Drinking Water with Saccharin Sodium Alters the Microbiota-Gut-Hypothalamus Axis in Guinea Pig
Abstract
:Simple Summary
Abstract
1. Instruction
2. Materials and Methods
2.1. Animal, Diets, and Management
2.2. Sample Collection
2.3. Analysis and Observation of Intestinal Morphological
2.4. Lactic Acid and SCFA Detection in the Ileal Digesta
2.5. RNA Isolation
2.6. RNA-Seq Analysis
2.7. Genes Identification
2.8. qRT-PCR
2.9. 16 S Ribosomal DNA Gene Sequencing
2.10. Statistical Analyses
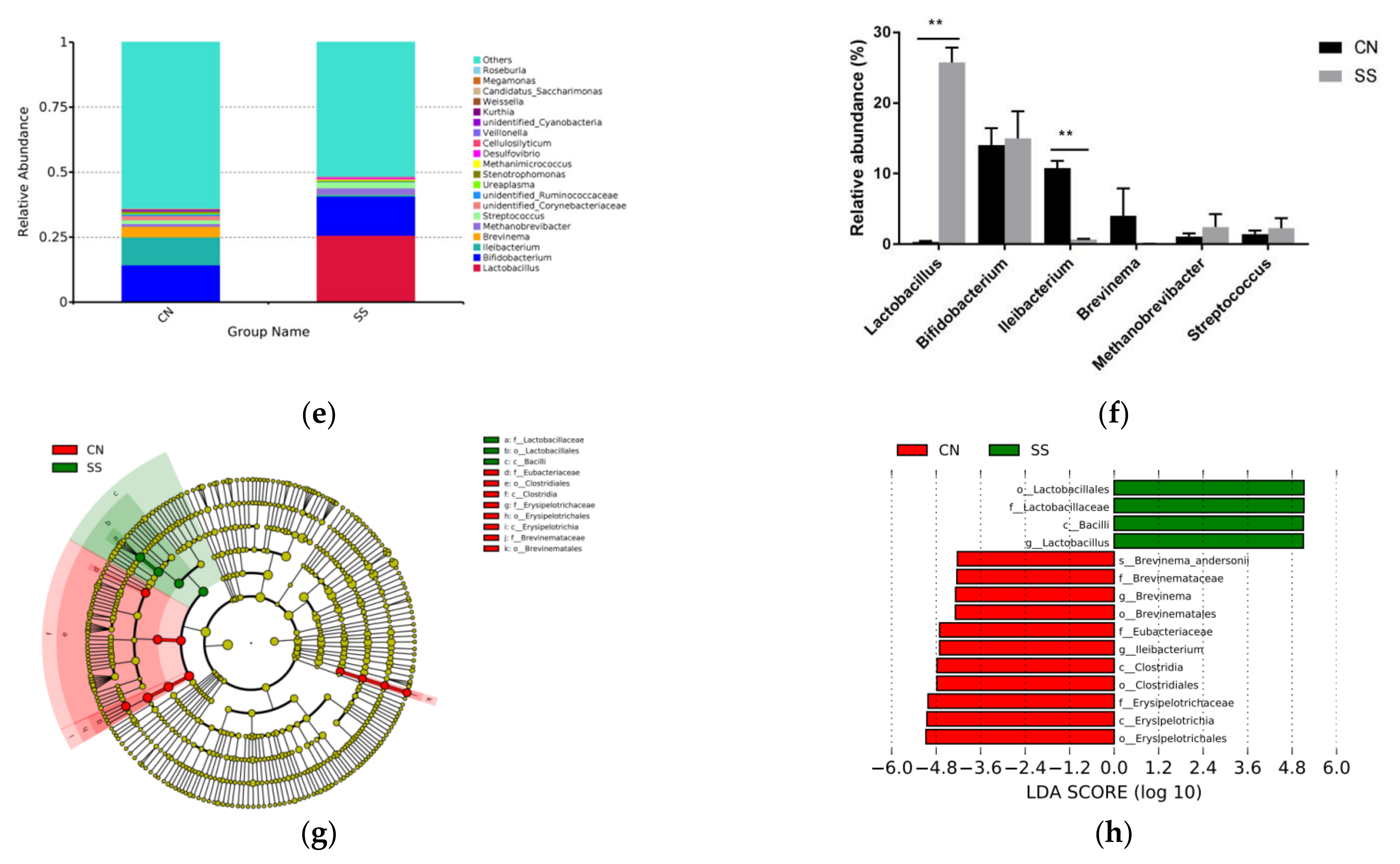
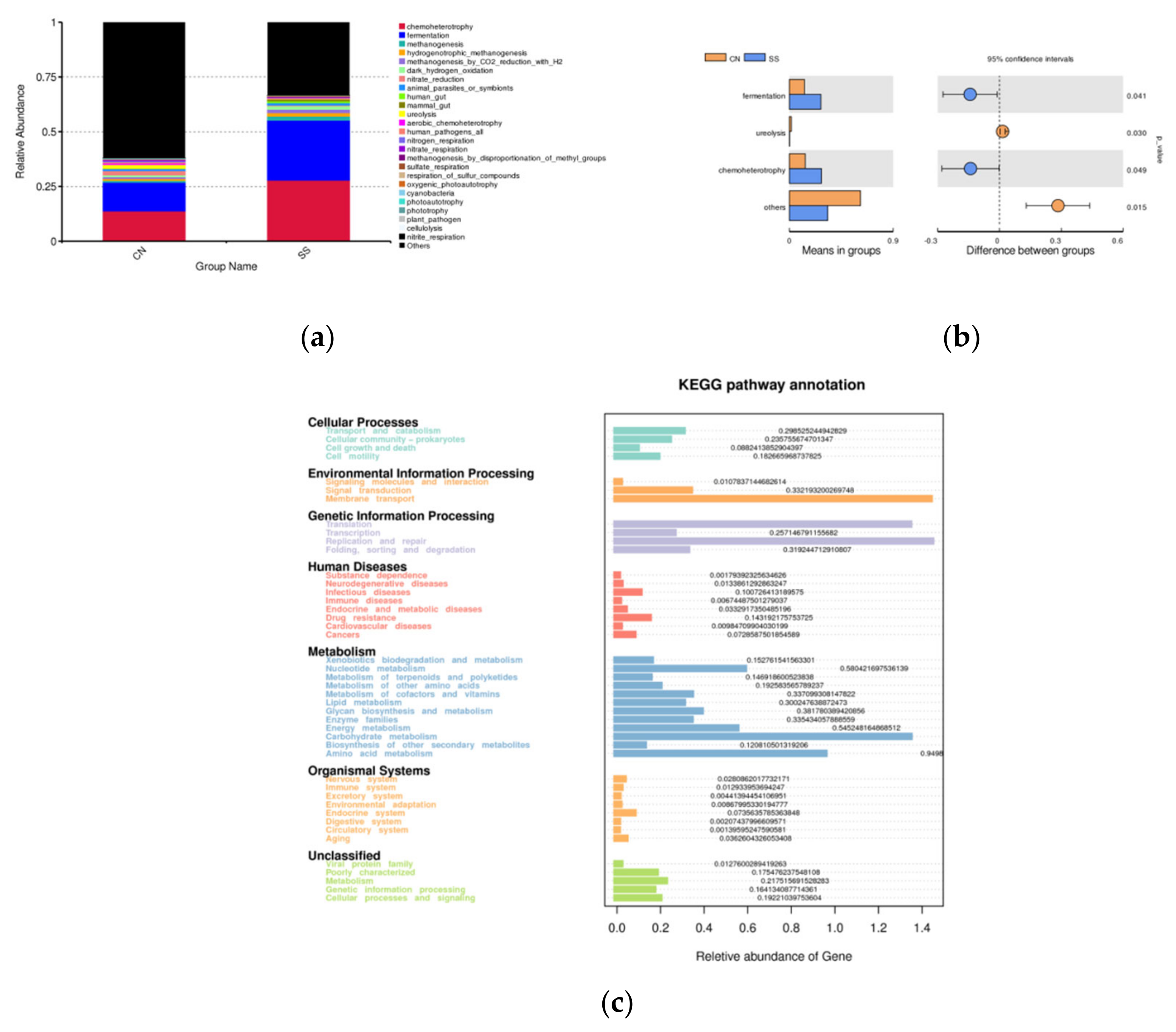
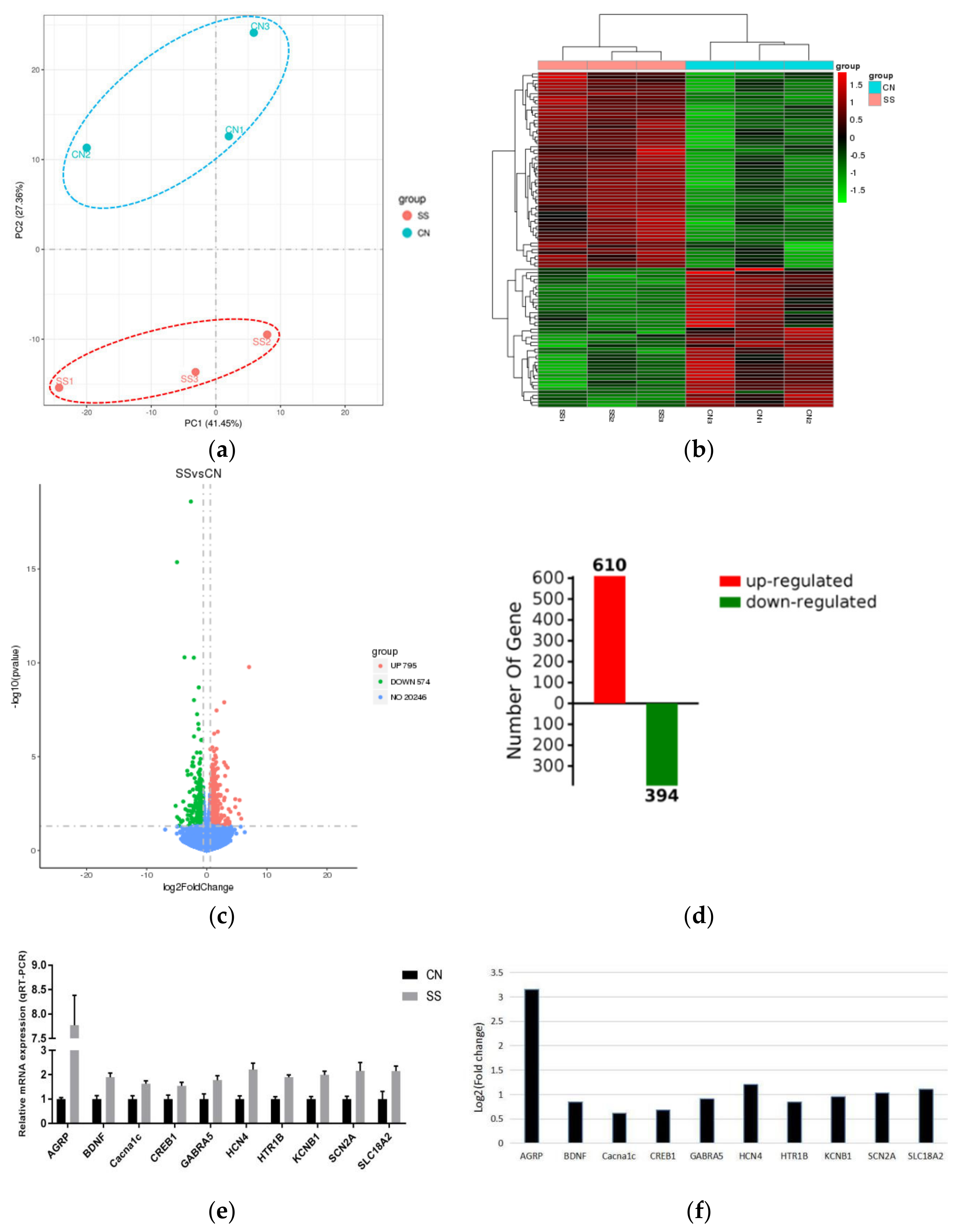
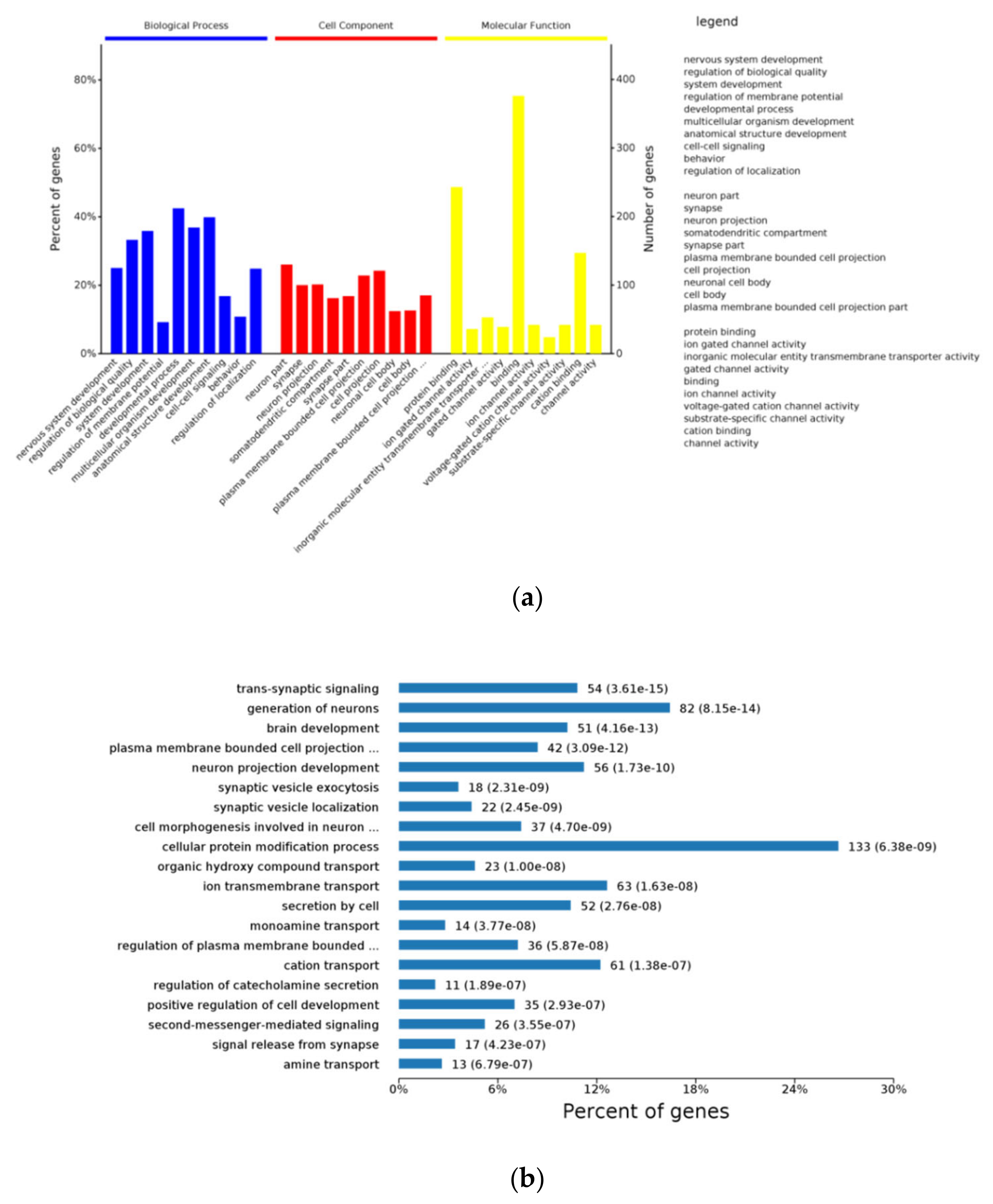
3. Results
3.1. Serum Biochemical Indexes
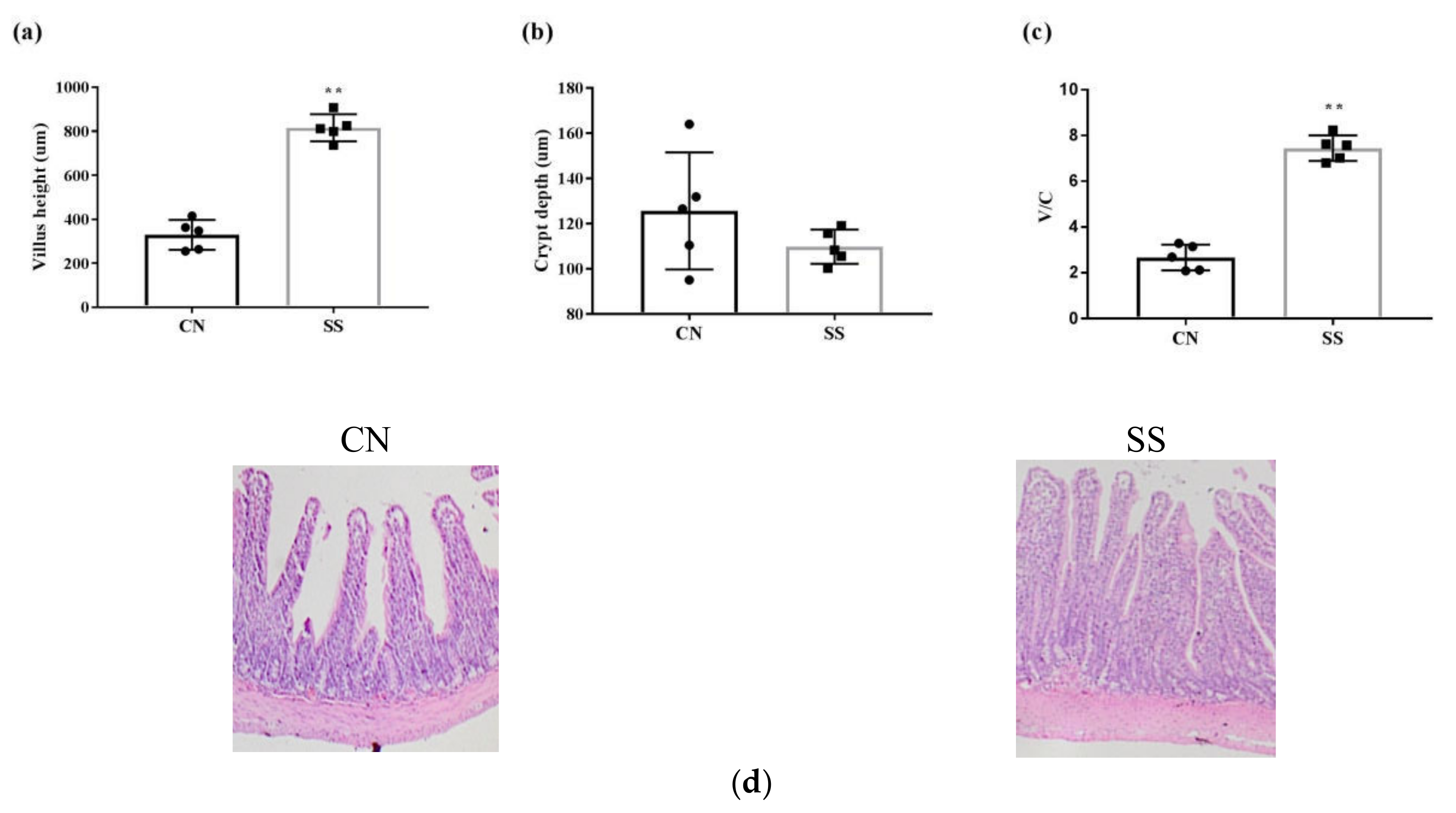
3.2. Morphological Analysis of the Ileum Villus
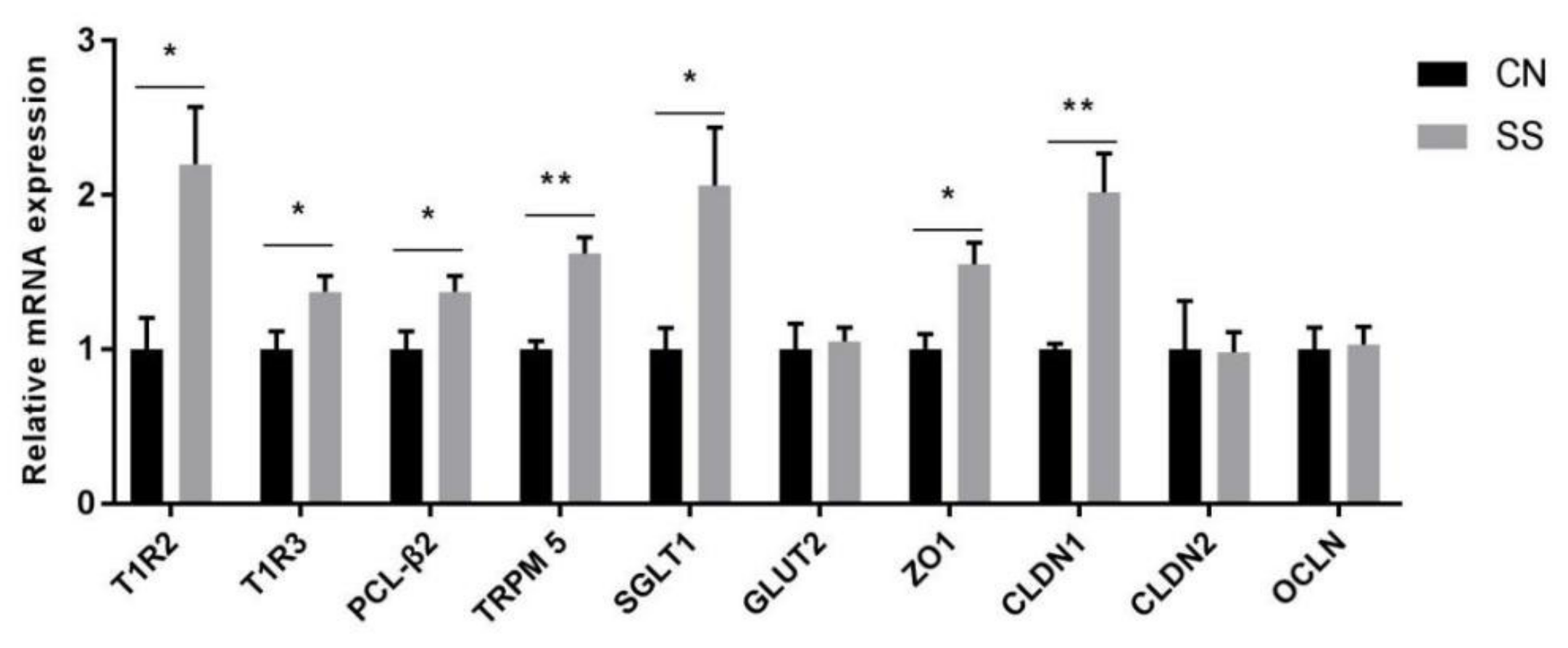
3.3. Gene Expressions of Taste Receptors, Glucose Transporter and Tight Junction in the Ileal Mucosa
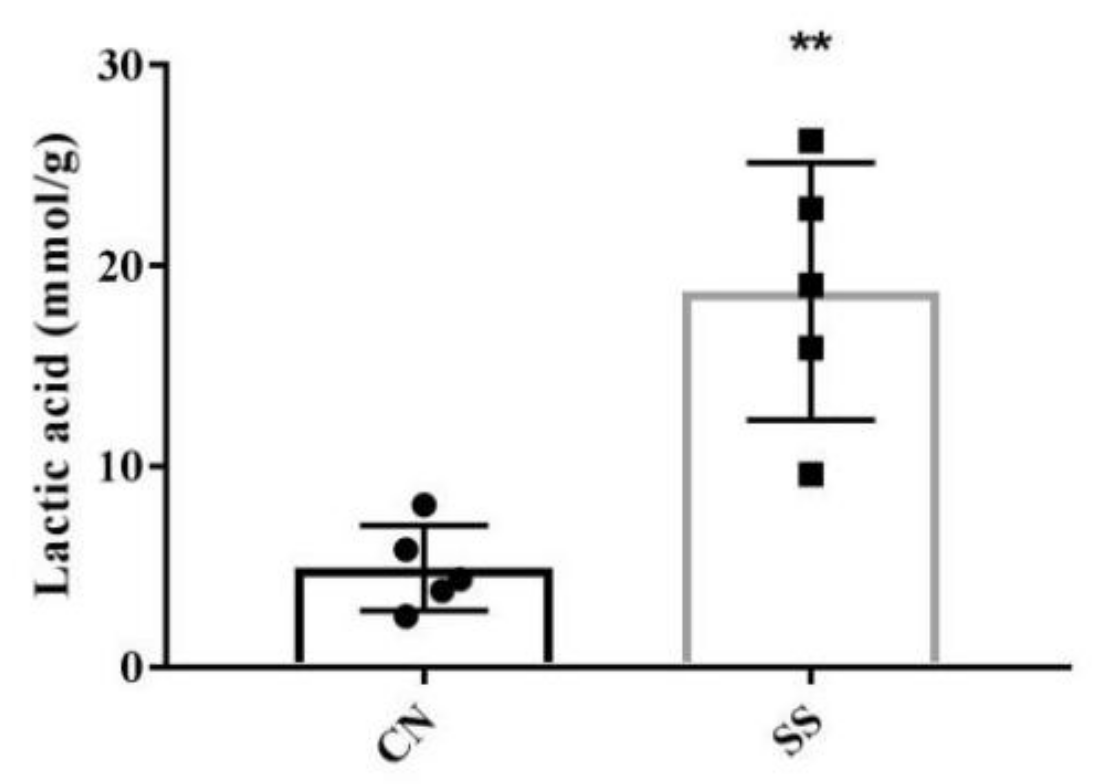
3.4. The Levels of Lactic Acid and SCFAs in the Ileal Digesta
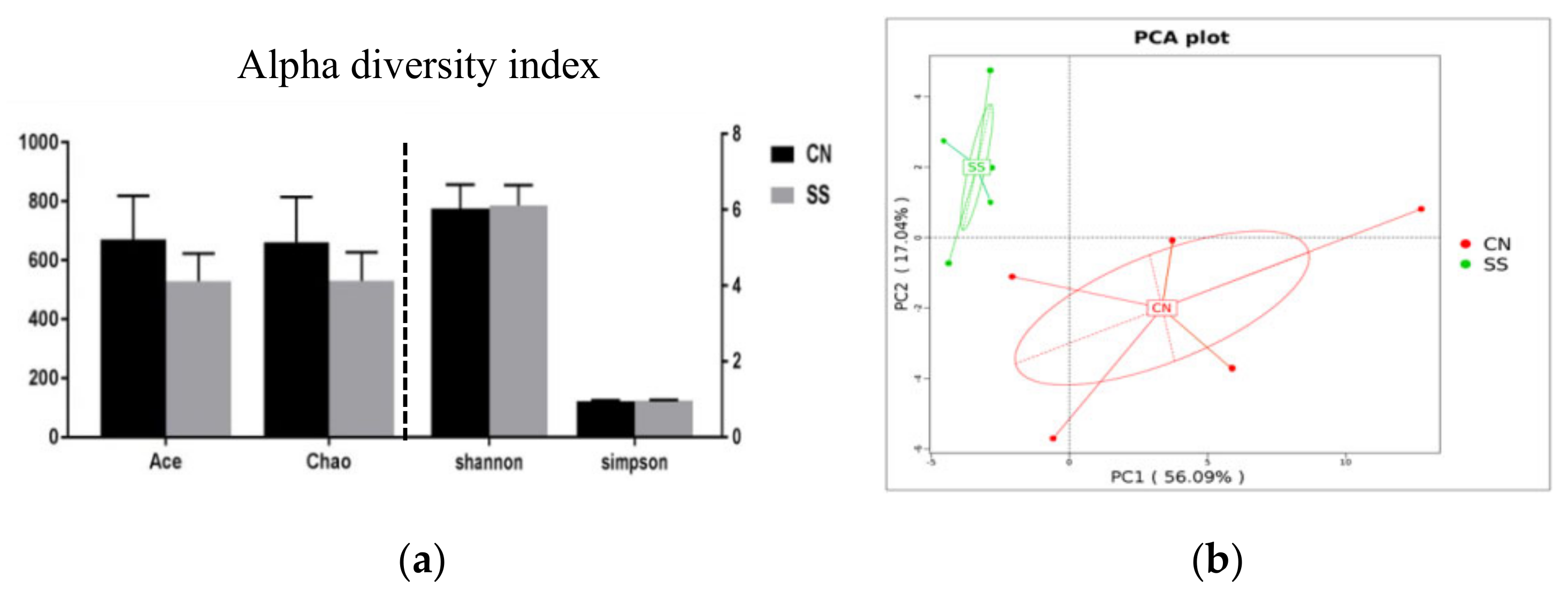
3.5. Effects of the Saccharin Sodium Intervention
3.6. Hypothalamic RNA Sequencing Data and Identification of DEGs in the Hypothalamus between Groups
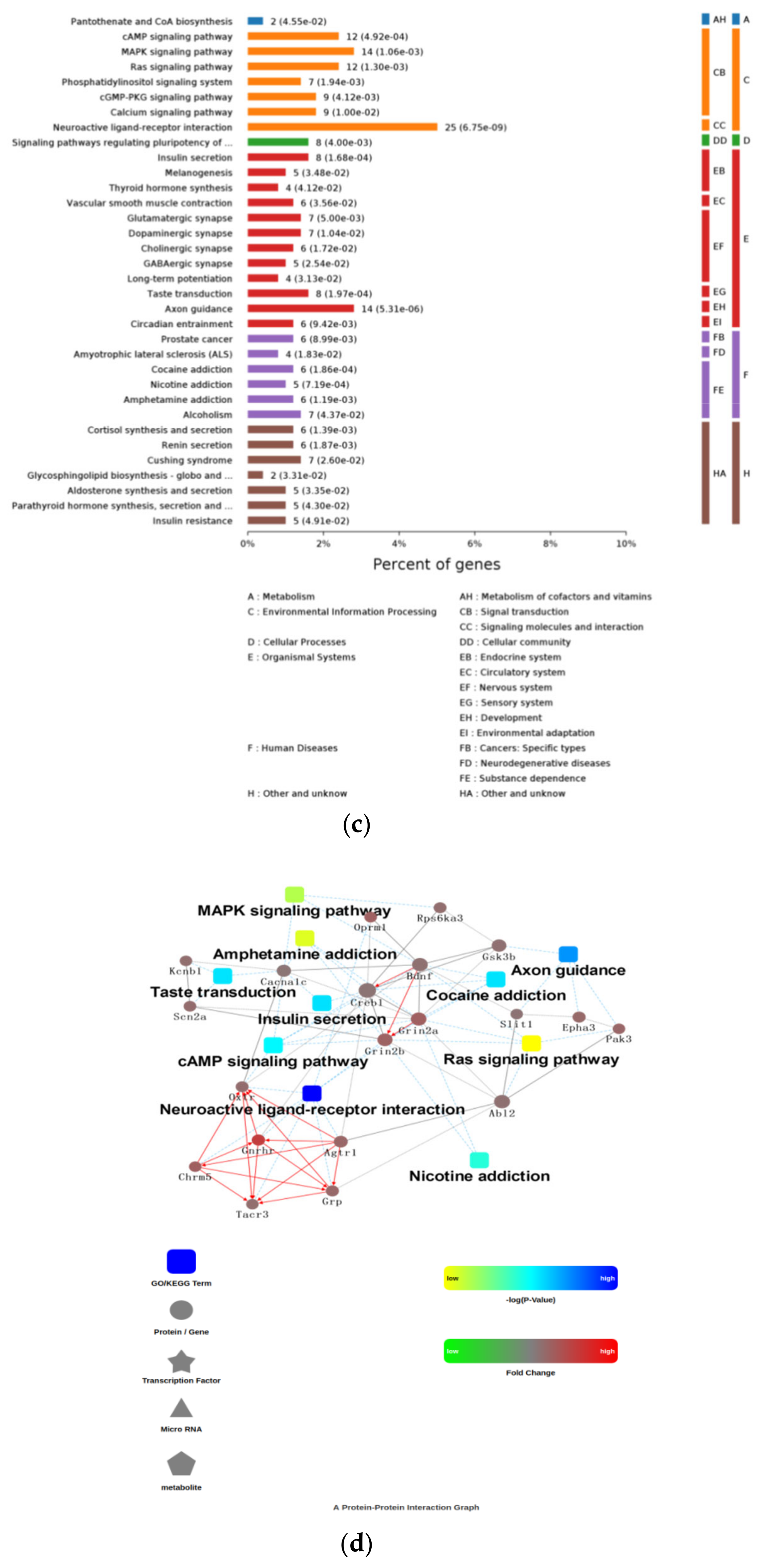
3.7. Function Enrichment Using DEGs of Hypothalamus RNA-Seq
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- DuBois, G.E.; Prakash, I. Non-Caloric Sweeteners, Sweetness Modulators, and Sweetener Enhancers. In Annual Review of Food Science and Technology; Doyle, M.P., Klaenhammer, T.R., Eds.; Annual Review of Food Science and Technology: Palo Alto, CA, USA, 2012; Volume 3, pp. 353–380. [Google Scholar]
