Detection of Porcine Circovirus 1/2/3 and Genetic Analysis of Porcine Circovirus 2 in Wild Boar from Jiangxi Province of China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Wild Boars Samples Collection Area
2.3. Viral DNA Extraction
2.4. Differential Detection of PCVs
2.5. Full Genome of PCV2 Amplification and Sequencing
2.6. Wild Boars PCV2 Sequence Alignment and Phylogenetic Analysis
3. Results
3.1. Prevalence of PCVs in Wild Boars
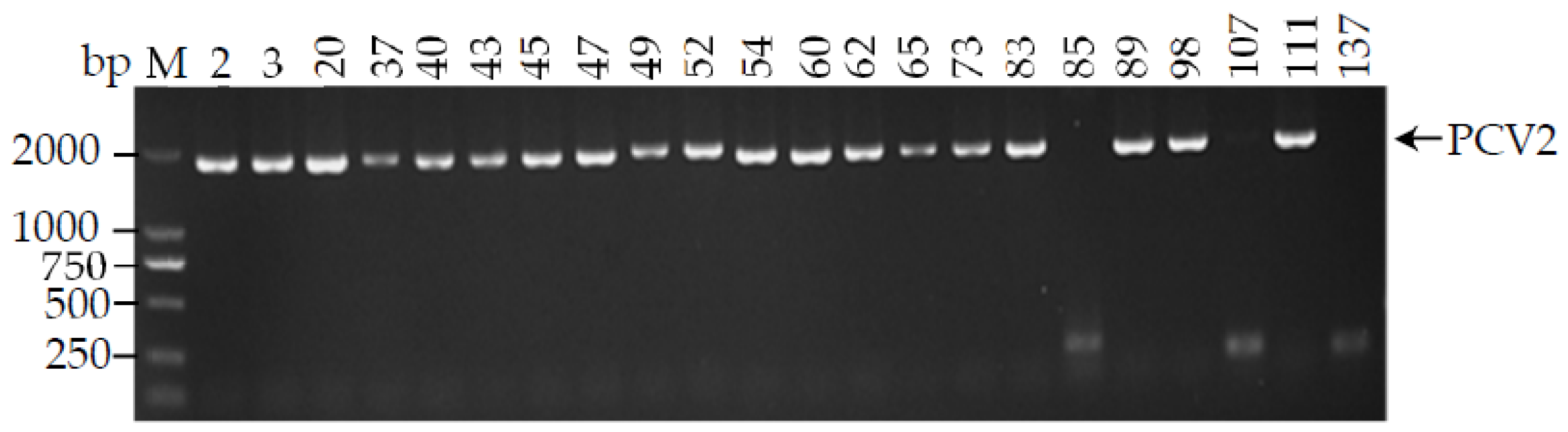
3.2. Amplification of PCV2 Complete Genome
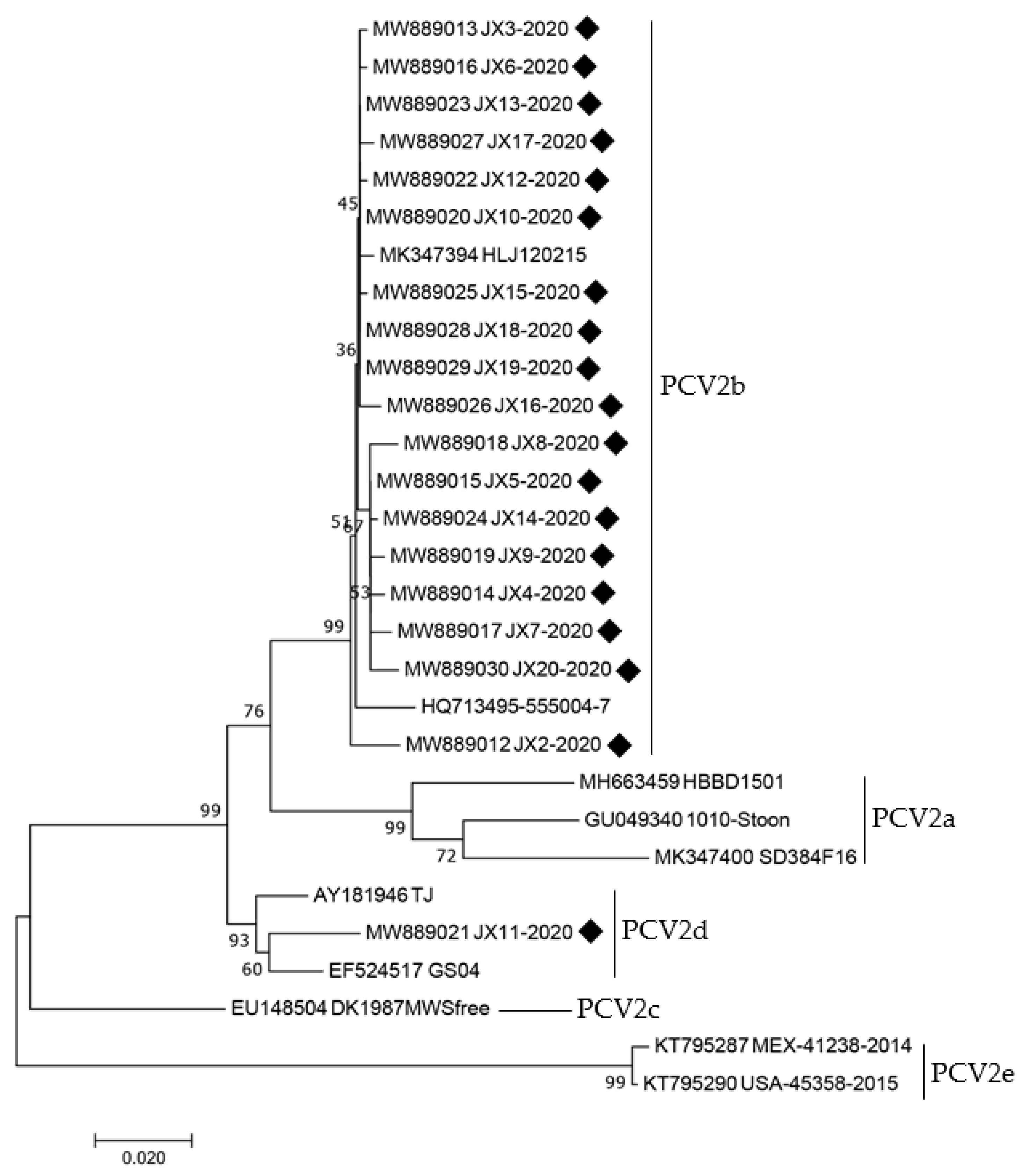
3.3. Genetic Analysis of PCV2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sibila, M.; Rocco, C.; Franzo, G.; Huerta, E.; Domingo, M.; Nunez, J.I.; Segales, J. Genotyping of Porcine Circovirus 2 (PCV-2) in Vaccinated Pigs Suffering from PCV-2-Systemic Disease between 2009 and 2020 in Spain. Pathogens 2021, 10, 1016. [Google Scholar] [CrossRef]
- Rakibuzzaman, A.; Pineyro, P.; Pillatzki, A.; Ramamoorthy, S. Harnessing the Genetic Plasticity of Porcine Circovirus Type 2 to Target Suicidal Replication. Viruses 2021, 13, 1676. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Li, X.; Li, S.; Xie, N.; Yan, X.; Li, X.; Zhu, J. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 2021, 68, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Han, H.H.; Karkada, N.; Jayadeva, G.; Dubin, G. Serologic response to porcine circovirus type 1 (PCV1) in infants vaccinated with the human rotavirus vaccine, Rotarix: A retrospective laboratory analysis. Hum. Vaccines Immunother. 2017, 13, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Shaheduzzaman, S.; Willliams, D.K.; Gao, Y.; Khan, A.S. Investigations of porcine circovirus type 1 (PCV1) in vaccine-related and other cell lines. Vaccine 2011, 29, 8429–8437. [Google Scholar] [CrossRef]
- An, D.J.; Song, D.S.; Park, J.Y.; Park, B.K. A DNA miniarray system for simultaneous visual detection of porcine circovirus type 1 (PCV1) and 2 (PCV2) in pigs. Vet. Res. Commun. 2009, 33, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Csagola, A.; Kiss, I.; Tuboly, T. Detection and analysis of porcine circovirus type 1 in Hungarian wild boars: Short communication. Acta Vet. Hung. 2008, 56, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Sirisereewan, C.; Thanawongnuwech, R.; Kedkovid, R. Current Understanding of the Pathogenesis of Porcine Circovirus 3. Pathogens 2022, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef]
