Whole-Genome Survey and Microsatellite Marker Detection of Antarctic Crocodile Icefish, Chionobathyscus dewitti
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genome Sequencing
2.2. Data Analysis, Genome Assembly, and Microsatellite Detection
3. Results and Discussion
3.1. Genome Sequencing Data Analysis and Genome Survey
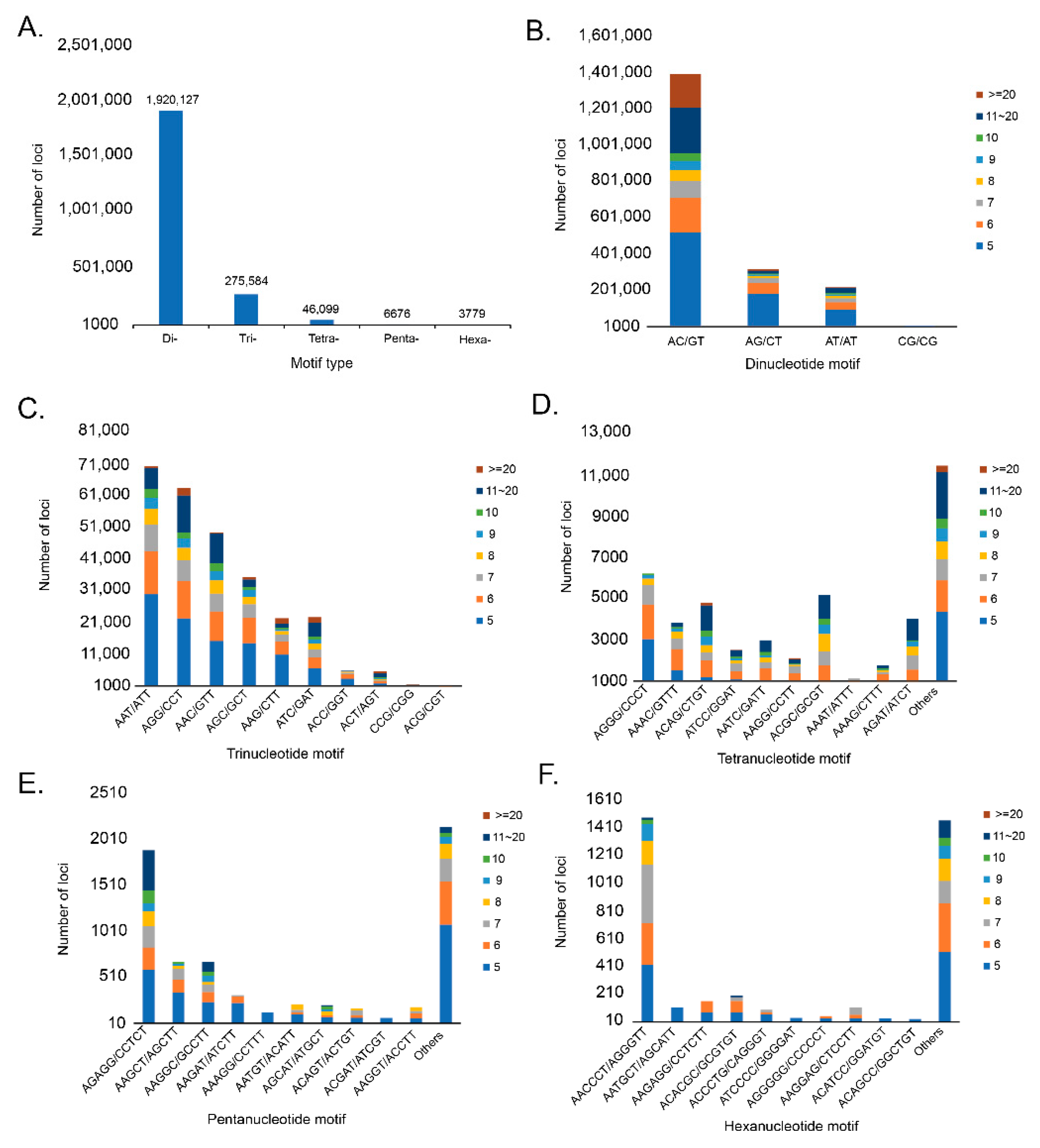
3.2. Genome Assembly and Microsatellite Detection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Prisco, G.; Eastman, J.T.; Giordano, D.; Parisi, E.; Verde, C. Biogeography and adaptation of Notothenioid fish: Hemoglobin function and globin–gene evolution. Gene 2007, 398, 143–155. [Google Scholar] [CrossRef]
- Scher, H.D.; Martin, E.E. Timing and Climatic Consequences of the Opening of Drake Passage. Science 2006, 312, 428–430. [Google Scholar] [CrossRef]
- Toggweiler, J.R.; Bjornsson, H. Drake Passage and palaeoclimate. J. Quat. Sci. 2000, 15, 319–328. [Google Scholar] [CrossRef]
- Verde, C.; Giordano, D.; Russo, R.; di Prisco, G. BLOOD|Erythropoiesis in Fishes. In Encyclopedia of Fish Physiology; Farrell, A.P., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 992–997. [Google Scholar]
- Kim, K.-Y.; Yoon, M.; Yoo, J.S.; Lee, D.-S. The full-length mitochondrial genome of the crocodile icefish, Chionobathyscus dewitti (Teleostei: Perciformes: Channichthyidae). Mitochondrial DNA Part B Resour. 2019, 4, 2371–2372. [Google Scholar] [CrossRef]
- Wells, R.M.G.; Ashby, M.D.; Duncan, S.J.; Macdonald, J.A. Comparative study of the erythrocytes and haemoglobins in nototheniid fishes from Antarctica. J. Fish Biol. 1980, 17, 517–527. [Google Scholar] [CrossRef]
- Di Prisco, G. A study of hemoglobin in antarctic fishes: Purification and characterization of hemoglobins from four species. Comp. Biochem. Physiol. Part B Comp. Biochem. 1988, 90, 631–637. [Google Scholar] [CrossRef]
- Yoon, T.-H.; Kang, H.-E.; Lee, S.R.; Lee, J.-B.; Baeck, G.W.; Park, H.; Kim, H.-W. Metabarcoding analysis of the stomach contents of the Antarctic Toothfish (Dissostichus mawsoni) collected in the Antarctic Ocean. PeerJ 2017, 5, e3977. [Google Scholar] [CrossRef]
- Sutton, C.; Manning, M. Biological parameters for icefish (Chionobathyscus dewitti) in the Ross Sea, Antarctica. CCAMLR Sci. 2008, 15, 139–165. [Google Scholar]
- Agostini, C.; Patarnello, T.; Ashford, J.R.; Torres, J.J.; Zane, L.; Papetti, C. Genetic differentiation in the ice-dependent fishPleuragramma antarcticaalong the Antarctic Peninsula. J. Biogeogr. 2015, 42, 1103–1113. [Google Scholar] [CrossRef]
- Agostini, C.; Papetti, C.; Patarnello, T.; Mark, F.C.; Zane, L.; Marino, I.A.M. Putative selected markers in the Chionodraco genus detected by interspecific outlier tests. Polar Biol. 2013, 36, 1509–1518. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef]