- Fernstrom, J.D. Non-nutritive sweeteners and obesity. In Annual Review of Food Science and Technology; Doyle, M.P., Klaenhammer, T.R., Eds.; Annual Review of Food Science and Technology: Palo Alto, CA, USA, 2015; Volume 6, pp. 119–136. [Google Scholar]
- Dyer, J.; Wood, I.S.; Palejwala, A.; Ellis, A.; Shirazi-Beechey, S. Expression of monosaccharide transporters in intestine of diabetic humans. Am. J. Physiol. Liver Physiol. 2002, 282, G241–G248. [Google Scholar] [CrossRef] [Green Version]
- Fowler, S.P.; Williams, K.; Resendez, R.G.; Hunt, K.J.; Hazuda, H.P.; Stern, M.P. Fueling the Obesity Epidemic? Artificially Sweetened Beverage Use and Long-term Weight Gain. Obesity 2008, 16, 1894–1900. [Google Scholar] [CrossRef] [PubMed]
- Egan, J.M.; Margolskee, R.F. Taste Cells of the Gut and Gastrointestinal Chemosensation. Mol. Interv. 2008, 8, 78–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Anderson, R.; Kirkland, J. The effect of sodium saccharin in the diet on caecal microflora. Food Cosmet. Toxicol. 1980, 18, 353–355. [Google Scholar] [CrossRef]
- Jha, R.; Fouhse, J.M.; Tiwari, U.P.; Li, L.; Willing, B.P. Dietary Fiber and Intestinal Health of Monogastric Animals. Front. Vet. Sci. 2019, 6, 48. [Google Scholar] [CrossRef] [Green Version]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-E.; Sisti, A.C.; Jin, H.; Vignovich, M.; Villavicencio, M.; Tsang, K.S.; Goffer, Y.; Zuker, C.S. The gut-brain axis mediates sugar preference. Nat. Cell Biol. 2020, 580, 511–516. [Google Scholar] [CrossRef]
- Patten, G.S.; Bird, A.R.; Topping, D.L.; Abeywardena, M.Y. Effects of convenience rice congee supplemented diets on guinea pig whole animal and gut growth, caecal digesta SCFA and in vitro ileal contractility. Asia Pac. J. Clin. Nutr. 2004, 13, 92–100. [Google Scholar]
