Introducing the Software CASE (Cluster and Analyze Sound Events) by Comparing Different Clustering Methods and Audio Transformation Techniques Using Animal Vocalizations
Abstract
Simple Summary
Abstract
1. Introduction
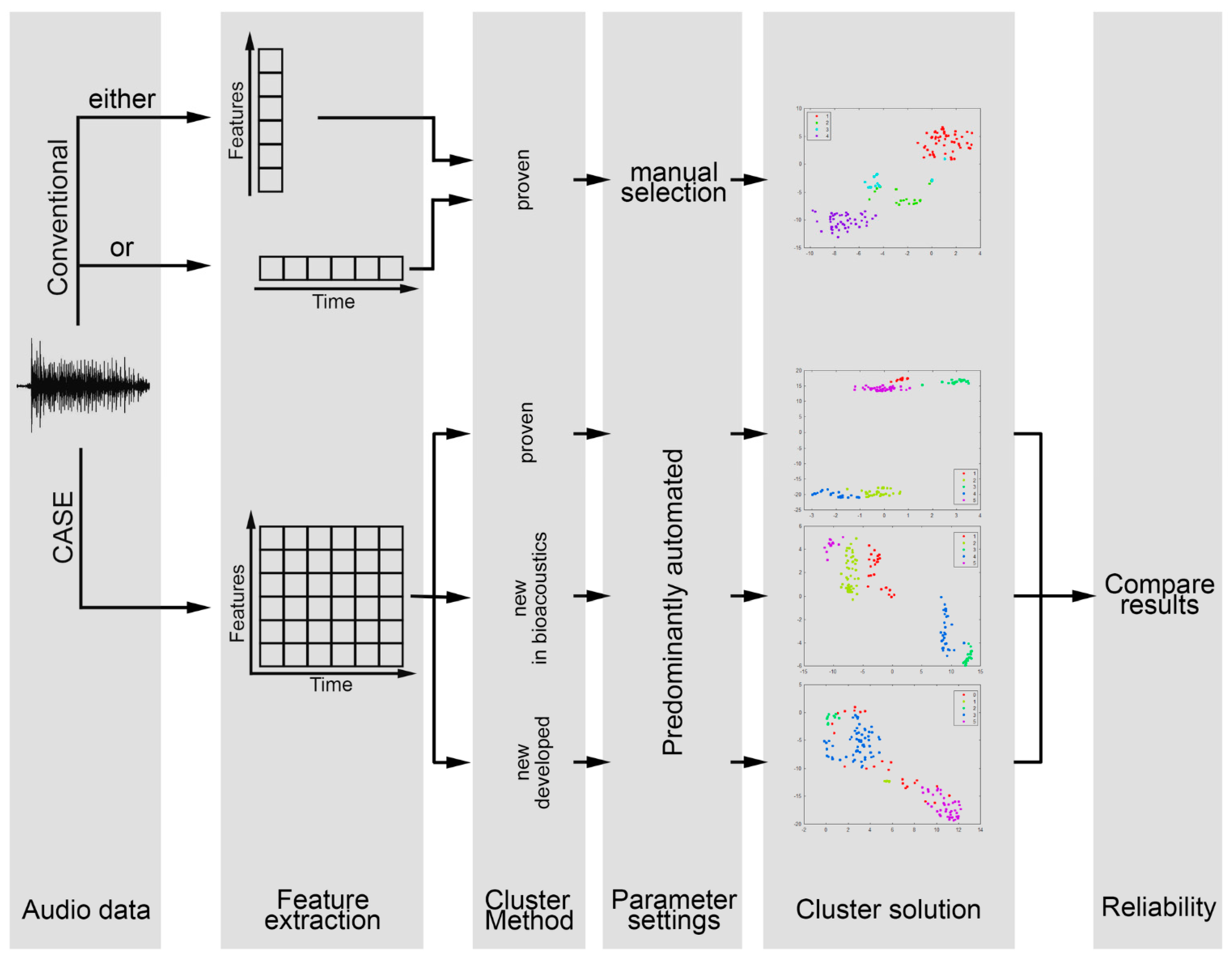
2. Materials and Methods
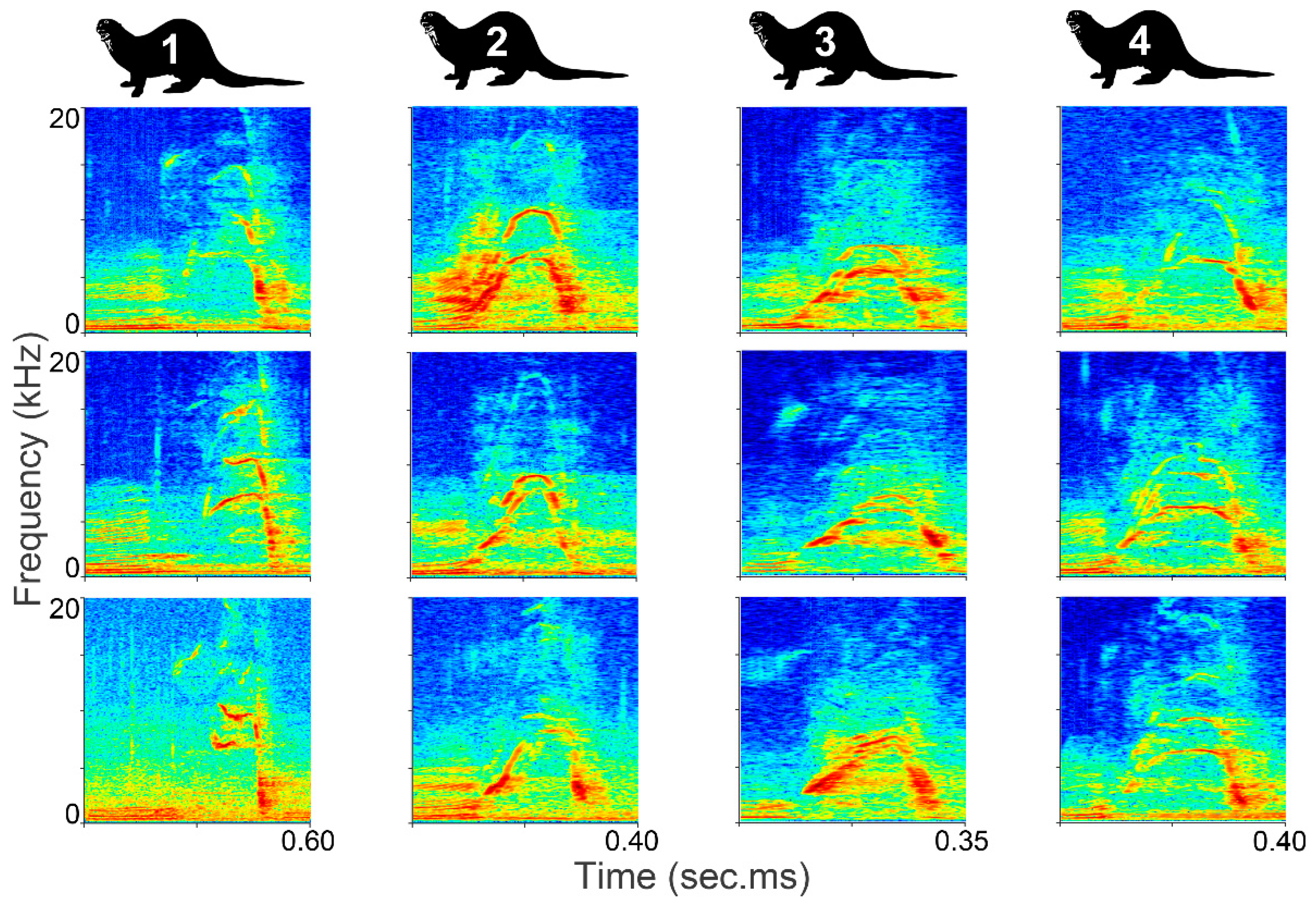
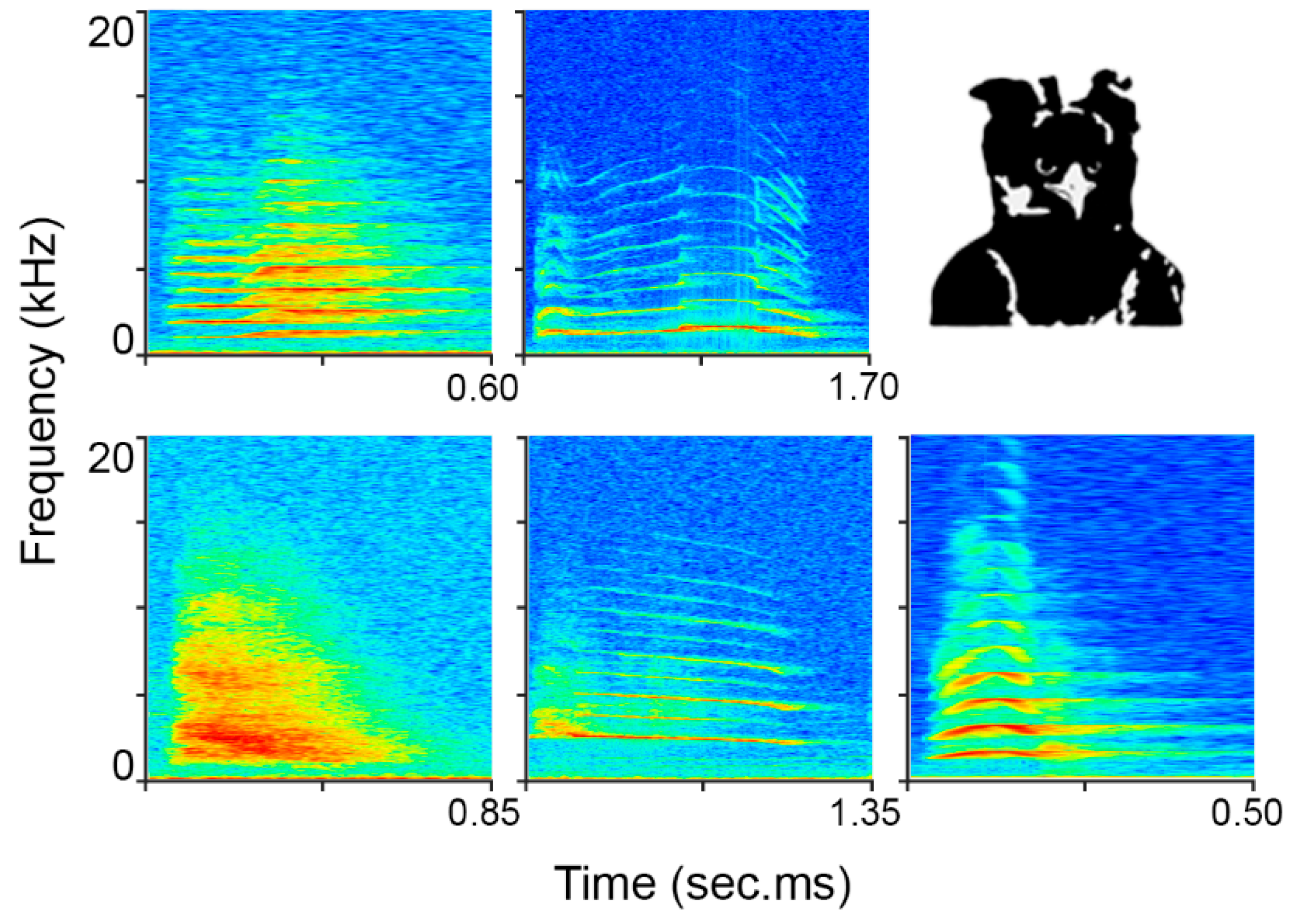
2.1. Data Collection and Study Site
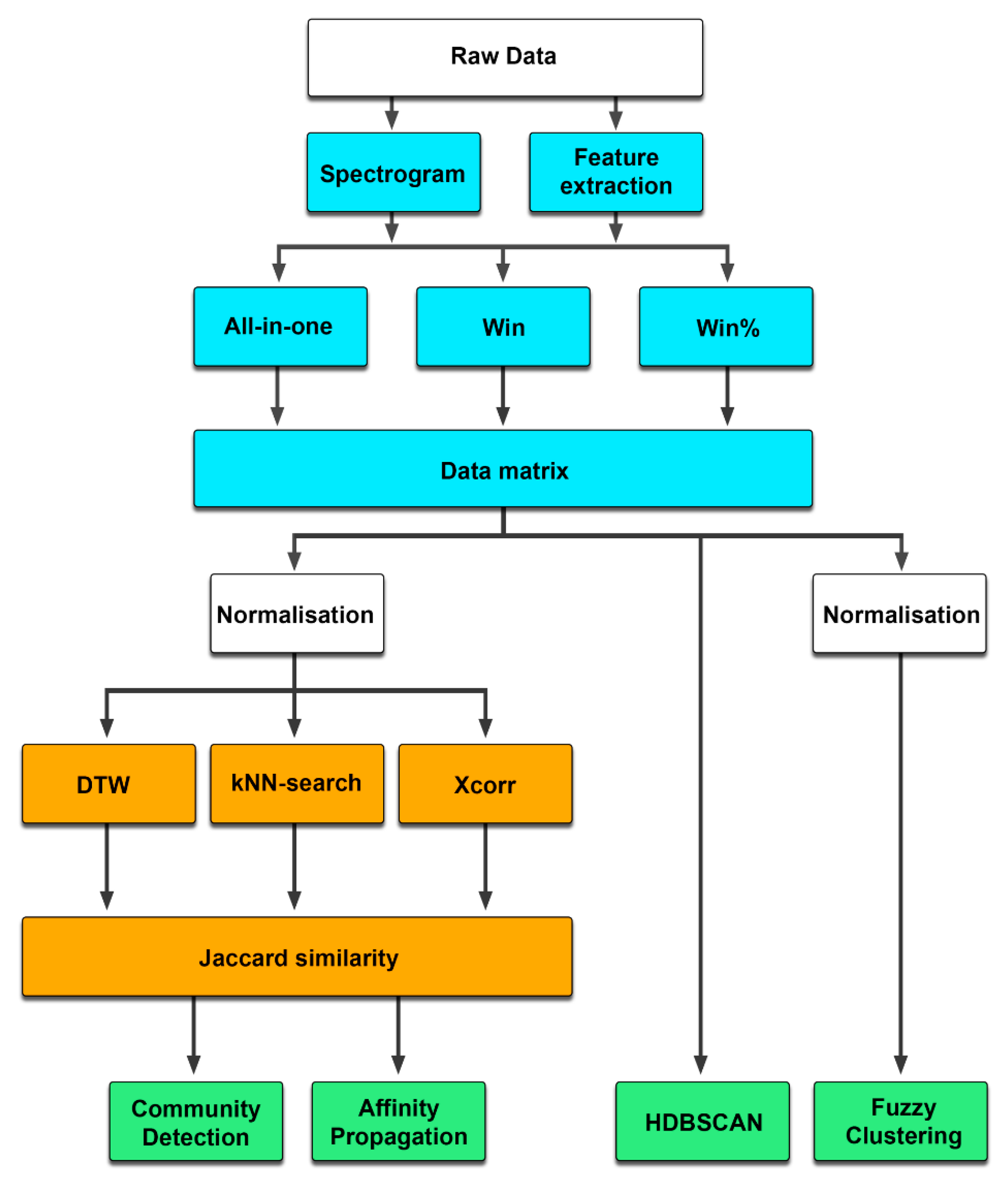
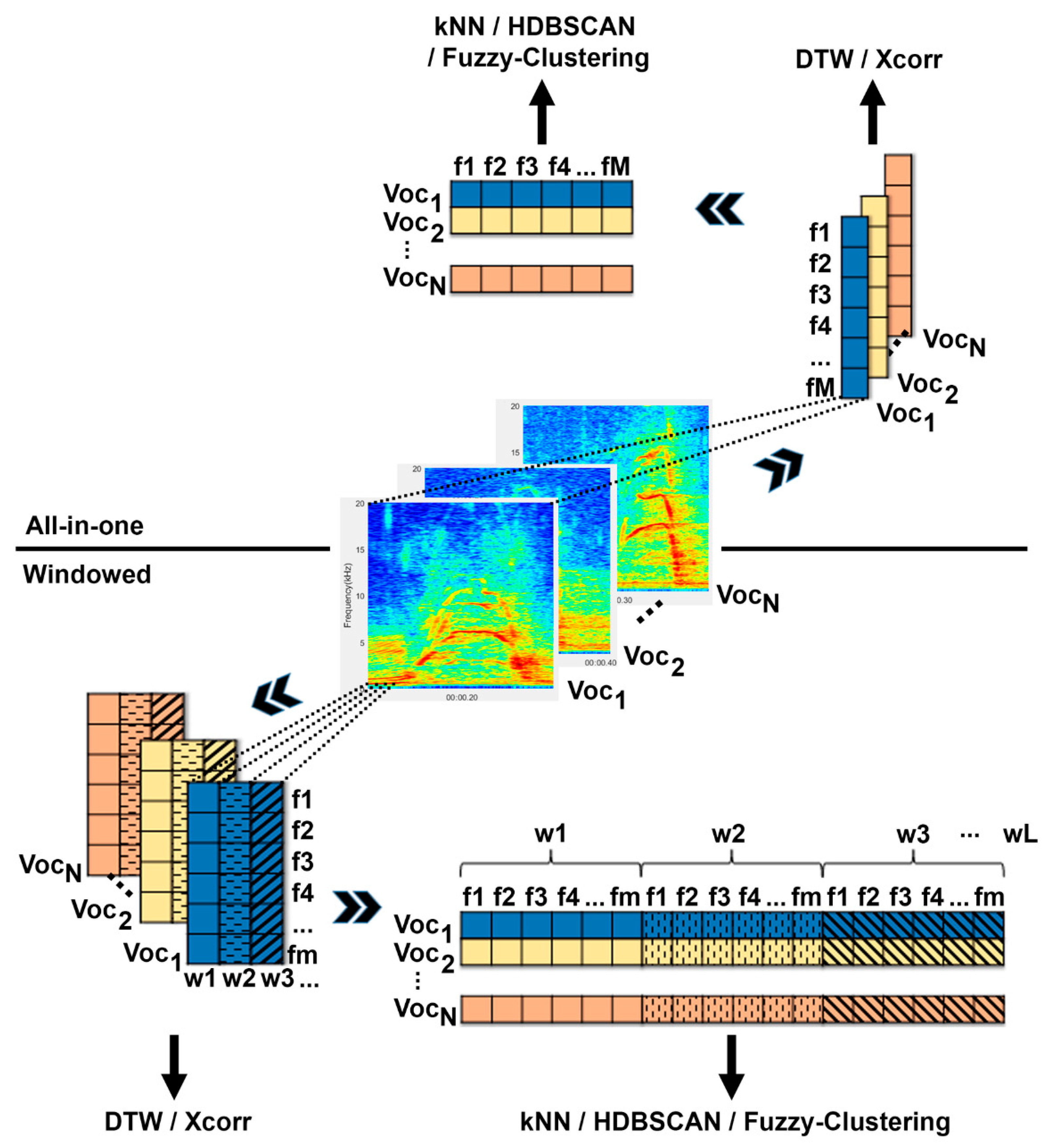
2.2. Data Transformation
2.2.1. All-in-One
2.2.2. Windowed
2.2.3. Feature Extraction
2.3. Clustering Methods
2.3.1. Similarity Matrices
2.3.2. Dynamic Time Warping
2.3.3. k-Nearest Neighbor Search
2.3.4. Cross-Correlation
2.3.5. Community Detection
2.3.6. Affinity Propagation
2.3.7. HDBSCAN
2.3.8. Fuzzy Clustering
2.4. Verifying Cluster Quality
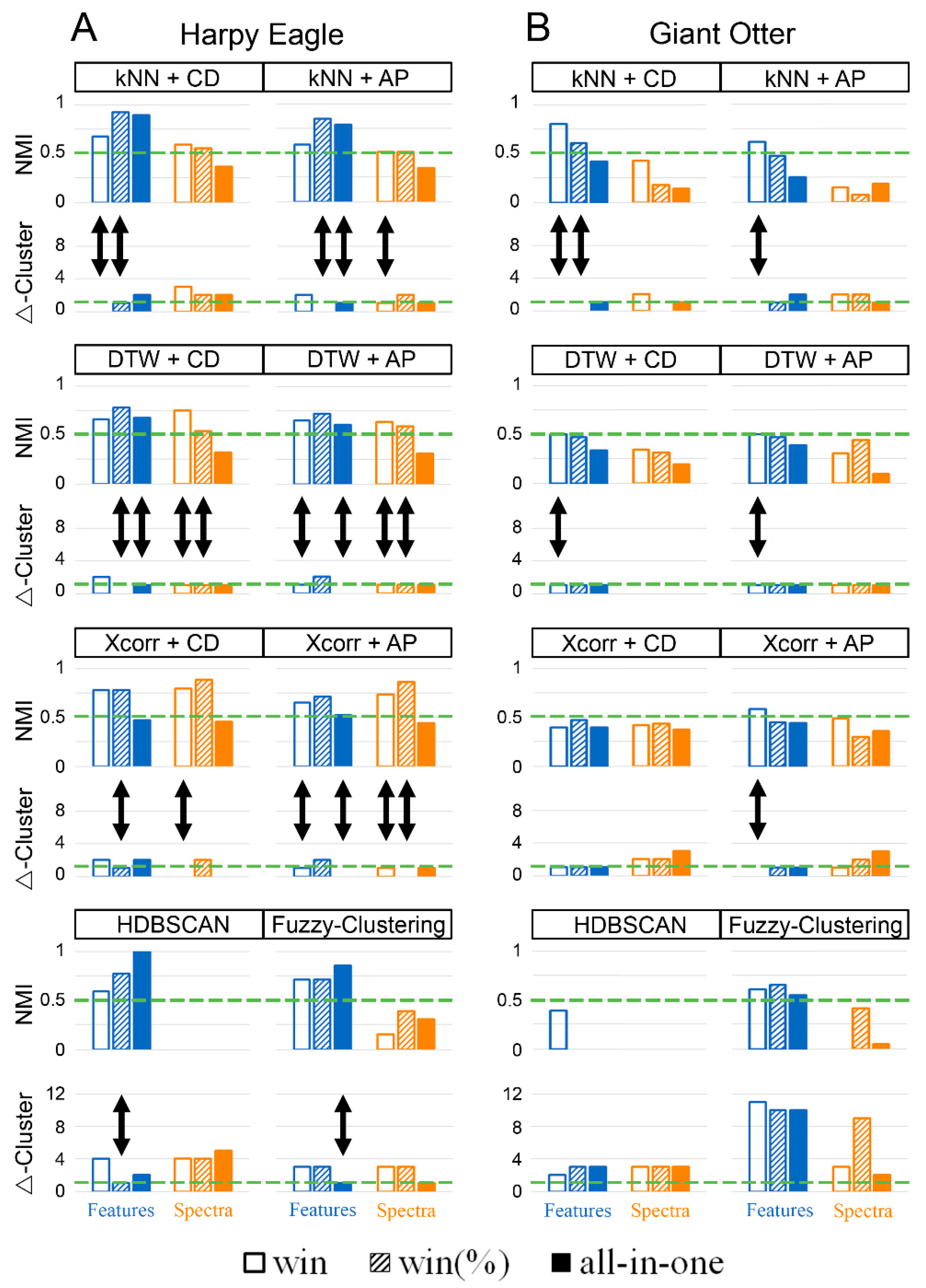
3. Results
4. Discussion
4.1. All-in-One vs. Windowed
4.2. Dimensions Reduction
4.3. Reliability of Resulting Cluster Solutions
4.4. Limitations and Future Opportunities
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Mcloughlin, M.P.; Stewart, R.; McElligott, A.G. Automated bioacoustics: Methods in ecology and conservation and their potential for animal welfare monitoring. J. R. Soc. Interface 2019, 16, 20190225. [Google Scholar] [CrossRef] [PubMed]
- Bardeli, R.; Wolff, D.; Kurth, F.; Koch, M.; Tauchert, K.-H.; Frommolt, K.-H. Detecting bird sounds in a complex acoustic environment and application to bioacoustic monitoring. Pattern Recognit. Lett. 2010, 31, 1524–1534. [Google Scholar] [CrossRef]
- Jones, K.E.; Russ, J.A.; Bashta, A.-T.; Bilhari, Z.; Catto, C.; Csősz, I.; Gorbachev, A.; Győrfi, P.; Hughes, A.; Ivashkiv, I.; et al. Indicator Bats Program: A System for the Global Acoustic Monitoring of Bats. In Biodiversity Monitoring and Conservation; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 211–247. [Google Scholar]
- Marques, T.A.; Thomas, L.; Martin, S.W.; Mellinger, D.K.; Ward, J.A.; Moretti, D.J.; Harris, D.; Tyack, P.L. Estimating animal population density using passive acoustics. Biol. Rev. Camb. Philos. Soc. 2013, 88, 287–309. [Google Scholar] [CrossRef]
- Aide, T.M.; Corrada-Bravo, C.; Campos-Cerqueira, M.; Milan, C.; Vega, G.; Alvarez, R. Real-time bioacoustics monitoring and automated species identification. PeerJ 2013, 1, e103. [Google Scholar] [CrossRef]
- Stevenson, B.C.; Borchers, D.L.; Altwegg, R.; Swift, R.J.; Gillespie, D.M.; Measey, G.J. A general framework for animal density estimation from acoustic detections across a fixed microphone array. Methods Ecol. Evol. 2015, 6, 38–48. [Google Scholar] [CrossRef]
- Frommolt, K.-H. Information obtained from long-term acoustic recordings: Applying bioacoustic techniques for monitoring wetland birds during breeding season. J. Ornithol. 2017, 158, 659–668. [Google Scholar] [CrossRef]
