Molecular Assessments, Statistical Effectiveness Parameters and Genetic Structure of Captive Populations of Tursiops truncatus Using 15 STRs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotyping Analysis
2.3. Statistical Analysis
3. Results
3.1. Microsatellite Diversity Parameters and Paternity Effectiveness
3.2. Null Alleles, Hardy–Weinberg Expectation, and Linkage Disequilibrium
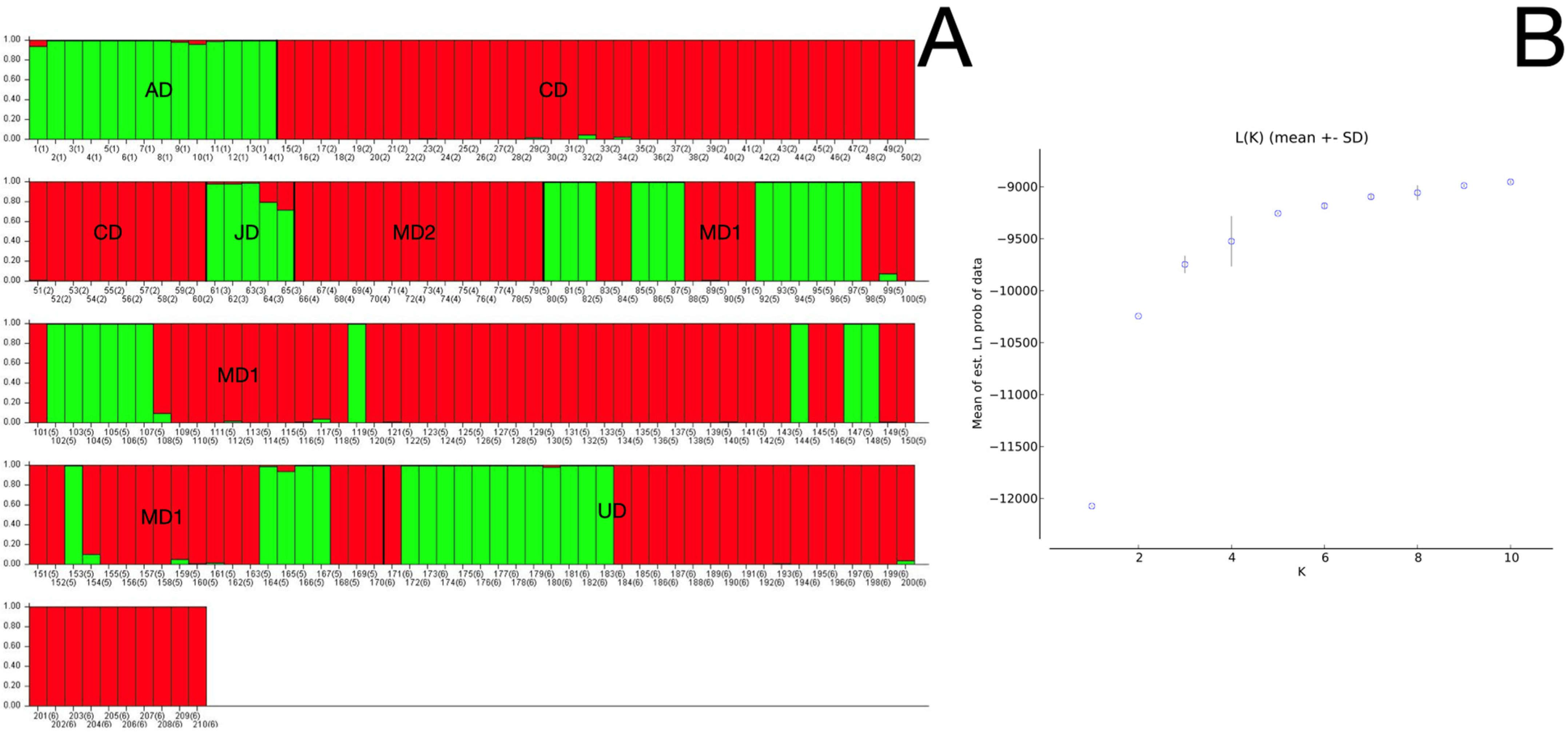
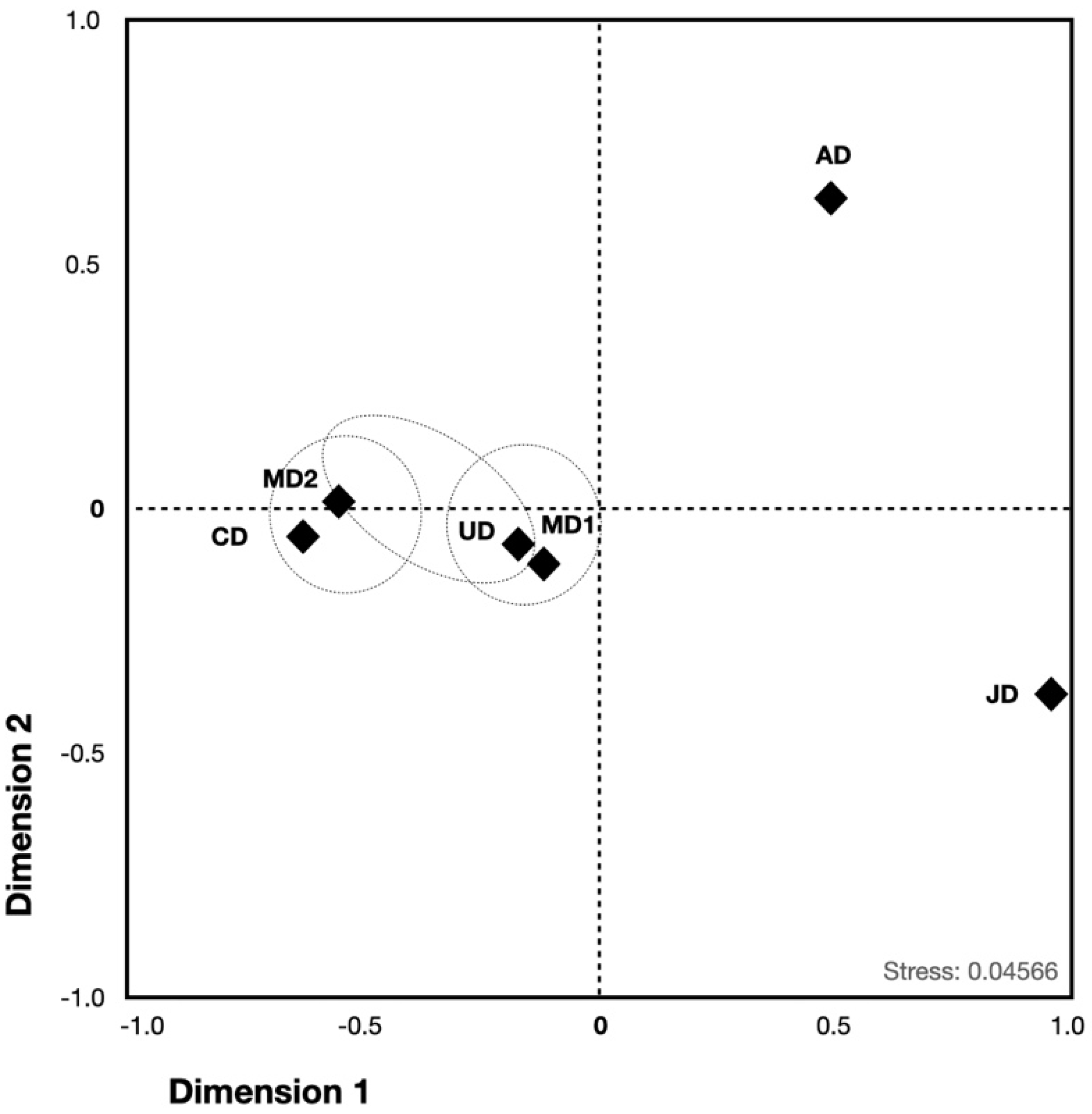
3.3. Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- The Society for Marine Mammalogy List of Marine Mammal Species and Subspecies. Available online: https://marinemammalscience.org/science-and-publications/list-marine-mammal-species-subspecies (accessed on 2 June 2022).