- Mallett, A.; Rowland, I.; Bearne, C. Modification of rat caecal microbial biotransformation activities by dietary saccharin. Toxicology 1985, 36, 253–262. [Google Scholar] [CrossRef]
- Jiang, J.; Qi, L.; Wei, Q.; Shi, F. Effects of daily exposure to saccharin sodium and rebaudioside A on the ovarian cycle and steroidogenesis in rats. Reprod. Toxicol. 2018, 76, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shen, T.; Shi, F.; Fu, Y. Influences of non-nutritive sweeteners on ovarian and uterine expression of T1R2 and T1R3 in peripubertal female guinea pigs. Anim. Sci. J. 2020, 91, e13348. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Hu, C.; Jiang, J.; Jin, S.; Wei, Q.; Wei, X.; Yu, D.; Shi, F. Effects of High-Dose Genistein on the Hypothalamic RNA Profile and Intestinal Health of Female Chicks. J. Agric. Food Chem. 2019, 67, 13737–13750. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, Y.; Xiong, Y.; Chen, Q.; Liang, H.; Niu, M.; Guo, B.; Li, M.; Zhang, X.; Li, Y.; et al. Selection and Validation of Novel RT-qPCR Reference Genes under Hormonal Stimuli and in Different Tissues of Santalum album. Sci. Rep. 2018, 8, 17511. [Google Scholar] [CrossRef] [Green Version]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]
- Gassmann, B. Sweeteners and metabolic syndrome. Ernahr. Umsch. 2005, 52, 476. [Google Scholar]
- Palmnäs, M.S.A.; Cowan, T.E.; Bomhof, M.R.; Su, J.; Reimer, R.A.; Vogel, H.J.; Hittel, D.S.; Shearer, J. Low-Dose Aspartame Consumption Differentially Affects Gut Microbiota-Host Metabolic Interactions in the Diet-Induced Obese Rat. PLoS ONE 2014, 9, e109841. [Google Scholar] [CrossRef]
