix attachment regions (MARs) of the immunoglobulin heavy chain enhancer. Represses T-cell receptor beta enhancer function by binding to MARbeta. | UniProtKB/Swiss-Prot |
| CIRI_circ_008531 | up | COL3A1 | ||
| CIRI_circ_0011293 | down | CANX | Influenza virus RNA transcription and replication, and the innate immune system. | UniProtKB/Swiss-Prot |
| CIRI_circ_003013 | down | COL3A1 | ||
| CIRI_circ_008959 | down | - | ||
| CIRI_circ_002330 | up | AZIN1 | ||
| CIRI_circ_0095 | down (6:0); up (12:0; 20:0) | - | ||
| CIRI_circ_005370 | down | - | ||
| CIRI_circ_007453 | up | - | ||
| CIRI_circ_00136 | down (6:0; 12:0); up (20:0) | - | ||
| CIRI_circ_004926 | up | TTC28 | ||
| CIRI_circ_005366 | up | AKIRIN2 | Required for the innate immune response. | UniProtKB/Swiss-Prot |
| CIRI_circ_0013703 | up | MAFG | ||
| CIRI_circ_003547 | up | RPS6KA5 | Activates TLR4 signaling and CNTF signaling. | UniProtKB/Swiss-Prot |
| CIRI_circ_003861 | down | VNN2 | ||
| CIRI_circ_00491 | down (6:0); up (12:0; 20:0) | DROSHA | ||
| CIRI_circ_0013304 | down | IGF1R | ||
| CIRI_circ_0014096 | down | - | ||
| CIRI_circ_004792 | down | TCP1 | ||
| CIRI_circ_0010535 | down | - | ||
| CIRI_circ_003044 | up | ATRNL1 | ||
| CIRI_circ_008077 | up | - | ||
| CIRI_circ_0012305 | up | CD2AP | May play a role in receptor clustering and cytoskeletal polarity in the junction between T-cell and antigen-presenting cell. | UniProtKB/Swiss-Prot |
| CIRI_circ_008738 | down | LARGE1 | ||
| CIRI_circ_002188 | up | DNAJB6 | ||
| CIRI_circ_007562 | up | QKI | HIV Life Cycle and Oncogenic MAPK signaling | GeneCards |
| CIRI_circ_002400 | up | TMEM214 | ||
| CIRI_circ_009497 | up | MPP6 | ||
| CIRI_circ_0014262 | down | - |
| Group | Enriched Pathways | Enriched Genes | p-Value |
|---|---|---|---|
| 6-h infected group | Dorsoventral axis formation | 3 | 3.40 × 10−2 |
| Focal adhesion | 7 | 3.50 × 10−2 | |
| Wnt signaling pathway | 5 | 7.40 × 10−2 | |
| FoxO signaling pathway | 5 | 7.60 × 10−2 | |
| 12-h infected group | Focal adhesion | 11 | 1.20 × 10−2 |
| Adherens junction | 6 | 2.20 × 10−2 | |
| SNARE interactions in vesicular transport | 4 | 3.10 × 10−2 | |
| Tight junction | 6 | 3.40 × 10−2 | |
| Cell cycle | 7 | 4.60 × 10−2 | |
| Ribosome biogenesis in eukaryotes | 5 | 7.10 × 10−2 | |
| ECM–receptor interaction | 5 | 9.50 × 10−2 | |
| 20-h infected group | MAPK signaling pathway | 20 | 8.60 × 10−4 |
| Oocyte meiosis | 12 | 8.90 × 10−4 | |
| Endocytosis | 17 | 1.60 × 10−2 | |
| Focal adhesion | 15 | 1.80 × 10−2 | |
| Adherens junction | 8 | 2.00 × 10−2 | |
| Ubiquitin mediated proteolysis | 11 | 2.80 × 10−2 | |
| Regulation of actin cytoskeleton | 14 | 2.80 × 10−2 | |
| Progesterone-mediated oocyte maturation | 8 | 3.30 × 10−2 | |
| Hedgehog signaling pathway | 4 | 5.20 × 10−2 | |
| FoxO signaling pathway | 10 | 5.60 × 10−2 | |
| mRNA surveillance pathway | 7 | 6.20 × 10−2 | |
| Herpes simplex infection | 11 | 6.20 × 10−2 | |
| Adrenergic signaling in cardiomyocytes | 9 | 8.70 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Li, G.; Tian, Y.; Zeng, T.; Xu, W.; Gu, T.; Lu, L. RNA Sequencing Reveals circRNA Expression Profiles in Chicken DF1 Cells Infected with H5N1 Influenza Virus. Animals 2022, 12, 158. https://doi.org/10.3390/ani12020158
Chen L, Li G, Tian Y, Zeng T, Xu W, Gu T, Lu L. RNA Sequencing Reveals circRNA Expression Profiles in Chicken DF1 Cells Infected with H5N1 Influenza Virus. Animals. 2022; 12(2):158. https://doi.org/10.3390/ani12020158
Chicago/Turabian StyleChen, Li, Guoqin Li, Yong Tian, Tao Zeng, Wenwu Xu, Tiantian Gu, and Lizhi Lu. 2022. "RNA Sequencing Reveals circRNA Expression Profiles in Chicken DF1 Cells Infected with H5N1 Influenza Virus" Animals 12, no. 2: 158. https://doi.org/10.3390/ani12020158
APA StyleChen, L., Li, G., Tian, Y., Zeng, T., Xu, W., Gu, T., & Lu, L. (2022). RNA Sequencing Reveals circRNA Expression Profiles in Chicken DF1 Cells Infected with H5N1 Influenza Virus. Animals, 12(2), 158. https://doi.org/10.3390/ani12020158