- Barratclough, A.; Wells, R.S.; Schwacke, L.H.; Rowles, T.K.; Gomez, F.M.; Fauquier, D.A.; Sweeney, J.C.; Townsend, F.I.; Hansen, L.J.; Zolman, E.S.; et al. Health Assessments of Common Bottlenose Dolphins (Tursiops truncatus): Past, Present, and Potential Conservation Applications. Front. Vet. Sci. 2019, 6, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randall, S.; Wells, M.D.S. Bottlenose Dolphin, Tursiops Truncates, Common Bottlenose Dolphin. In Encyclopedia of Marine Mammals, 3rd ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 249–255. [Google Scholar]
- Klatsky, L.J.; Wells, R.S.; Sweeney, J.C. Offshore Bottlenose Dolphins (Tursiops truncatus): Movement and Dive Behavior Near the Bermuda Pedestal. J. Mammal. 2007, 88, 59–66. [Google Scholar] [CrossRef] [Green Version]
- McCormack, M.A.; Battaglia, F.; McFee, W.E.; Dutton, J. Mercury concentrations in blubber and skin from stranded bottlenose dolphins (Tursiops truncatus) along the Florida and Louisiana coasts (Gulf of Mexico, USA) in relation to biological variables. Environ. Res. 2020, 180, 108886. [Google Scholar] [CrossRef] [PubMed]
- Tóth, B.; Khosravi, R.; Ashrafzadeh, M.R.; Bagi, Z.; Fehér, M.; Bársony, P.; Kovács, G.; Kusza, S. Genetic Diversity and Structure of Common Carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: Implications for Conservation. Genes 2020, 11, 1268. [Google Scholar] [CrossRef]
- Probert, R.; Bastian, A.; Elwen, S.H.; James, B.S.; Gridley, T. Vocal correlates of arousal in bottlenose dolphins (Tursiops spp.) in human care. PLoS ONE 2021, 16, e0250913. [Google Scholar] [CrossRef]
- Harley, H.E.; Fellner, W.; Stamper, M.A. Cognitive research with dolphins (Tursiops truncatus) at Disney’s The Seas: A program for enrichment, science education, and conservation. Int. J. Comp. Psychol. 2010, 23, 331–343. [Google Scholar]
- O’Brien, J.K.; Robeck, T.R. The Value of Ex Situ Cetacean Populations in Understanding Reproductive Physiology and Developing Assisted Reproductive Technology for Ex Situ and In Situ Species Management and Conservation Efforts. Int. J. Comp. Psychol. 2010, 223, 227–248. [Google Scholar]
- Giménez, J.; Ramírez, F.; Almunia, J.; Forero, M.G.; de Stephanis, R. From the pool to the sea: Applicable isotope turnover rates and diet to skin discrimination factors for bottlenose dolphins (Tursiops truncatus). J. Exp. Mar. Biol. Ecol. 2016, 475, 54–61. [Google Scholar] [CrossRef]
- Ley General de la Vida Silvestre. Available online: https://www.profepa.gob.mx/innovaportal/file/5779/1/ley_general_de_vida_silvestre.pdf. (accessed on 9 July 2022).