- Swithers, S.E.; Laboy, A.F.; Clark, K.; Cooper, S.; Davidson, T. Experience with the high-intensity sweetener saccharin impairs glucose homeostasis and GLP-1 release in rats. Behav. Brain Res. 2012, 233, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Collison, K.S.; Makhoul, N.J.; Zaidi, M.; Saleh, S.M.; Andres, B.; Inglis, A.; Al-Rabiah, R.; Al-Mohanna, F.A. Gender Dimorphism in Aspartame-Induced Impairment of Spatial Cognition and Insulin Sensitivity. PLoS ONE 2012, 7, e31570. [Google Scholar] [CrossRef] [Green Version]
- Sternini, C.; Anselmi, L.; Rozengurt, E. Enteroendocrine cells: A site of ‘taste’ in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 2008, 15, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Margolskee, R.F.; Dyer, J.; Kokrashvili, Z.; Salmon, K.S.H.; Ilegems, E.; Daly, K.; Maillet, E.; Ninomiya, Y.; Mosinger, B.; Shirazi-Beechey, S.P. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc. Natl. Acad. Sci. USA 2007, 104, 15075–15080. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Cui, M.; Zhao, B.; Liu, Z.; Snyder, L.A.; Benard, L.M.J.; Osman, R.; Margolskee, R.F.; Max, M. Lactisole Interacts with the Transmembrane Domains of Human T1R3 to Inhibit Sweet Taste. J. Biol. Chem. 2005, 280, 15238–15246. [Google Scholar] [CrossRef] [Green Version]
- Al Massadi, O.; Nogueiras, R.; Dieguez, C.; Girault, J.-A. Ghrelin and food reward. Neuropharmacology 2019, 148, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, H.-R. Vagal and hormonal gut-brain communication: From satiation to satisfaction. Neurogastroenterol. Motil. 2008, 20, 64–72. [Google Scholar] [CrossRef]