- Sun, W.; Du, Q.; Han, Z.; Bi, J.; Lan, T.; Wang, W.; Zheng, M. Detection and genetic characterization of porcine circovirus 4 (PCV4) in Guangxi, China. Gene 2021, 773, 145384. [Google Scholar] [CrossRef]
- Nguyen, V.G.; Do, H.Q.; Huynh, T.M.; Park, Y.H.; Park, B.K.; Chung, H.C. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound. Emerg. Dis. 2022, 69, 538–548. [Google Scholar] [CrossRef]
- Franzo, G.; Ruiz, A.; Grassi, L.; Sibila, M.; Drigo, M.; Segales, J. Lack of Porcine circovirus 4 Genome Detection in Pig Samples from Italy and Spain. Pathogens 2020, 9, 433. [Google Scholar] [CrossRef]
- Tian, R.B.; Zhao, Y.; Cui, J.T.; Zheng, H.H.; Xu, T.; Hou, C.Y.; Wang, Z.Y.; Li, X.S.; Zheng, L.L.; Chen, H.Y. Molecular detection and phylogenetic analysis of Porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound. Emerg. Dis. 2021, 68, 276–282. [Google Scholar] [CrossRef]
- Niu, G.; Zhang, X.; Ji, W.; Chen, S.; Li, X.; Yang, L.; Zhang, L.; Ouyang, H.; Li, C.; Ren, L. Porcine circovirus 4 rescued from an infectious clone is replicable and pathogenic in vivo. Transbound. Emerg. Dis. 2022, 24, 1–10. [Google Scholar] [CrossRef]
- Molini, U.; Coetzee, L.M.; Hemberger, M.Y.; Khaiseb, S.; Cattoli, G.; Dundon, W.G.; Franzo, G. The Oryx Antelope (Oryx gazella): An Unexpected Host for Porcine Circovirus-2 (PCV-2). Pathogens 2021, 10, 1402. [Google Scholar] [CrossRef]
- Yu, W.; Sun, Y.; He, Q.; Sun, C.; Dong, T.; Zhang, L.; Zhan, Y.; Wang, N.; Yang, Y.; Sun, Y. Mitochondrial Localization Signal of Porcine Circovirus Type 2 Capsid Protein Plays a Critical Role in Cap-Induced Apoptosis. Vet. Sci. 2021, 8, 272. [Google Scholar] [CrossRef]
- Kang, S.J.; Bae, S.M.; Lee, H.J.; Jeong, Y.J.; Lee, M.A.; You, S.H.; Lee, H.S.; Hyun, B.H.; Lee, N.; Cha, S.H. Porcine Circovirus (PCV) Genotype 2d-Based Virus-like Particles (VLPs) Induced Broad Cross-Neutralizing Antibodies against Diverse Genotypes and Provided Protection in Dual-Challenge Infection of a PCV2d Virus and a Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV). Pathogens 2021, 10, 1145. [Google Scholar]