- Lima, A.; Sébilleau, M.; Boye, M.; Durand, C.; Hausberger, M.; Lemasson, A. Captive bottlenose dolphins do discriminate human-made sounds both underwater and in the air. Front. Psychol. 2018, 9, 55. [Google Scholar] [CrossRef] [Green Version]
- Clegg, I.L.K.; Delfour, F. Cognitive judgement bias is associated with frequency of anticipatory behavior in bottlenose dolphins. Zoo Biol. 2018, 37, 67–73. [Google Scholar] [CrossRef]
- Fahlman, A.; Brodsky, M.; Miedler, S.; Dennison, S.; Ivančić, M.; Levine, G.; Rocho-Levine, J.; Manley, M.; Rocabert, J.; Borque-Espinosa, A. Ventilation and gas exchange before and after voluntary static surface breath-holds in clinically healthy bottlenose dolphins, Tursiops truncatus. J. Exp. Biol. 2019, 222, jeb192211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, I.; Nishida, S.; Yang, W.-C.; Isobe, T.; Tajima, Y.; Hoelzel, A.R. Genetic diversity of bottlenose dolphin (Tursiops sp.) populations in the western North Pacific and the conservation implications. Mar. Biol. 2017, 164, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas-Fonseca, O.A.; Yates, P.; Kirkman, S.P.; Pistorius, P.A.; Moore, D.M.; Natoli, A.; Cockcroft, V.; Hoelzel, A.R. Population structure associated with bioregion and seasonal prey distribution for Indo-Pacific bottlenose dolphins (Tursiops aduncus) in South Africa. Mol. Ecol. 2021, 30, 4642–4659. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Johansson, M.L.; Heath, D.D. Role of genomics and transcriptomics in selection of reintroduction source populations. Conserv. Biol. 2016, 30, 1010–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escorza-Treviño, S.; Dizon, A.E. Phylogeography, intraspecific structure and sex-biased dispersal of Dall’s porpoise, Phocoenoides dalli, revealed by mitochondrial and microsatellite DNA analyses. Mol. Ecol. 2000, 9, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Butler, I.A.; Siletti, K.; Oxley, P.R.; Kronauer, D.J.C. Conserved microsatellites in ants enable population genetic and colony pedigree studies across a wide range of species. PLoS ONE 2014, 9, e107334. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, E.C.; Ferrari, S.F.; Bastos, H.B.; Wajntal, A.; Aleixo, A.; Schneider, M.P.C. Comparative genetic diversity of wild and captive populations of the bare-faced curassow (Crax fasciolata) based on cross-species microsatellite markers: Implications for conservation and management. Biochem. Genet. 2010, 48, 472–479. [Google Scholar] [CrossRef]
- Salas, M.; Fernández-Fontelo, A.; Martínez-Nevado, E.; Fernández-Morán, J.; López-Goya, A.; Manteca, X. Caretaker Score Reliability for Personality Assessment of Bottlenose Dolphin (Tursiops truncatus). Animals 2021, 11, 2073. [Google Scholar] [CrossRef]
- Rosel, P.E.; Hansen, L.; Hohn, A.A. Restricted dispersal in a continuously distributed marine species: Common bottlenose dolphins Tursiops truncatus in coastal waters of the western North Atlantic. Mol. Ecol. 2009, 18, 5030–5045. [Google Scholar] [CrossRef]
- Krützen, M.; Barré, L.M.; Connor, R.C.; Mann, J.; Sherwin, W.B. ‘O father: Where art thou?’—Paternity assessment in an open fission-fusion society of wild bottlenose dolphins (Tursiops sp.) in Shark Bay, Western Australia. Mol. Ecol. 2004, 13, 1975–1990. [Google Scholar] [CrossRef]
- Krützen, M.; Valsecchi, E.; Connor, R.C.; Sherwin, W.B. Characterization of microsatellite loci in Tursiops aduncus. Mol. Ecol. Notes 2001, 1, 170–172. [Google Scholar] [CrossRef]
- Rosel, P.E.; Forgetta, V.; Dewark, K. Isolation and characterization of twelve polymorphic microsatellite markers in bottlenose dolphins (Tursiops truncatus). Mol. Ecol. Notes 2005, 5, 830–833. [Google Scholar] [CrossRef]
- Rooney, A.P.; Honeycutt, R.L.; Davis, S.K.; Derr, J.N. Evaluating a putative bottleneck in a population of bowhead whales from patterns of microsatellite diversity and genetic disequilibria. J. Mol. Evol. 1999, 49, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, M.; Domingo-Roura, X.; Takenaka, O. Microsatellites in the bottlenose dolphin Tursiops truncatus. Mol. Ecol. 1997, 6, 695–696. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, E.; Amos, W. Microsatellite markers for the study of cetacean populations. Mol. Ecol. 1996, 5, 151–156. [Google Scholar] [CrossRef]
- Hoelzel, A.R.; Potter, C.W.; Best, P.B. Genetic differentiation between parapatric “nearshore” and “offshore” populations of the bottlenose dolphin. Proc. Biol. Sci. 1998, 265, 1177–1183. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3. 2001. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 11 January 2022).