- Pillay, R.; Fletcher, R.J.; Sieving, K.E.; Udell, B.J.; Bernard, H. Bioacoustic monitoring reveals shifts in breeding songbird populations and singing behaviour with selective logging in tropical forests. J. Appl. Ecol. 2019, 56, 2482–2492. [Google Scholar] [CrossRef]
- Revilla-Martín, N.; Budinski, I.; Puig-Montserrat, X.; Flaquer, C.; López-Baucells, A. Monitoring cave-dwelling bats using remote passive acoustic detectors: A new approach for cave monitoring. Bioacoustics 2020, 30, 527–542. [Google Scholar] [CrossRef]
- Pamula, H.; Pocha, A.; Klaczynski, M. Towards the acoustic monitoring of birds migrating at night. Biodivers. Inf. Sci. Stand. 2019, 3, e36589. [Google Scholar] [CrossRef]
- Marin-Cudraz, T.; Muffat-Joly, B.; Novoa, C.; Aubry, P.; Desmet, J.-F.; Mahamoud-Issa, M.; Nicolè, F.; van Niekerk, M.H.; Mathevon, N.; Sèbe, F. Acoustic monitoring of rock ptarmigan: A multi-year comparison with point-count protocol. Ecol. Indic. 2019, 101, 710–719. [Google Scholar] [CrossRef]
- Teixeira, D.; Maron, M.; Rensburg, B.J. Bioacoustic monitoring of animal vocal behavior for conservation. Conserv. Sci. Pract. 2019, 1, e72. [Google Scholar] [CrossRef]
- Eens, M.; Pinxten, R.; Verheyen, R.F. Function of the song and song repertoire in the european starling (Sturnus vulgaris): An aviary experiment. Behaviour 1993, 125, 51–66. [Google Scholar] [CrossRef]
- Hasselquist, D.; Bensch, S.; Schantz, T. Correlation between male song repertoire, extra-pair paternity and offspring survival in the great reed warbler. Nature 1996, 381, 229–232. [Google Scholar] [CrossRef]
- Freeberg, T.M.; Dunbar, R.I.M.; Ord, T.J. Social complexity as a proximate and ultimate factor in communicative complexity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1785–1801. [Google Scholar] [CrossRef] [PubMed]
- Ripley, B.D. Pattern Recognition and Neural Networks; Cambridge Univ. Press: Cambridge, UK, 2009; ISBN 9780521717700. [Google Scholar]
- Kershenbaum, A.; Blumstein, D.T.; Roch, M.A.; Akçay, Ç.; Backus, G.; Bee, M.A.; Bohn, K.; Cao, Y.; Carter, G.; Cäsar, C.; et al. Acoustic sequences in non-human animals: A tutorial review and prospectus. Biol. Rev. Camb. Philos. Soc. 2016, 91, 13–52. [Google Scholar] [CrossRef] [PubMed]
- Kershenbaum, A. Entropy rate as a measure of animal vocal complexity. Bioacoustics 2014, 23, 195–208. [Google Scholar] [CrossRef]
- Jones, A.E.; ten Cate, C.; Bijleveld, C.C. The interobserver reliability of scoring sonagrams by eye: A study on methods, illustrated on zebra finch songs. Anim. Behav. 2001, 62, 791–801. [Google Scholar] [CrossRef][Green Version]
- Clink, D.J.; Klinck, H. Unsupervised acoustic classification of individual gibbon females and the implications for passive acoustic monitoring. Methods Ecol. Evol. 2020, 12, 328–341. [Google Scholar] [CrossRef]
- Adi, K.; Johnson, M.T.; Osiejuk, T.S. Acoustic censusing using automatic vocalization classification and identity recognition. J. Acoust. Soc. Am. 2010, 127, 874–883. [Google Scholar] [CrossRef]