- Um, H.; Yang, S.; Oh, T.; Cho, H.; Park, K.H.; Suh, J.; Chae, C. A field efficacy trial of a trivalent vaccine containing porcine circovirus type 2a and 2b, and Mycoplasma hyopneumoniae in three herds. Vet. Med. Sci. 2022, 8, 578–590. [Google Scholar] [CrossRef]
- Ariyama, N.; Aguero, B.; Valdes, V.; Berrios, F.; Bucarey, S.; Mor, S.; Brito, B.; Neira, V. Update of Genetic Diversity of Porcine Circovirus Type 2 in Chile Evidences the Emergence of PCV2d Genotype. Front. Vet. Sci. 2021, 8, 789491. [Google Scholar] [CrossRef]
- Feng, H.; Fu, J.; Zhang, B.; Xue, T.; Liu, C. A Novel Virus-Like Agent Originated From Genome Rearrangement of Porcine Circovirus Type 2 (PCV2) Enhances PCV2 Replication and Regulates Intracellular Redox Status In Vitro. Front. Cell. Infect. Microbiol. 2022, 12, 855920. [Google Scholar] [CrossRef]
- Jia, Y.; Zhu, Q.; Xu, T.; Chen, X.; Li, H.; Ma, M.; Zhang, Y.; He, Z.; Chen, H. Detection and genetic characteristics of porcine circovirus type 2 and 3 in Henan province of China. Mol. Cell. Probes 2022, 61, 101790. [Google Scholar] [CrossRef]
- Park, K.H.; Cho, H.; Oh, T.; Yang, S.; Chae, C. Multiplex polymerase chain reaction for the detection and differentiation of 4 porcine circovirus 2 genotypes (PCV-2a, -2b, -2d, and -2e) in clinical samples. Can. J. Vet. Res. 2022, 86, 153–156. [Google Scholar]
- Bao, F.; Mi, S.; Luo, Q.; Guo, H.; Tu, C.; Zhu, G.; Gong, W. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound. Emerg. Dis. 2018, 65, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Rajkhowa, T.K.; Lalnunthanga, P.; Rao, P.L.; Subbiah, M.; Lalrohlua, B. Emergence of porcine circovirus 2g (PCV2g) and evidence for recombination between genotypes 2g, 2b and 2d among field isolates from non-vaccinated pigs in Mizoram, India. Infect. Genet. Evol. 2021, 90, 104775. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Wang, T.; Deng, J.; Chen, Y.; Yan, Q.; Wang, D.; Zhu, Y. Genomic analysis of porcine circovirus type 2 from southern China. Vet. Med. Sci. 2020, 6, 875–889. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lee, J.Y.; Oh, T.; Park, K.H.; Cho, H.; Suh, J.; Min, K.D.; Ham, H.J.; Chae, C. Comparative growth performance of 3 types of combination vaccines containing porcine circovirus 2 and Mycoplasma hyopneumoniae under field conditions. Can. J. Vet. Res. 2022, 86, 93–101. [Google Scholar]