- Van Ossterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Tereba, A. PowerStats v1.2 Tools for Analysis of Population Statistics, Profiles DNA vol. 3 1999. Available online: http://www.promega.com/geneticidtools/powerstats (accessed on 11 January 2022).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Duricki, D.A.; Soleman, S.; Moon, L.D.F. Analysis of longitudinal data from animals with missing values using SPSS. Nat. Protoc. 2016, 11, 1112–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noris, G.; Santana, C.; Meraz-Ríos, M.A.; de Lourdes Munoz, M.; Majluf-Cruz, A.; Magaña, J.J.; Granados, J.; Quezada, R.; Revilla, M.C.; Martínez-Salas, S.; et al. Mexican mestizo population sub-structure: Effects on genetic and forensic statistical parameters. Mol. Biol. Rep. 2012, 39, 10139–10156. [Google Scholar] [CrossRef]
- Viana, J.M.S.; Garcia, A.A.F. Significance of linkage disequilibrium and epistasis on genetic variances in noninbred and inbred populations. BMC Genom. 2022, 23, 286. [Google Scholar] [CrossRef]
- National Research Council (US); Committee on DNA Forensic Science. The Evaluation of Forensic DNA Evidence; An Update; Press, N.A., Ed.; National Academy of Sciences: Washington, DC, USA, 1996. Available online: https://www.ncbi.nlm.nih.gov/books/NBK232610/ (accessed on 29 June 2022)ISBN 0-309-05395-1.
- Frankham, R. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 2008, 17, 325–333. [Google Scholar] [CrossRef]
- Marshall, I.R.; Brauer, C.J.; Wedderburn, S.D.; Whiterod, N.S.; Hammer, M.P.; Barnes, T.C.; Attard, C.R.M.; Möller, L.M.; Beheregaray, L.B. Longitudinal monitoring of neutral and adaptive genomic diversity in a reintroduction. Conserv. Biol. 2022, e13889. [Google Scholar] [CrossRef]
- Grueber, C.E.; Hogg, C.J.; Ivy, J.A.; Belov, K. Impacts of early viability selection on management of inbreeding and genetic diversity in conservation. Mol. Ecol. 2015, 24, 1645–1653. [Google Scholar] [CrossRef]
- Marine Mammal Commission. Status of Marine Mammal Species and Populations. Available online: https://www.mmc.gov/priority-topics/species-of-concern/status-of-marine-mammal-species-and-populations/ (accessed on 11 June 2022).