- Fernandez-Silva, I.; Whitney, J.; Wainwright, B.; Andrews, K.R.; Ylitalo-Ward, H.; Bowen, B.W.; Toonen, R.J.; Goetze, E.; Karl, S.A. Microsatellites for Next-Generation Ecologists: A Post-Sequencing Bioinformatics Pipeline. PLoS ONE 2013, 8, e55990. [Google Scholar] [CrossRef]
- Meglécz, E.; Pech, N.; Gilles, A.; Dubut, V.; Hingamp, P.; Trilles, A.; Grenier, R.; Martin, J.-F. QDD version 3.1: A user-friendly computer program for microsatellite selection and primer design revisited: Experimental validation of variables determining genotyping success rate. Mol. Ecol. Resour. 2014, 14, 1302–1313. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA genome assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Sollars, E.S.A.; Harper, A.L.; Kelly, L.J.; Sambles, C.M.; Ramirez-Gonzalez, R.H.; Swarbreck, D.; Kaithakottil, G.; Cooper, E.D.; Uauy, C.; Havlickova, L.; et al. Genome sequence and genetic diversity of European ash trees. Nature 2017, 541, 212–216. [Google Scholar] [CrossRef]
- Li, G.-Q.; Song, L.-X.; Jin, C.-Q.; Li, M.; Gong, S.-P.; Wang, Y.-F. Genome survey and SSR analysis of Apocynum venetum. Biosci. Rep. 2019, 39, BSR20190146. [Google Scholar] [CrossRef]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential Distribution of Simple Sequence Repeats in Eukaryotic Genome Sequences. Mol. Biol. Evol. 2001, 18, 1161–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidell, B.D.; Vayda, M.E.; Small, D.J.; Moylan, T.J.; Londraville, R.L.; Yuan, M.-L.; Rodnick, K.J.; Eppley, Z.A.; Costello, L. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. Proc. Natl. Acad. Sci. USA 1997, 94, 3420–3424. [Google Scholar] [CrossRef]
- Choi, E.; Lee, S.J.; Jo, E.; Kim, J.; Parker, S.J.; Kim, J.-H.; Park, H. Genomic Survey and Microsatellite Marker Investigation of Patagonian Moray Cod (Muraenolepis orangiensis). Animals 2022, 12, 1608. [Google Scholar] [CrossRef] [PubMed]
- Detrich, H.W.; Stuart, A.; Schoenborn, M.; Parker, S.K.; Methé, B.A.; Amemiya, C.T. Genome enablement of the notothenioidei: Genome size estimates from 11 species and BAC libraries from 2 Representative Taxa. J. Exp. Zool. Part B Mol. Dev. Evol. 2010, 314B, 369–381. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, J.-H.; Jo, E.; Choi, E.; Kim, J.; Choi, S.-G.; Chung, S.; Kim, H.-W.; Park, H. Chromosomal assembly of the Antarctic toothfish (Dissostichus mawsoni) genome using third-generation DNA sequencing and Hi-C technology. Zool. Res. 2021, 42, 124–129. [Google Scholar] [CrossRef]
- Chen, L.; Lu, Y.; Li, W.; Ren, Y.; Yu, M.; Jiang, S.; Fu, Y.; Wang, J.; Peng, S.; Bilyk, K.T.; et al. The genomic basis for colonizing the freezing Southern Ocean revealed by Antarctic toothfish and Patagonian robalo genomes. GigaScience 2019, 8, giz016. [Google Scholar] [CrossRef]