- Konturek, S.J.; Konturek, J.W.; Pawlik, T.; Brzozowski, T. Brain-gut axis and its role in the control of food intake. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2004, 55, 137–154. [Google Scholar]
- Han, Y.; Xia, G.; Wu, Q. Functional Interrogation of the AgRP Neural Circuits in Control of Appetite, Body Weight, and Behaviors. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: Berlin, Germany, 2018; Volume 1090, pp. 1–16. [Google Scholar]
- Liu, S.; Marcelin, G.; Blouet, C.; Jeong, J.H.; Jo, Y.-H.; Schwartz, G.J.; Chua, S. A gut-brain axis regulating glucose metabolism mediated by bile acids and competitive fibroblast growth factor actions at the hypothalamus. Mol. Metab. 2018, 8, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nat. Cell Biol. 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Wu, H.; Lu, L.; Jiang, L.; Yu, Q. Lactobacillus reuteri Promotes Intestinal Development and Regulates Mucosal Immune Function in Newborn Piglets. Front. Veter Sci. 2020, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Nii, T.; Kakuya, H.; Isobe, N.; Yoshimura, Y. Lactobacillus reuteri Enhances the Mucosal Barrier Function against Heat-killed Salmonella Typhimurium in the Intestine of Broiler Chicks. J. Poult. Sci. 2020, 57, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Qi, C.; Zhu, H.; Yu, R.; Xie, C.; Peng, Y.; Yin, S.-W.; Fan, J.; Zhao, S.; Sun, J. Lactobacillus reuteri improves gut barrier function and affects diurnal variation of the gut microbiota in mice fed a high-fat diet. Food Funct. 2019, 10, 4705–4715. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Gálvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, L.M.; Sohn, J.; Tyrrell, K.L.; Citron, D.M.; Lawson, P.A.; Patel, N.B.; Iizumi, T.; Perez-Perez, G.I.; Goldstein, E.J.C.; Blaser, M.J. Corrigendum: Description of two novel members of the family Erysipelotrichaceae: Ileibacterium valens gen. nov., sp. nov. and Dubosiella newyorkensis, gen. nov., sp. nov., from the murine intestine, and emendation to the description of Faecalibacterium rodentium. Int. J. Syst. Evol. Microbiol. 2017, 67, 4289. [Google Scholar] [CrossRef] [PubMed]
- Woting, A.; Pfeiffer, N.; Loh, G.; Klaus, S.; Blaut, M. Clostridium ramosum Promotes High-Fat Diet-Induced Obesity in Gnotobiotic Mouse Models. MBio 2014, 5, e01514–e01530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.; Leung, J.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, B.; Bäckhed, B.O.S.F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Grunewald, K.K.; Mitchell, L.K. Growth of Mice Fed Milk Fermented with Lactobacillus acidophilus1. J. Food Prot. 1983, 46, 315–317. [Google Scholar] [CrossRef]
- Schwarzer, M.; Makki, K.; Storelli, G.; Machuca-Gayet, I.; Srutkova, D.; Hermanova, P.; Martino, M.E.; Balmand, S.; Hudcovic, T.; Heddi, A.; et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 2016, 351, 854–857. [Google Scholar] [CrossRef]
- Zhang, Q.; Koser, S.L.; Donkin, S.S. Propionate induces mRNA expression of gluconeogenic genes in bovine calf hepatocytes. J. Dairy Sci. 2016, 99, 3908–3915. [Google Scholar] [CrossRef] [Green Version]
- Rios-Covian, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, H.N.; Collins, M.D. Proposal To Restrict the Genus Bacteroides (Castellani and Chalmers) to Bacteroides fragilis and Closely Related Species. Int. J. Syst. Bacteriol. 1989, 39, 85–87. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [Green Version]