- Caballero, S.; Islas-Villanueva, V.; Tezanos-Pinto, G.; Duchene, S.; Delgado-Estrella, A.; Sanchez-Okrucky, R.; Mignucci-Giannoni, A.A. Phylogeography, genetic diversity and population structure of common bottlenose dolphins in the Wider Caribbean inferred from analyses of mitochondrial DNA control region sequences and microsatellite loci: Conservation and management implications. Anim. Conserv. 2012, 15, 95–112. [Google Scholar] [CrossRef]
- Louis, M.; Fontaine, M.C.; Spitz, J.; Schlund, E.; Dabin, W.; Deaville, R.; Caurant, F.; Cherel, Y.; Guinet, C.; Simon-Bouhet, B. Ecological opportunities and specializations shaped genetic divergence in a highly mobile marine top predator. Proc. Biol. Sci. 2014, 281, 20141558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farías-Curtidor, N.; Barragán-Barrera, D.C.; Chávez-Carreño, P.A.; Jiménez-Pinedo, C.; Palacios, D.M.; Caicedo, D.; Trujillo, F.; Caballero, S. Range extension for the common dolphin (Delphinus sp.) to the Colombian Caribbean, with taxonomic implications from genetic barcoding and phylogenetic analyses. PLoS ONE 2017, 12, e0171000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milián-García, Y.; Ramos-Targarona, R.; Pérez-Fleitas, E.; Sosa-Rodríguez, G.; Guerra-Manchena, L.; Alonso-Tabet, M.; Espinosa-López, G.; Russello, M.A. Genetic evidence of hybridization between the critically endangered Cuban crocodile and the American crocodile: Implications for population history and in situ/ex situ conservation. Heredity 2015, 114, 272–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oudejans, M.G.; Visser, F.; Englund, A.; Rogan, E.; Ingram, S.N. Evidence for distinct coastal and offshore communities of bottlenose dolphins in the north east Atlantic. PLoS ONE 2015, 10, e0122668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellas, A.B.; Wells, R.S.; Rosel, P.E. Mitochondrial and nuclear DNA analyses reveal fine scale geographic structure in bottlenose dolphins (Tursiops truncatus) in the Gulf of Mexico. Conserv. Genet. 2005, 6, 715–728. [Google Scholar] [CrossRef]
- Richards, V.P.; Greig, T.W.; Fair, P.A.; McCulloch, S.D.; Politz, C.; Natoli, A.; Driscoll, C.A.; Hoelzel, A.R.; David, V.; Bossart, G.D.; et al. Patterns of population structure for inshore bottlenose dolphins along the eastern United States. J. Hered. 2013, 104, 765–778. [Google Scholar] [CrossRef] [Green Version]
- Segura-García, I.; Rojo-Arreola, L.; Rocha-Olivares, A.; Heckel, G.; Gallo-Reynoso, J.P.; Hoelzel, R. Eco-Evolutionary Processes Generating Diversity Among Bottlenose Dolphin, Tursiops truncatus, Populations off Baja California, Mexico. Evol. Biol. 2018, 45, 223–236. [Google Scholar] [CrossRef] [Green Version]
- Quérouil, S.; Silva, M.A.; Freitas, L.; Prieto, R.; Magalhães, S.; Dinis, A.; Alves, F.; Matos, J.A.; Mendonça, D.; Hammond, P.S.; et al. High gene flow in oceanic bottlenose dolphins (Tursiops truncatus) of the North Atlantic. Conserv. Genet. 2007, 8, 1405. [Google Scholar] [CrossRef] [Green Version]
- Tezanos-Pinto, G.; Baker, C.S.; Russell, K.; Martien, K.; Baird, R.W.; Hutt, A.; Stone, G.; Mignucci-Giannoni, A.A.; Caballero, S.; Endo, T.; et al. A worldwide perspective on the population structure and genetic diversity of bottlenose dolphins (Tursiops truncatus) in New Zealand. J. Hered. 2009, 100, 11–24. [Google Scholar] [CrossRef] [Green Version]