- Rama Rao, K.; Garg, S.; Montgomery, J. Investigation of unsupervised models for biodiversity assessment. In AI 2018: Advances in Artificial Intelligence; Mitrovic, T., Xue, B., Li, X., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 160–171. ISBN 978-3-030-03990-5. [Google Scholar]
- Phillips, Y.F.; Towsey, M.; Roe, P. Revealing the ecological content of long-duration audio-recordings of the environment through clustering and visualisation. PLoS ONE 2018, 13, e0193345. [Google Scholar] [CrossRef]
- Ruff, Z.J.; Lesmeister, D.B.; Appel, C.L.; Sullivan, C.M. Workflow and convolutional neural network for automated identification of animal sounds. Ecol. Indic. 2021, 124, 107419. [Google Scholar] [CrossRef]
- Wadewitz, P.; Hammerschmidt, K.; Battaglia, D.; Witt, A.; Wolf, F.; Fischer, J. Characterizing vocal repertoires--hard vs. soft classification approaches. PLoS ONE 2015, 10, e0125785. [Google Scholar] [CrossRef] [PubMed]
- Riondato, I.; Cissello, E.; Papale, E.; Friard, O.; Gamba, M.; Giacoma, C. Unsupervised acoustic analysis of the vocal repertoire of the gray-shanked douc langur (Pygathrix cinerea). J. Comp. Acous. 2017, 25, 1750018. [Google Scholar] [CrossRef]
- Schneider, S.; Goettlich, S.; Diercks, C.; Dierkes, P.W. Discrimination of acoustic stimuli and maintenance of graded alarm call structure in captive meerkats. Animals 2021, 11, 3064. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, D.; Karagiannis, A.; Gallopin, T.; Gutch, H.W.; Cauli, B. Beyond the frontiers of neuronal types. Front. Neural Circuits 2013, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef]
- Hsu, H.-H.; Hsieh, C.-W. Feature selection via correlation coefficient clustering. J. Softw. 2010, 5, 1371–1377. [Google Scholar] [CrossRef]
- Matthews, J. Detection of frequency-modulated calls using a chirp model. Can. Acoust. 2004, 32, 66–75. [Google Scholar]
- Fischer, J.; Wegdell, F.; Trede, F.; Dal Pesco, F.; Hammerschmidt, K. Vocal convergence in a multi-level primate society: Insights into the evolution of vocal learning. Proc. Biol. Sci. 2020, 287, 20202531. [Google Scholar] [CrossRef]
- Kershenbaum, A.; Sayigh, L.S.; Janik, V.M. The encoding of individual identity in dolphin signature whistles: How much information is needed? PLoS ONE 2013, 8, e77671. [Google Scholar] [CrossRef]
- Odom, K.J.; Araya-Salas, M.; Morano, J.L.; Ligon, R.A.; Leighton, G.M.; Taff, C.C.; Dalziell, A.H.; Billings, A.C.; Germain, R.R.; Pardo, M.; et al. Comparative bioacoustics: A roadmap for quantifying and comparing animal sounds across diverse taxa. Biol. Rev. Camb. Philos. Soc. 2021, 96, 1135–1159. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.W.; Markler, P.; Beeman, K. Quantitative analysis of animal vocal phonology: An application to swamp sparrow song. Ethology 1987, 76, 101–115. [Google Scholar] [CrossRef]
- Han, N.C.; Muniandy, S.V.; Dayou, J. Acoustic classification of australian anurans based on hybrid spectral-entropy approach. Appl. Acoust. 2011, 72, 639–645. [Google Scholar] [CrossRef]