- Park, K.H.; Oh, T.; Cho, H.; Yang, S.; Chae, C. The first isolation of porcine circovirus type 2e from a Korean pig. Arch. Virol. 2020, 165, 2927–2930. [Google Scholar] [CrossRef]
- Cho, H.; Kang, I.; Oh, T.; Yang, S.; Park, K.H.; Min, K.D.; Ham, H.J.; Chae, C. Comparative study of the virulence of 3 major Korean porcine circovirus type 2 genotypes (a, b, and d). Can. J. Vet. Re.s 2020, 84, 235–240. [Google Scholar]
- Klaumann, F.; Franzo, G.; Sohrmann, M.; Correa-Fiz, F.; Drigo, M.; Nunez, J.I.; Sibila, M.; Segales, J. Retrospective detection of Porcine circovirus 3 (PCV-3) in pig serum samples from Spain. Transbound. Emerg. Dis. 2018, 65, 1290–1296. [Google Scholar] [CrossRef]
- Reiner, G.; Bronnert, B.; Hohloch, C.; Reinacher, M.; Willems, H. Distribution of ORF2 and ORF3 genotypes of porcine circovirus type 2 (PCV-2) in wild boars and domestic pigs in Germany. Vet. Microbiol. 2011, 148, 372–376. [Google Scholar] [CrossRef][Green Version]
- Dei Giudici, S.; Lo Presti, A.; Bonelli, P.; Angioi, P.P.; Sanna, G.; Zinellu, S.; Balzano, F.; Salis, F.; Ciccozzi, M.; Oggiano, A. Phylogenetic analysis of porcine circovirus type 2 in Sardinia, Italy, shows genotype 2d circulation among domestic pigs and wild boars. Infect. Genet. Evol. 2019, 71, 189–196. [Google Scholar] [CrossRef]
- Faccini, S.; Barbieri, I.; Gilioli, A.; Sala, G.; Gibelli, L.R.; Moreno, A.; Sacchi, C.; Rosignoli, C.; Franzini, G.; Nigrelli, A. Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound. Emerg. Dis. 2017, 64, 1661–1664. [Google Scholar] [CrossRef]
- Feng, H.; Segales, J.; Wang, F.; Jin, Q.; Wang, A.; Zhang, G.; Franzo, G. Comprehensive Analysis of Codon Usage Patterns in Chinese Porcine Circoviruses Based on Their Major Protein-Coding Sequences. Viruses 2022, 14, 81. [Google Scholar] [CrossRef]
- Han, L.; Yuan, G.F.; Chen, S.J.; Dai, F.; Hou, L.S.; Fan, J.H.; Zuo, Y.Z. Porcine circovirus type 2 (PCV2) infection in Hebei Province from 2016 to 2019: A retrospective study. Arch. Virol. 2021, 166, 2159–2171. [Google Scholar] [CrossRef]
- Karuppannan, A.K.; Opriessnig, T. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Porter, E.; Stoy, C.; Dong, J.; Anderson, J.; Fu, J.; Pogranichniy, R.; Woodworth, J.; Peddireddi, L.; et al. Development of a differential multiplex real-time PCR assay for porcine circovirus type 2 (PCV2) genotypes PCV2a, PCV2b and PCV2d. J. Virol. Methods 2020, 286, 113971. [Google Scholar] [CrossRef]