- Ahn, D.-H.; Shin, S.C.; Kim, B.-M.; Kang, S.; Kim, J.-H.; Ahn, I.; Park, J.; Park, H. Draft genome of the Antarctic dragonfish, Parachaenichthys charcoti. GigaScience 2017, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.-S.; Down, T.A.; Latorre, I.; Ahringer, J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 2011, 39, e103. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hu, Y.; Sui, Z.; Fu, F.; Wang, J.; Chang, L.; Guo, W.; Li, B. Genome Survey Sequencing and Genetic Background Characterization of Gracilariopsis lemaneiformis (Rhodophyta) Based on Next-Generation Sequencing. PLoS ONE 2013, 8, e69909. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Gao, Z.; Luo, W.; Liu, X.; Wang, W. Characteristics of microsatellites in blunt snout bream (Megalobrama amblycephala) EST sequences using 454FLX. Acta Hydrobiol. Sin. 2013, 37, 982–988. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Dai, G.; Cao, Y.; Chen, X.; Chen, L.; Shangguan, J.; Ning, Y. Isolation and characterization of eleven microsatellite loci in the marbled rockfish, Sebastiscus marmoratus (Scorpaenidae). Conserv. Genet. Resour. 2013, 6, 53–55. [Google Scholar] [CrossRef]
- Lei, Y.; Zhou, Y.; Price, M.; Song, Z. Genome-wide characterization of microsatellite DNA in fishes: Survey and analysis of their abundance and frequency in genome-specific regions. BMC Genom. 2021, 22, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.-Y.; Song, N.; Xiao, S.-J.; Gao, T.-X. Whole genome survey analysis and microsatellite motif identification of Sebastiscus marmoratus. Biosci. Rep. 2020, 40, BSR20192252. [Google Scholar] [CrossRef] [PubMed]
- Papetti, C.; Harms, L.; Jürgens, J.; Sandersfeld, T.; Koschnick, N.; Windisch, H.S.; Knust, R.; Pörtner, H.-O.; Lucassen, M. Microsatellite markers for the notothenioid fish Lepidonotothen nudifrons and two congeneric species. BMC Res. Notes 2016, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.K.; Touma, J.; Bravo, S.; Leiva, F.; Vargas-Chacoff, L.; Valenzuela, A.; Datagnan, P.; Amthauer, R.; Reyes, A.; Vidal, R. Novel microsatellite markers discovery in Patagonian toothfish (Dissostichus eleginoides) using high-throughput sequencing. Mol. Biol. Rep. 2019, 46, 5525–5530. [Google Scholar] [CrossRef]
- Papetti, C.; Windisch, H.S.; La Mesa, M.; Lucassen, M.; Marshall, C.; Lamare, M.D. Non-Antarctic notothenioids: Past phylogenetic history and contemporary phylogeographic implications in the face of environmental changes. Mar. Genom. 2016, 25, 1–9. [Google Scholar] [CrossRef]


| Raw Data (bp) | Total Reads | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|
| 60,426,455,804 | 400,175,204 | 96.1 | 91.0 | 49.9 |
| MaSuRCA | |
|---|---|
| Number of scaffolds | 695,247 |