- Besten, G.D.; Lange, K.; Havinga, R.; Van Dijk, T.H.; Gerding, A.; Van Eunen, K.; Müller, M.; Groen, A.K.; Hooiveld, G.; Bakker, B.M.; et al. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am. J. Physiol. Liver Physiol. 2013, 305, G900–G910. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Kunisawa, J.; Kiyono, H. Dietary and Microbial Metabolites in the Regulation of Host Immunity. Front. Microbiol. 2017, 8, 2171. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.E.B.; Hedemann, M.S.; Lærke, H.N. The role of carbohydrates in intestinal health of pigs. Anim. Feed Sci. Technol. 2012, 173, 41–53. [Google Scholar] [CrossRef]
- Keeney, K.M.; Finlay, B.B. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr. Opin. Microbiol. 2011, 14, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matheus, V.; Monteiro, L.; Oliveira, R.; Maschio, D.; Collares-Buzato, C. Butyrate reduces high-fat diet-induced metabolic alterations, hepatic steatosis and pancreatic beta cell and intestinal barrier dysfunctions in prediabetic mice. Exp. Biol. Med. 2017, 242, 1214–1226. [Google Scholar] [CrossRef] [Green Version]

| Indexes | CN | SS | p-Value |
|---|---|---|---|
| TG (mmol/L) | 0.75 ± 0.09 | 0.83 ± 0.07 | 0.488 |
| ALT (U/L) | 71.3 ± 7.0 | 86.9 ± 8.2 | 0.168 |
| AST (U/L) | 105 ± 11 | 114 ± 14 | 0.614 |
| CRE (μmol/L) | 17.1 ± 3.9 | 11.6 ± 3.2 | 0.282 |
| ALP (U/L) | 262 ± 26 | 295 ± 27 | 0.392 |
| UA (μmol/L) * | 86.4 ± 14.1 | 121.3 ± 8.7 | 0.05 |
| GLU (mmol/L) * | 10.2 ± 0.3 | 12.4 ± 0.4 | 0.001 |
| CHO (mmol/L) | 0.83 ± 0.04 | 0.80 ± 0.04 | 0.646 |
| TP (mmol/L) | 48.1 ± 2.2 | 49.5 ± 3.4 | 0.73 |
| CCK (pg/mL) | 38.8 ± 5.2 | 43.2 ± 5.3 | 0.136 |
| PPY(pg/mL) | 35.8 ± 2.9 | 34.0 ± 5.6 | 0.456 |