- Qu, T.; Li, R.; Yan, M.; Luo, B.; Yang, T.; Yu, X. High prevalence of PCV2d in Hunan province, China: A retrospective analysis of samples collected from 2006 to 2016. Arch. Virol. 2018, 163, 1897–1906. [Google Scholar] [CrossRef]
- Fanelli, A.; Pellegrini, F.; Camero, M.; Catella, C.; Buonavoglia, D.; Fusco, G.; Martella, V.; Lanave, G. Genetic Diversity of Porcine Circovirus Types 2 and 3 inWild Boar in Italy. Animals 2022, 12, 953. [Google Scholar] [CrossRef]
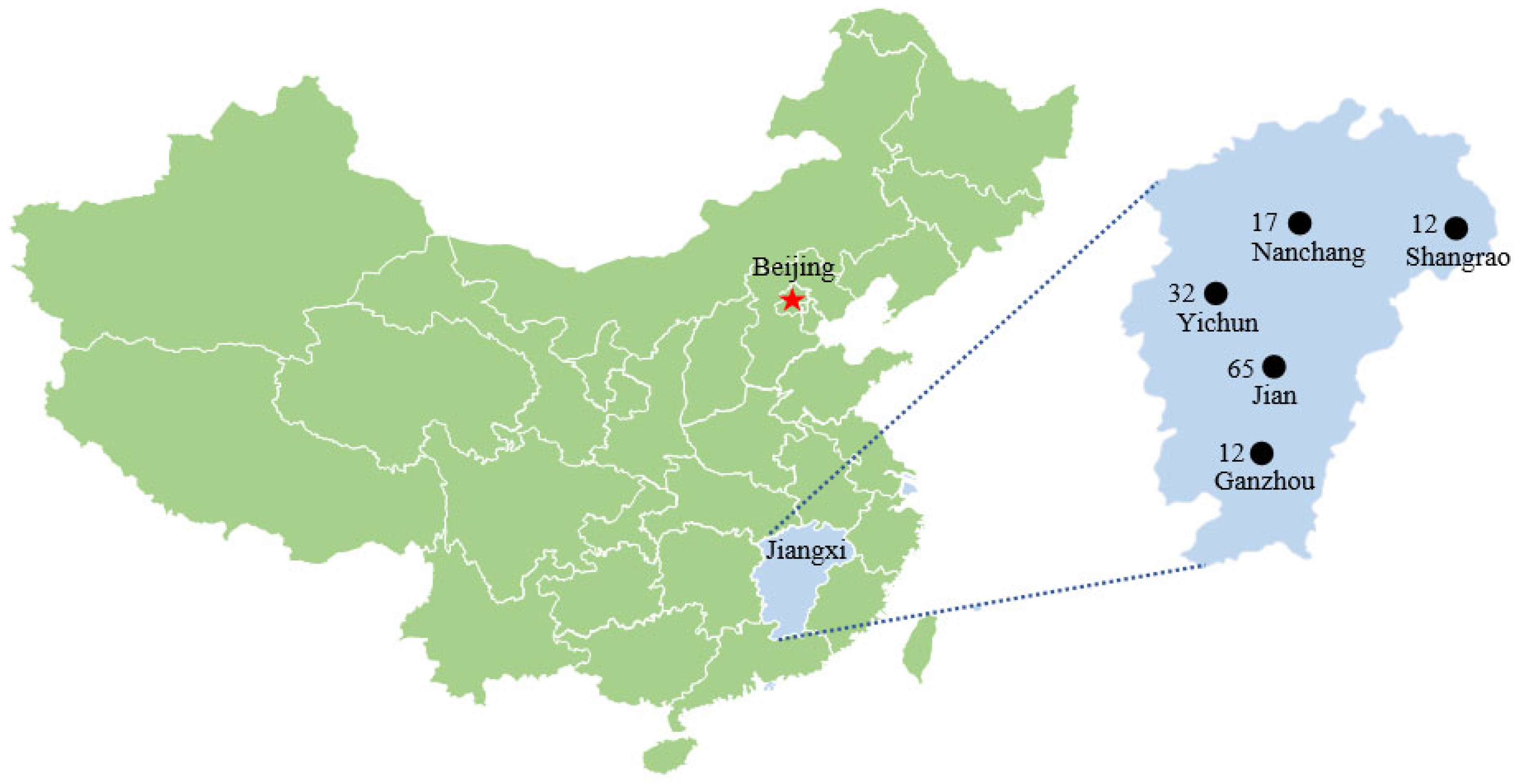

| Shangrao | JiAn | Yichun | Ganzhou | Nanchang | Total |
|---|---|---|---|---|---|
| 12 | 65 | 32 | 12 | 17 | 138 |
| Assays | Primers /Probes | Sequences (5′-3′) | Genes | Amplicons |
|---|---|---|---|---|
| Duplex real-time PCR | PCV1F | AACCCCATAAGAGGTGGGTGTT | ORF1 | 129 bp |
| PCV1R | TTCTACCCTCTTCCAAACCTTCCT | |||
| PCV1-P | VIC-TCCGAGGAGGAGAAAAACAA AATACGGGA-BHQ1 | |||
| PCV2F | CTGAGTCTTTTTTATCACTTCGTAATGGT | ORF1-ORF2 | 146 bp | |
| PCV2R | ACTGCGTTCGAAAACAGTATATACGA | |||
| PCV2-P | FAM-TTAAGTGGGGGGTCTTTAAGATTA AATTC TCTGAATTGT-BHQ1 | |||
| Real-time PCR | PCV3F | ACTGCGTTCGAAAACAGTATATACGA | ORF2 | 111 bp |
| PCV3R | CATAAATGCTCCAAAGCAGTGCT | |||
| PCV3-P | VIC-ATATGTGTTGAGCCATGGGGTGGG TCT-BHQ1 | |||
| Traditional PCR | PCV2FF | TAATAAAAACCATTACGAAGTGATA | Full genome | 2000 bp |
| PCV2FR | GGTTTTTATTATTCATTAAGGGTTA |
| No. | Accession Number | Strains | Country | Year | Host | Length | Genotype | Organs |
|---|---|---|---|---|---|---|---|---|
| 1 | MW889012 | JX2-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 2 | MW889013 | JX3-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 3 | MW889014 | JX4-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | kidney |
| 4 | MW889015 | JX5-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 5 | MW889016 | JX6-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 6 | MW889017 | JX7-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | spleen |