| Total size of scaffolds | 897,784,561 |
| Longest scaffold | 51,375 |
| Number of scaffolds > 1 K nt | 222,128 (31.9%) |
| Number of scaffolds > 10 K nt | 6622 (1.0%) |
| N50 scaffold length | 2401 |
| L50 scaffold count | 87,813 |
| GC contents (%) | 41.72 |
| Primer No. | Motif | PCR Product Size (bp) | Forward Primer (5′ → 3′) | Tm (°C) | Reverse Primer (5′ → 3′) | Tm (°C) |
|---|---|---|---|---|---|---|
| 1 | AT 9 | 105 | AACTGCACAGAACAGGGTCA | 59.823 | AAGCCTCGGTATGCAGAGTG | 59.454 |
| 2 | AC 14 | 120 | GTGAAGAACTTGGTGCTGCC | 59.897 | CAGGTACGCTAAGCCAGGAG | 59.688 |
| 3 | AC 7 | 128 | AAGTGGGCTGGTGAACTTGT | 59.187 | TCCCGTGATGTCTTACCGAC | 59.741 |
| 4 | AC 8 | 133 | TAAACCCAGGAGCGTAAGGC | 59.824 | GCCTCTTGCTCCGACACATA | 59.749 |
| 5 | AC 7 | 138 | CCCTGAATGGCTTCCAACAC | 59.964 | ACGAGCCTTAATGCACACCA | 59.109 |
| 6 | AC 11 | 141 | AGCATGTGTTCTTGCCAGGA | 59.307 | CACAGGTGGAGGCTACCTTT | 59.889 |
| 7 | AC 10 | 149 | TGTGTTAAGAGCCGTGCTGT | 59.821 | CCTCGCTGCCTTTGAGGTAT | 59.894 |
| 8 | AC 17 | 159 | CACCAGGCAGCTCGTTACTA | 59.061 | GTGTTTACGAACAGCGGGTT | 59.469 |
| 9 | AC 12 | 161 | ACAGCCGGTTGACTGAAACT | 59.179 | ATCTCCAGTTCTCTGCGTCC | 59.82 |
| 10 | AT 9 | 163 | CATGAGCAATGTTCCGTCGG | 59.046 | CCTGTTGGAGACACAAAGCC | 59.625 |
| 11 | AC 11 | 166 | CCACAGATGTTGACTTGGCG | 59.746 | GGCCCGTAACACCCTGTATT | 59.481 |
| 12 | AC 22 | 171 | AGGCATTTAACCTCGGCACA | 59.451 | GGCTCCTATTTCACCCAGCT | 59.962 |
| 13 | AC 12 | 190 | GAAAGCAGGCACTCAGATGC | 59.753 | TCATTCCAGCACACTCTCCG | 59.549 |
| 14 | AC 14 | 192 | GTGCATCTTTCTACCGCTGC | 59.537 | CTGCTCGACCCTGATGACAT | 59.625 |
| 15 | AC 9 | 194 | CGGGTAAACGCTATGGAGGT | 59.751 | CGGACTCCATACTGTTGGCA | 59.537 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, S.-J.; Jo, E.; Choi, E.; Cho, M.; Choi, S.; Kim, J.-H.; Park, H. Whole-Genome Survey and Microsatellite Marker Detection of Antarctic Crocodile Icefish, Chionobathyscus dewitti. Animals 2022, 12, 2598. https://doi.org/10.3390/ani12192598
Kim J, Lee S-J, Jo E, Choi E, Cho M, Choi S, Kim J-H, Park H. Whole-Genome Survey and Microsatellite Marker Detection of Antarctic Crocodile Icefish, Chionobathyscus dewitti. Animals. 2022; 12(19):2598. https://doi.org/10.3390/ani12192598
Chicago/Turabian StyleKim, Jinmu, Seung-Jae Lee, Euna Jo, Eunkyung Choi, Minjoo Cho, Soyun Choi, Jeong-Hoon Kim, and Hyun Park. 2022. "Whole-Genome Survey and Microsatellite Marker Detection of Antarctic Crocodile Icefish, Chionobathyscus dewitti" Animals 12, no. 19: 2598. https://doi.org/10.3390/ani12192598
APA StyleKim, J., Lee, S.-J., Jo, E., Choi, E., Cho, M., Choi, S., Kim, J.-H., & Park, H. (2022). Whole-Genome Survey and Microsatellite Marker Detection of Antarctic Crocodile Icefish, Chionobathyscus dewitti. Animals, 12(19), 2598. https://doi.org/10.3390/ani12192598