- Fruet, P.F.; Secchi, E.R.; Daura-Jorge, F.; Vermeulen, E.; Flores, P.A.C.; Simões-Lopes, P.C.; Genoves, R.C.; Laporta, P.; Di Tullio, J.C.; Freitas, T.R.O.; et al. Remarkably low genetic diversity and strong population structure in common bottlenose dolphins (Tursiops truncatus) from coastal waters of the Southwestern Atlantic Ocean. Conserv. Genet. 2014, 15, 879–895. [Google Scholar] [CrossRef]
- Malone, E.W.; Perkin, J.S.; Leckie, B.M.; Kulp, M.A.; Hurt, C.R.; Walker, D.M. Which species, how many, and from where: Integrating habitat suitability, population genomics, and abundance estimates into species reintroduction planning. Glob. Chang. Biol. 2018, 24, 3729–3748. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.E.; Shreves, K.; Pilot, M.; Andrews, K.R.; Moore, D.M.; Kishida, T.; Möller, L.; Natoli, A.; Gaspari, S.; McGowen, M.; et al. Phylogenomics of the genus Tursiops and closely related Delphininae reveals extensive reticulation among lineages and provides inference about eco-evolutionary drivers. Mol. Phylogenet. Evol. 2020, 146, 106756. [Google Scholar] [CrossRef] [PubMed]
- Kardos, M.; Armstrong, E.E.; Fitzpatrick, S.W.; Hauser, S.; Hedrick, P.W.; Miller, J.M.; Tallmon, D.A.; Funk, W.C. The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. USA 2021, 118, e2104642118. [Google Scholar] [CrossRef] [PubMed]
- Pratt, E.A.L.; Beheregaray, L.B.; Bilgmann, K.; Zanardo, N.; Diaz-Aguirre, F.; Brauer, C.; Sandoval-Castillo, J.; Möller, L.M. Seascape genomics of coastal bottlenose dolphins along strong gradients of temperature and salinity. Mol. Ecol. 2022, 31, 2223–2241. [Google Scholar] [CrossRef]
- Louis, M.; Viricel, A.; Lucas, T.; Peltier, H.; Alfonsi, E.; Berrow, S.; Brownlow, A.; Covelo, P.; Dabin, W.; Deaville, R.; et al. Habitat-driven population structure of bottlenose dolphins, Tursiops truncatus, in the North-East Atlantic. Mol. Ecol. 2014, 23, 857–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minterr, B.A.; Collins, J.P. Guidelines for Reintroductions and Other Conservation Translocations; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2010; Volume 20, ISBN 9782831716091. [Google Scholar]
- Cunningham-Smith, P.; Colbert-Luke, D.; Wells, R.; Speakman, T. Evaluation of Human Interactions with a Provisioned Wild Bottlenose Dolphin (Tursiops truncatus) near Sarasota Bay, Florida, and Efforts to Curtail the Interactions. Aquat. Mamm. 2006, 32, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Maity, S.; Singh, S.K.; Yadav, V.K.; Chandra, K.; Sharma, L.K.; Thakur, M. DNA matchmaking in captive facilities: A case study with tigers. Mol. Biol. Rep. 2022, 49, 4107–4114. [Google Scholar] [CrossRef]
- Miller, L.J.; Zeigler-Hill, V.; Mellen, J.; Koeppel, J.; Greer, T.; Kuczaj, S. DolphinShows and Interaction Programs: Benefits for Conservation Education? Zoo Biol. 2013, 32, 45–53. [Google Scholar] [CrossRef]
| Locus | Gene Bank Access Number | Tandem Repeat | Sequence primer (5′–3′) | Allele Size | Annealing T | Reference | |
|---|---|---|---|---|---|---|---|
| D08 | NA | (TG)n | F | GATCCATCATATTGTCAAGTT | 94–122 | 58 | [27] |
| R | TCCTGGGTGATGAGTCTTC | ||||||