| 7 | MW889018 | JX8-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 8 | MW889019 | JX9-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 9 | MW889020 | JX10-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 10 | MW889021 | JX11-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 11 | MW889022 | JX12-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 12 | MW889023 | JX13-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 13 | MW889024 | JX14-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 14 | MW889025 | JX15-2020 | China | 2020 | Wild boars | 1767 bp | PCV2d | lymph node |
| 15 | MW889026 | JX16-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 16 | MW889027 | JX17-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | lymph node |
| 17 | MW889028 | JX18-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | kidney |
| 18 | MW889029 | JX19-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | kidney |
| 19 | MW889030 | JX20-2020 | China | 2020 | Wild boars | 1767 bp | PCV2b | kidney |
| Porcine Circovirus Genotype | |||||
|---|---|---|---|---|---|
| PCV1 | PCV2 | PCV3 | PCV1 + PCV2 | PCV2 + PCV3 | |
| Positive (n) | 30 | 31 | 8 | 10 | 5 |
| Total (n) | 138 | 138 | 138 | 138 | 138 |
| Rate (%) | 21.73 | 22.46 | 5.79 | 7.25 | 3.62 |
| No. | Organ | 1 | 2 | 3 | No. | Organ | 1 | 2 | 3 | No. | Organ | 1 | 2 | 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | liver | − | − | − | 48 | lymph node | + | − | − | 94 | lung | + | − | − |
| 2 | kidney | − | + | + | 49 | kidney | − | + | − | 95 | kidney | − | − | − |
| 3 | spleen | − | + | − | 50 | lymph node | − | − | − | 96 | lung | − | − | − |
| 4 | brain | − | − | − | 51 | liver | − | − | − | 97 | kidney | − | − | − |
| 5 | lymph node | + | − | − | 52 | kidney | − | + | − | 98 | kidney | + | + | − |
| 6 | kidney | − | − | − | 53 | Liver | − | − | − | 99 | lung | − | − | − |
| 7 | kidney | − | − | − | 54 | lymph node | − | + | − | 100 | kidney | − | − | − |
| 8 | lung | + | − | − | 55 | kidney | − | − | − | 101 | brain | − | − | − |
| 9 | lung | − | − | − | 56 | liver | − | − | − | 102 | kidney | − | − | − |
| 10 | lung | + | − | − | 57 | liver | − | − | − | 103 | lung | + | − | − |
| 11 | spleen | − | − | − | 58 | brain | − | − | − | 104 | kidney | − | − | − |
| 12 | spleen | + | − | − | 59 | spleen | − | − | − | 105 | lymph node | + | − | − |
| 13 | spleen | − | − | − | 60 | lymph node | − | + | − | 106 | lung | − | − | − |