- Xie, J.; Towsey, M.; Eichinski, P.; Zhang, J.; Roe, P. Acoustic feature extraction using perceptual wavelet packet decomposition for frog call classification. In Proceedings of the 2015 IEEE 11th International Conference on e-Science (e-Science), Munich, Germany, 31 August–4 September 2015; pp. 237–242, ISBN 978-1-4673-9325-6. [Google Scholar]
- Abu Alfeilat, H.A.; Hassanat, A.B.A.; Lasassmeh, O.; Tarawneh, A.S.; Alhasanat, M.B.; Eyal Salman, H.S.; Prasath, V.B.S. Effects of distance measure choice on k-nearest neighbor classifier performance: A review. Big Data 2019, 7, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, S. Community detection in graphs. Phys. Rep. 2010, 486, 75–174. [Google Scholar] [CrossRef]
- Malliaros, F.D.; Vazirgiannis, M. Clustering and community detection in directed networks: A survey. Phys. Rep. 2013, 533, 95–142. [Google Scholar] [CrossRef]
- Newman, M.E.J. Fast algorithm for detecting community structure in networks. Phys. Rev. E 2004, 69, 66133. [Google Scholar] [CrossRef]
- Peel, L.; Larremore, D.B.; Clauset, A. The ground truth about metadata and community detection in networks. Sci. Adv. 2017, 3, e1602548. [Google Scholar] [CrossRef]
- Campello, R.J.G.B.; Moulavi, D.; Sander, J. Density-based clustering based on hierarchical density estimates. In Advances in Knowledge Discovery and Data Mining; Pei, J., Tseng, V.S., Cao, L., Motoda, H., Xu, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 160–172. ISBN 978-3-642-37456-2. [Google Scholar]
- Sainburg, T.; Thielk, M.; Gentner, T.Q. Finding, visualizing, and quantifying latent structure across diverse animal vocal repertoires. PLoS Comput. Biol. 2020, 16, e1008228. [Google Scholar] [CrossRef]
- Poupard, M.; Best, P.; Schlüter, J.; Symonds, H.; Spong, P.; Glotin, H. Large-scale unsupervised clustering of orca vocalizations: A model for describing orca communication systems. PeerJ Prepr. 2019, 7, e27979v1. [Google Scholar] [CrossRef]
- Mumm, C.A.; Urrutia, M.C.; Knörnschild, M. Vocal individuality in cohesion calls of giant otters, Pteronura brasiliensis. Anim. Behav. 2014, 88, 243–252. [Google Scholar] [CrossRef]
- Schrader, L.; Hammerschmidt, K. Computer-aided analysis of acoustic parameters in animal vocalisations: A multi-parametric approach. Bioacoustics 1997, 7, 247–265. [Google Scholar] [CrossRef]
- Paliwal, K.K.; Agarwal, A.; Sinha, S.S. A modification over Sakoe and Chiba’s dynamic time warping algorithm for isolated word recognition. Signal Processing 1982, 4, 329–333. [Google Scholar] [CrossRef]
- Sakoe, H.; Chiba, S. Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans. Acoust. Speech Signal Processing 1978, ASSP-26, 43–49. [Google Scholar] [CrossRef]
- Friedman, J.H.; Bentley, J.L.; Finkler, R.A. An algorithm for finding best matches in logarithmic expected time. ACM Trans. Math. Softw. 1977, 3, 209–226. [Google Scholar] [CrossRef]