| GHRP(pg/mL) * | 54.8 ± 3.5 | 60.8 ± 4.3 | 0.014 |
| GLP1(pg/mL) | 4.50 ± 0.72 | 4.15 ± 0.75 | 0.347 |
| Groups | Acetic Acid * (mg/kg) | Propionic Acid * (mg/kg) | Isobutyric Acid (mg/kg) | N-Butyric Acid (mg/kg) | Isovaleric Acid (mg/kg) | N-Valeric Acid * (mg/kg) | N-Hexanoic Acid (mg/kg) |
|---|---|---|---|---|---|---|---|
| CN | 800 ± 31 | 484 ± 23 | 26.2 ± 0.7 | 587 ± 148 | 18.2 ± 1.5 | 47.8 ± 1.3 | 2.60 ± 0.28 |
| SS | 1021 ± 71 | 625 ± 28 | 33.9 ± 7.0 | 695 ± 77 | 24.3 ± 6.9 | 60.8 ± 4.3 | 2.77 ± 0.59 |
| p-value | <0.001 | <0.001 | 0.069 | 0.183 | 0.092 | <0.001 | 0.589 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhu, S.; Lv, Z.; Dai, H.; Wang, Z.; Wei, Q.; Hamdard, E.; Mustafa, S.; Shi, F.; Fu, Y. Drinking Water with Saccharin Sodium Alters the Microbiota-Gut-Hypothalamus Axis in Guinea Pig. Animals 2021, 11, 1875. https://doi.org/10.3390/ani11071875
Li J, Zhu S, Lv Z, Dai H, Wang Z, Wei Q, Hamdard E, Mustafa S, Shi F, Fu Y. Drinking Water with Saccharin Sodium Alters the Microbiota-Gut-Hypothalamus Axis in Guinea Pig. Animals. 2021; 11(7):1875. https://doi.org/10.3390/ani11071875
Chicago/Turabian StyleLi, Junrong, Shanli Zhu, Zengpeng Lv, Hongjian Dai, Zhe Wang, Quanwei Wei, Enayatullah Hamdard, Sheeraz Mustafa, Fangxiong Shi, and Yan Fu. 2021. "Drinking Water with Saccharin Sodium Alters the Microbiota-Gut-Hypothalamus Axis in Guinea Pig" Animals 11, no. 7: 1875. https://doi.org/10.3390/ani11071875
APA StyleLi, J., Zhu, S., Lv, Z., Dai, H., Wang, Z., Wei, Q., Hamdard, E., Mustafa, S., Shi, F., & Fu, Y. (2021). Drinking Water with Saccharin Sodium Alters the Microbiota-Gut-Hypothalamus Axis in Guinea Pig. Animals, 11(7), 1875. https://doi.org/10.3390/ani11071875