| 14 | kidney | − | − | − | 61 | spleen | − | − | − | 107 | lymph node | − | + | + |
| 15 | kidney | − | − | − | 62 | lymph node | + | + | − | 108 | lymph node | + | + | − |
| 17 | brain | − | − | − | 63 | spleen | − | − | − | 109 | lung | − | − | − |
| 18 | brain | − | − | − | 64 | kidney | − | − | − | 110 | lymph node | − | − | − |
| 19 | kidney | − | − | − | 65 | kidney | − | + | − | 111 | spleen | − | + | − |
| 20 | lymph node | − | + | − | 66 | brain | − | − | − | 112 | spleen node | − | − | − |
| 21 | lymph node | − | − | − | 67 | liver | − | − | − | 113 | lymph node | − | + | − |
| 22 | lymph node | − | − | − | 68 | lymph node | − | − | + | 114 | lymph node | − | − | − |
| 23 | kidney | + | − | − | 69 | lymph node | − | − | + | 115 | lung | − | − | − |
| 24 | kidney | − | − | − | 70 | liver | − | − | − | 116 | lymph node | + | − | − |
| 25 | spleen | − | − | − | 71 | kidney | − | − | − | 117 | spleen | − | − | − |
| 26 | brain | − | − | − | 72 | kidney | − | − | − | 118 | lung | − | − | − |
| 27 | spleen | + | − | − | 73 | lymph node | + | + | − | 119 | kidney | + | − | − |
| 28 | kidney | + | − | − | 74 | liver | − | − | − | 120 | spleen | + | − | − |
| 29 | spleen | − | − | − | 75 | kidney | − | − | + | 121 | liver | − | − | − |
| 30 | kidney | − | − | − | 76 | liver | − | − | − | 122 | brain | − | − | − |
| 31 | liver | − | − | − | 77 | lung | − | − | − | 123 | lymph node | − | + | − |
| 32 | liver | − | − | − | 78 | brain | − | − | − | 124 | liver | − | − | − |
| 33 | spleen | − | − | − | 79 | liver | − | − | − | 125 | lymph node | − | + | − |
| 34 | lymph node | + | − | − | 80 | kidney | − | − | − | 126 | liver | − | − | − |
| 35 | liver | − | − | − | 81 | kidney | + | − | − | 127 | liver | − | − | − |
| 36 | kidney | − | − | − | 82 | kidney | − | − | − | 128 | lymph node | − | + | − |
| 37 | lymph node | − | + | 83 | lymph node | + | + | − | 129 | liver | − | − | − | |
| 38 | lymph node | + | − | − | 84 | lung | − | − | − | 130 | spleen | − | − | − |
| 39 | kidney | − | − | − | 85 | lymph node | + | + | − | 131 | liver | − | − | − |
| 40 | lymph node | + | + | 86 | lung | − | − | − | 132 | spleen | − | + | − | |
| 41 | lymph node | + | − | − | 87 | lymph node | − | − | − | 133 | lymph node | − | − | − |
| 42 | kidney | − | − | − | 88 | lung | + | − | − | 134 | lymph node | − | + | + |