- Sarvaiya, J.N.; Patnaik, S.; Bombaywala, S. Image registration by template matching using normalized cross-correlation. In Proceedings of the 2009 International Conference on Advances in Computing, Control, & Telecommunication Technologies (ACT 2009), Trivandrum, Kerala, 28–29 December 2009; pp. 819–822, ISBN 978-1-4244-5321-4. [Google Scholar]
- Wang, k.; Zhang, J.; Li, D.; Zhang, X.; Guo, T. Adaptive affinity propagation clustering. Acta Autom. Sin. 2007, 33, 1242–1246. [Google Scholar]
- Campello, R.J.G.B.; Moulavi, D.; Zimek, A.; Sander, J. Hierarchical density estimates for data clustering, visualization, and outlier detection. ACM Trans. Knowl. Discov. Data 2015, 10, 1–51. [Google Scholar] [CrossRef]
- Zadeh, L.A. Fuzzy sets. Inf. Control 1965, 8, 338–353. [Google Scholar] [CrossRef]
- Zadeh, L.A. Is there a need for fuzzy logic? Inf. Sci. 2008, 178, 2751–2779. [Google Scholar] [CrossRef]
- Schneider, S.; Dierkes, P.W. Localize Animal Sound Events Reliably (LASER): A new software for sound localization in zoos. J. Zool. Bot. Gard. 2021, 2, 146–163. [Google Scholar] [CrossRef]
- Gamba, M.; Friard, O.; Riondato, I.; Righini, R.; Colombo, C.; Miaretsoa, L.; Torti, V.; Nadhurou, B.; Giacoma, C. Comparative analysis of the vocal repertoire of eulemur: A dynamic time warping approach. Int. J. Primatol. 2015, 36, 894–910. [Google Scholar] [CrossRef]
- van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Winkler, R.; Klawonn, F.; Kruse, R. Problems of fuzzy c-means clustering and similar algorithms with high dimensional data sets. In Challenges at the Interface of Data Analysis, Computer Science, and Optimization; Gaul, W.A., Geyer-Schulz, A., Schmidt-Thieme, L., Kunze, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 79–87. ISBN 978-3-642-24465-0. [Google Scholar]
- Glotin, H.; Razik, J.; Paris, S.; Halkias, X. Sparse coding for large scale bioacoustic similarity function improved by multiscale scattering. Proc. Meet. Acoust. 2013, 19, 10015. [Google Scholar] [CrossRef]
- Fischer, J.; Hammerschmidt, K.; Cheney, D.L.; Seyfarth, R.M. Acoustic features of male baboon loud calls: Influences of context, age, and individuality. J. Acoust. Soc. Am. 2002, 111, 1465–1474. [Google Scholar] [CrossRef]
- Erb, W.M.; Hodges, J.K.; Hammerschmidt, K. Individual, contextual, and age-related acoustic variation in Simakobu (Simias concolor) loud calls. PLoS ONE 2013, 8, e83131. [Google Scholar] [CrossRef]
- Meise, K.; Keller, C.; Cowlishaw, G.; Fischer, J. Sources of acoustic variation: Implications for production specificity and call categorization in chacma baboon (Papio ursinus) grunts. J. Acoust. Soc. Am. 2011, 129, 1631–1641. [Google Scholar] [CrossRef]
- Sharan, R.V.; Moir, T.J. Acoustic event recognition using cochleagram image and convolutional neural networks. Appl. Acoust. 2019, 148, 62–66. [Google Scholar] [CrossRef]