| EV37 | NA | (AC)n | F | AGCTTGATTTGGAAGTCATGA | 189–241 | 56 | [28] |
| R | TAGTAGAGCCGTGATAAAGTGC | ||||||
| KWM2 | NA | (AC)n | F | GCTGTGAAAATTAAATGT | 138–160 | 47 | [29] |
| R | CACTGTGGACAAATGTAA | ||||||
| KWM9 | NA | (AC)n | F | TGTCACCAGGCAGGACCC | 170–188 | 59 | [29] |
| R | GGGAGGGGCATGTTTCTG | ||||||
| KWM12 | NA | (AC)n | F | CCATACAATCCAGCAGTC | 160–186 | 50 | [29] |
| R | CACTGCAGAATGATGACC | ||||||
| MK6 | AF237891 | (GT)n | F | GTCCTCTTTCCAGGTGTAGCC | 147–187 | 51 | [24] |
| R | GCCCACTAAGTATGTTGCAGC | ||||||
| MK8 | AF237892 | (CA)n | F | TCCTGGAGCATCTTATAGTGGC | 80–114 | 58 | [24] |
| R | CTCTTTGACATGCCCTCACC | ||||||
| MK9 | AF237893 | (CA)n | F | CATAACAAAGTGGGATGACTCC | 161–175 | 54 | [24] |
| R | TTATCCTGTTGGCTGCAGTG | ||||||
| Ttr04 | DQ018982 | (CA)n | F | CTGACCAGGCACTTTCCAC | 103–127 | 65 | [25] |
| R | GTTTGTTTCCCAGGATTTTAGTGC | ||||||
| Ttr11 | DQ018981 | (CA)n | F | CTTTCAACCTGGCCTTTCTG | 193–219 | 61 | [25] |
| R | GTTTGGCCACTACAAGGGAGTGAA | ||||||
| Ttr19 | DQ018980 | (CA)n | F | TGGGTGGACCTCATCAAATC | 182–200 | 61 | [25] |
| R | GTTTAAGGGCTGTAAGAGG | ||||||
| Ttr58 | DQ018985 | (CA)n | F | TGGGTCTTGAGGGGTCTG | 166–194 | 62 | [25] |
| R | GTTTGCTGAGGCTCCTTGTTGG | ||||||
| Ttr63 | DQ018986 | (CA)n | F | CAGCTTACAGCCAAATGAGAG | 83–149 | 59 | [25] |
| R | GTTTCTCCATGGCTGAGTCATCA | ||||||
| TexVet5 | AF004905 | (CA)n | F | GATTGTGCAAATGGAGACA | 196–216 | 55 | [26] |
| R | TTGAGATGACTCCTGTGGG | ||||||
| TexVet7 | AF004907 | (CA)n | F | TGCACTGTAGGGTGTTCAGCAG | 155–169 | 64 | [26] |
| R | CTTAATTGGGGGCGATTTCAC | ||||||
| AD | CD | JD | MD | UD | |
|---|---|---|---|---|---|
| AD | - | ≤0.0001 | ≤0.0001 | ≤0.0001 | ≤0.0001 |
| CD | 0.27886 | - | ≤0.0001 | ≤0.0001 | ≤0.0001 |
| JD | 0.15746 | 0.23717 | - | ≤0.0001 | ≤0.0001 |
| MD | 0.17679 | 0.05293 | 0.11988 | - | 0.25225 ± 0.0264 |
| UD | 0.20180 | 0.03210 | 0.13256 | 0.00965 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez, R.; Neri-Bazán, R.M.; Posadas-Mondragon, A.; Vizcaíno-Dorado, P.A.; Magaña, J.J.; Aguilar-Faisal, J.L. Molecular Assessments, Statistical Effectiveness Parameters and Genetic Structure of Captive Populations of Tursiops truncatus Using 15 STRs. Animals 2022, 12, 1857. https://doi.org/10.3390/ani12141857
Gómez R, Neri-Bazán RM, Posadas-Mondragon A, Vizcaíno-Dorado PA, Magaña JJ, Aguilar-Faisal JL. Molecular Assessments, Statistical Effectiveness Parameters and Genetic Structure of Captive Populations of Tursiops truncatus Using 15 STRs. Animals. 2022; 12(14):1857. https://doi.org/10.3390/ani12141857
Chicago/Turabian StyleGómez, Rocío, Rocío M. Neri-Bazán, Araceli Posadas-Mondragon, Pablo A. Vizcaíno-Dorado, Jonathan J. Magaña, and José Leopoldo Aguilar-Faisal. 2022. "Molecular Assessments, Statistical Effectiveness Parameters and Genetic Structure of Captive Populations of Tursiops truncatus Using 15 STRs" Animals 12, no. 14: 1857. https://doi.org/10.3390/ani12141857
APA StyleGómez, R., Neri-Bazán, R. M., Posadas-Mondragon, A., Vizcaíno-Dorado, P. A., Magaña, J. J., & Aguilar-Faisal, J. L. (2022). Molecular Assessments, Statistical Effectiveness Parameters and Genetic Structure of Captive Populations of Tursiops truncatus Using 15 STRs. Animals, 12(14), 1857. https://doi.org/10.3390/ani12141857