| 43 | lymph node | + | + | − | 89 | lymph node | + | + | − | 135 | liver | − | − | − |
| 44 | kidney | − | − | − | 90 | liver | − | − | − | 136 | spleen | − | − | − |
| 45 | lymph node | − | + | + | 91 | liver | − | − | − | 137 | kidney | − | + | − |
| 46 | kidney | − | − | − | 92 | kidney | − | + | + | 138 | liver | − | − | − |
| 47 | lymph node | + | + | − | 93 | lung | + | + | − |
| No. | Accession Number | Strains | Genotype | Country | Year | Host |
|---|---|---|---|---|---|---|
| 1 | GU049340 | 1010-Stoon | PCV2a | Cannada | 2007 | wild boar |
| 2 | MH663459 | HBBD1501 | PCV2a | China | 2015 | domestic pig |
| 3 | MK347400 | SD384F16 | PCV2a | China | 2016 | domestic pig |
| 4 | HQ713495 | 5-55004-7 | PCV2b | USA | 2005 | domestic pig |
| 5 | MK347394 | HLJ120215 | PCV2b | China | 2015 | domestic pig |
| 6 | EU148504 | DK1987PMWSfree | PCV2c | Denmark | 2007 | domestic pig |
| 7 | EF524517 | GS04 | PCV2d | China | 2007 | wild boar |
| 8 | AY181946 | TJ | PCV2d | China | 2002 | domestic pig |
| 9 | KT795287 | MEX-41238-2014 | PCV2e | Mexico | 2014 | wild boar |
| 10 | KT795290 | USA-45358-2015 | PCV2e | USA | 2015 | wild boar |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Chen, Z.; Li, Y.; Ding, Z.; Zeng, Q.; Wan, T.; Wu, H. Detection of Porcine Circovirus 1/2/3 and Genetic Analysis of Porcine Circovirus 2 in Wild Boar from Jiangxi Province of China. Animals 2022, 12, 2021. https://doi.org/10.3390/ani12162021
Hu X, Chen Z, Li Y, Ding Z, Zeng Q, Wan T, Wu H. Detection of Porcine Circovirus 1/2/3 and Genetic Analysis of Porcine Circovirus 2 in Wild Boar from Jiangxi Province of China. Animals. 2022; 12(16):2021. https://doi.org/10.3390/ani12162021
Chicago/Turabian StyleHu, Xifeng, Zheng Chen, Yu Li, Zhen Ding, Qinghua Zeng, Tong Wan, and Huansheng Wu. 2022. "Detection of Porcine Circovirus 1/2/3 and Genetic Analysis of Porcine Circovirus 2 in Wild Boar from Jiangxi Province of China" Animals 12, no. 16: 2021. https://doi.org/10.3390/ani12162021
APA StyleHu, X., Chen, Z., Li, Y., Ding, Z., Zeng, Q., Wan, T., & Wu, H. (2022). Detection of Porcine Circovirus 1/2/3 and Genetic Analysis of Porcine Circovirus 2 in Wild Boar from Jiangxi Province of China. Animals, 12(16), 2021. https://doi.org/10.3390/ani12162021