- Fettiplace, R. Diverse mechanisms of sound frequency discrimination in the vertebrate cochlea. Trends Neurosci. 2020, 43, 88–102. [Google Scholar] [CrossRef]
- Wienicke, A.; Häusler, U.; Jürgens, U. Auditory frequency discrimination in the squirrel monkey. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 2001, 187, 189–195. [Google Scholar] [CrossRef]
- Oikarinen, T.; Srinivasan, K.; Meisner, O.; Hyman, J.B.; Parmar, S.; Fanucci-Kiss, A.; Desimone, R.; Landman, R.; Feng, G. Deep convolutional network for animal sound classification and source attribution using dual audio recordings. J. Acoust. Soc. Am. 2019, 145, 654. [Google Scholar] [CrossRef]
- Ko, K.; Park, S.; Ko, H. Convolutional feature vectors and support vector machine for animal sound classification. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 376–379. [Google Scholar] [CrossRef]
- Tacioli, L.; Toledo, L.; Medeiros, C. An architecture for animal sound identification based on multiple feature extraction and classification algorithms. In Anais do Brazilian e-Science Workshop (BreSci); Sociedade Brasileira de Computação-SBC: Porto Alegre, Brasil, 2017; pp. 29–36. [Google Scholar]
- Abbas, O.A. Comparisons between data clustering algorithms. Int. Arab. J. Inf. Technol. 2008, 5, 320–325. [Google Scholar]
- Shirkhorshidi, A.S.; Aghabozorgi, S.; Wah, T.Y.; Herawan, T. Big Data Clustering: A Review. In Computational Science and Its Applications-ICCSA 2014: 14th International Conference, Guimarães, Portugal, 30 June–3 July 2014; Proceedings; Murgante, B., Misra, S., Rocha, A.M.A.C., Torre, C., Rocha, J.G., Falcão, M.I., Taniar, D., Apduhan, B.O., Gervasi, O., Eds.; Springer: Cham, Switzerland, 2014; pp. 707–720. ISBN 978-3-319-09155-6. [Google Scholar]
- Hathaway, R.J.; Bezdek, J.C. Extending fuzzy and probabilistic clustering to very large data sets. Comput. Stat. Data Anal. 2006, 51, 215–234. [Google Scholar] [CrossRef]
| Acoustic Features | Definition of Features (All-in-One) | Definition of Features (Windowed) | Reference |
|---|---|---|---|
| F0 | Median fundamental frequency | Fundamental frequency for each window | Audio Toolbox “pitch” with NCF |
| Delta F0 | Median value of the difference between adjacent values for F0 per window | Value of the difference between adjacent values for F0 | |
| Dominant f | Frequency with highest amplitude in the spectrum | Frequency with highest amplitude in the spectrum of each window | |
| Min f | Lower bound of the 99% occupied bandwidth | Lower bound of the 99% occupied bandwidth | Signal Processing Toolbox “obw” |
| Max f | Upper bound of the 99% occupied bandwidth | Upper bound of the 99% occupied bandwidth | Signal Processing Toolbox “obw” |
| Bandwidth | 99% occupied bandwidth | 99% occupied bandwidth | Signal Processing Toolbox “obw” |
| Duration | Time between onset and offset of a vocalization in seconds. | Duration only determined for win%, not for win. | |
| 1st Quartile | 1st quartile of the energy distribution | 1st quartile of the energy distribution | [47] |
| 2nd Quartile | 2nd quartile of the energy distribution | 2nd quartile of the energy distribution | [47] |
| 3rd Quartile | 3rd quartile of the energy distribution | 3rd quartile of the energy distribution | [47] |
| Max Q1 | Frequency with highest amplitude in the 1st quartile | Frequency with highest amplitude in the 1st quartile | |
| Max Q2 | Frequency with highest amplitude in the 2nd quartile | Frequency with highest amplitude in the 2nd quartile | |
| Max Q3 | Frequency with highest amplitude in the 3rd quartile | Frequency with highest amplitude in the 3rd quartile | |
| Max Q4 | Frequency with highest amplitude in the 4th quartile | Frequency with highest amplitude in the 4th quartile | |
| FB1 | Median frequency of the 1st frequency band determined by LPC (Hz) | Frequency of the 1st frequency band determined by LPC | Signal Processing Toolbox “lpc” with 205 coefficients |
| FB2 | Median frequency of the 2nd frequency band | Frequency of the 2nd frequency band | LPC with 205 coefficients |
| FB3 | Median frequency of the 3rd frequency band | Frequency of the 3rd frequency band | LPC with 205 coefficients |
| BW FB1 | Median bandwidth of FB1 | Bandwidth of FB1 | “findpeaks” |
| BW FB2 | Median bandwidth of FB2 | Bandwidth of FB2 | “findpeaks” |
| BW FB3 | Median bandwidth of FB3 | Bandwidth of FB3 | “findpeaks” |
| Delta FB1-FB2 | Difference between FB1 and FB2 | Difference between FB1 and FB2 | |
| Delta FB2-FB3 | Difference between FB2 and FB3 | Difference between FB2 and FB3 | |
| Number of FB | Number of frequency bands determined by LPC | Number of frequency bands determined by LPC | Number calculated by LPC |
| Harmonic Ratio | Harmonic ratio is returned with values in the range of 0 to 1. A value of 0 represents low harmonicity, and a value of 1 represents high harmonicity | Harmonic ratio is returned with values in the range of 0 to 1. A value of 0 represents low harmonicity, and a value of 1 represents high harmonicity | Audio Toolbox “harmonicRatio” |
| Spectral Flatness | Measures how noisy a signal is. The higher the value, the noisier the signal | Measures how noisy a signal is. The higher the value, the noisier the signal | Audio Toolbox “spectralFlatness” |
| MFCC 1 | 1st mel frequency cepstral coefficient | 1st mel frequency cepstral coefficient | Audio Toolbox “mfcc” |
| MFCC 2 | 2nd mel frequency cepstral coefficient | 2nd mel frequency cepstral coefficient | Audio Toolbox “mfcc” |
| Clustering Methods | Acoustic Features | Spectra | Data Set | ||||
|---|---|---|---|---|---|---|---|
| Win | Win(%) | All-in-One | Win | Win(%) | All-in-One | ||
| kNN + CD | 0.80 | 0.61 | Giant Otter | ||||
| kNN + AP | 0.61 | ||||||
| DTW + CD | 0.50 | ||||||
| DTW + AP | 0.50 | ||||||
| Xcorr + CD | |||||||
| Xcorr + AP | 0.58 | ||||||
| HDBSCAN | |||||||
| fuzzy | |||||||
| Clustering Methods | Acoustic Features | Spectra | Data Set | ||||
|---|---|---|---|---|---|---|---|
| Win | Win(%) | All-in-One | Win | Win(%) | All-in-One | ||
| kNN + CD | 0.67 | 0.92 | Harpy Eagle | ||||
| kNN + AP | 0.85 | 0.79 | 0.51 | ||||
| DTW + CD | 0.78 | 0.67 | 0.75 | 0.54 | |||
| DTW + AP | 0.65 | 0.61 | 0.64 | 0.59 | |||
| Xcorr + CD | 0.78 | 0.79 | |||||
| Xcorr + AP | 0.65 | 0.74 | 0.87 | ||||
| HDBSCAN | 0.77 | ||||||
| fuzzy | 0.85 | ||||||
| Acoustic Features (Win%) | Spectrogram (Win%) | Acoustic Features (All-in-One) | ||||||
|---|---|---|---|---|---|---|---|---|
| kNN + CD | kNN + AP | DTW + CD | DTW + AP | HDBSCAN | Xcorr + CD | Xcorr + AP | Fuzzy | |
| kNN + AP | 0.90 | |||||||
| DTW + CD | 0.70 | 0.65 | ||||||
| DTW + AP | 0.64 | 0.63 | 0.86 | |||||
| HDBSCAN | 0.68 | 0.68 | 0.92 | 0.88 | ||||
| Xcorr + CD | 0.91 | 0.81 | 0.65 | 0.60 | 0.68 | |||
| Xcorr + AP | 0.88 | 0.79 | 0.68 | 0.62 | 0.64 | 0.84 | ||
| Fuzzy | 0.78 | 0.72 | 0.73 | 0.67 | 0.76 | 0.80 | 0.74 | |
| Acoustic Features (Win%) | Spectrogram (Win%) | Acoustic Features (All-in-One) | ||||||
|---|---|---|---|---|---|---|---|---|
| kNN + CD | kNN + AP | DTW + CD | DTW + AP | HDBSCAN | Xcorr + CD | Xcorr + AP | Fuzzy | |
| kNN + AP | 0.64 | |||||||
| DTW + CD | 0.56 | 0.45 | ||||||
| DTW + AP | 0.56 | 0.45 | 1 | |||||
| HDBSCAN | 0.4 | 0.42 | 0.10 | 0.10 | ||||
| Xcorr + CD | 0.37 | 0.46 | 0.41 | 0.41 | 0.46 | |||
| Xcorr + AP | 0.41 | 0.43 | 0.41 | 0.41 | 0.32 | 0.67 | ||
| Fuzzy | 0.57 | 0.59 | 0.51 | 0.51 | 0.47 | 0.63 | 0.42 | |
| Harpy Eagle | Giant Otter | |||
|---|---|---|---|---|
| Person | NMI | Δ-Cluster | NMI | Δ-Cluster |
| A | 0.9233 | 1 | 0.3013 | 1 |
| B | 0.9293 | 0 | 0.5079 | 3 |
| C | 0.8527 | 1 | 0.381 | 1 |
| D | 0.9545 | 0 | 0.3236 | 0 |
| E | 0.9233 | 1 | 0.3877 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schneider, S.; Hammerschmidt, K.; Dierkes, P.W. Introducing the Software CASE (Cluster and Analyze Sound Events) by Comparing Different Clustering Methods and Audio Transformation Techniques Using Animal Vocalizations. Animals 2022, 12, 2020. https://doi.org/10.3390/ani12162020
Schneider S, Hammerschmidt K, Dierkes PW. Introducing the Software CASE (Cluster and Analyze Sound Events) by Comparing Different Clustering Methods and Audio Transformation Techniques Using Animal Vocalizations. Animals. 2022; 12(16):2020. https://doi.org/10.3390/ani12162020
Chicago/Turabian StyleSchneider, Sebastian, Kurt Hammerschmidt, and Paul Wilhelm Dierkes. 2022. "Introducing the Software CASE (Cluster and Analyze Sound Events) by Comparing Different Clustering Methods and Audio Transformation Techniques Using Animal Vocalizations" Animals 12, no. 16: 2020. https://doi.org/10.3390/ani12162020
APA StyleSchneider, S., Hammerschmidt, K., & Dierkes, P. W. (2022). Introducing the Software CASE (Cluster and Analyze Sound Events) by Comparing Different Clustering Methods and Audio Transformation Techniques Using Animal Vocalizations. Animals, 12(16), 2020. https://doi.org/10.3390/ani12162020







