A First Insight into the Gonad Transcriptome of Hong Kong Catfish (Clarias fuscus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and RNA Extraction
2.3. Full-Length Transcriptome Sequencing
2.4. Illumina Transcriptome Sequencing
2.5. Transcriptome Gene Annotation
2.6. Identification of lncRNAs and Simple Sequence Repeats (SSRs)
2.7. Differential Expression Gene Analysis
2.8. Gene Expression Validation
3. Results
3.1. Transcriptome Data from the Gonads of Hong Kong Catfish
3.2. Transcriptome Annotation
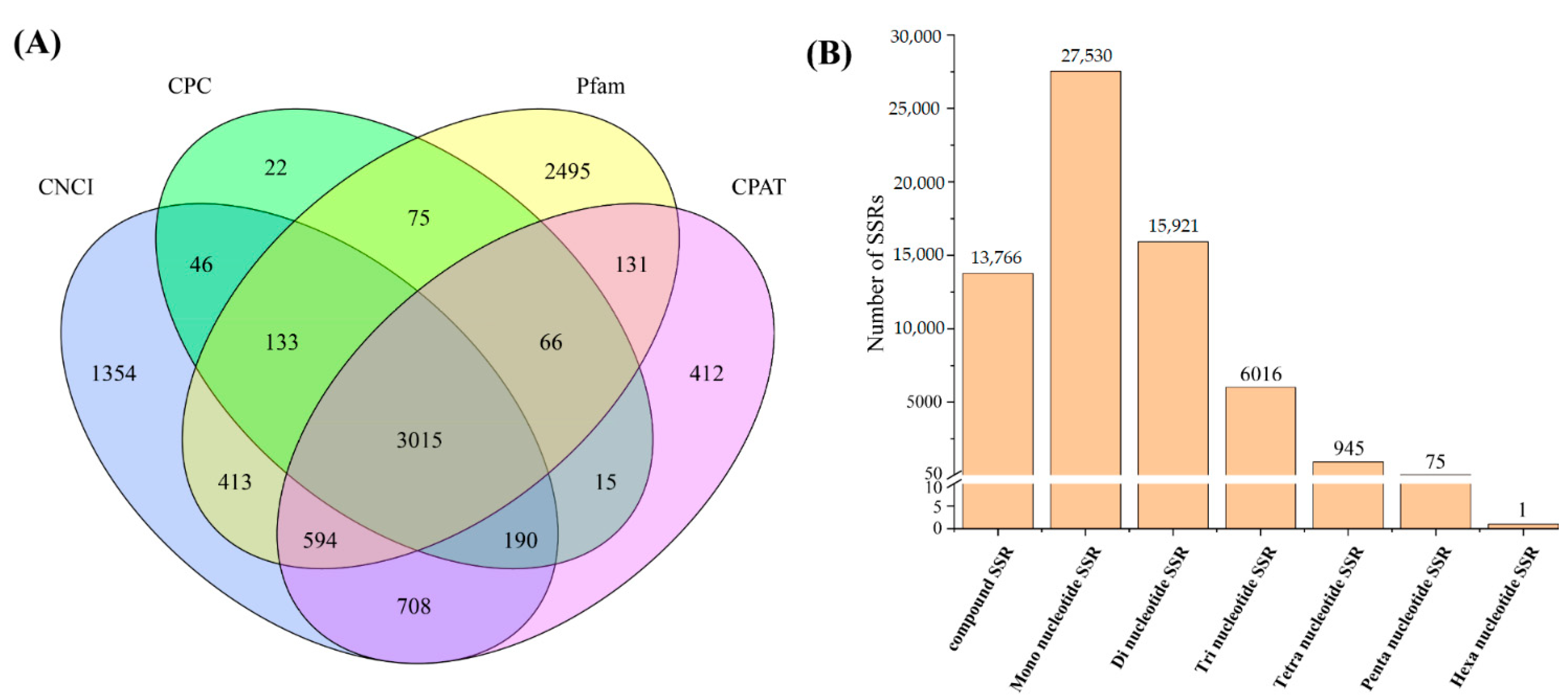
3.3. Identification of lncRNAs and SSRs
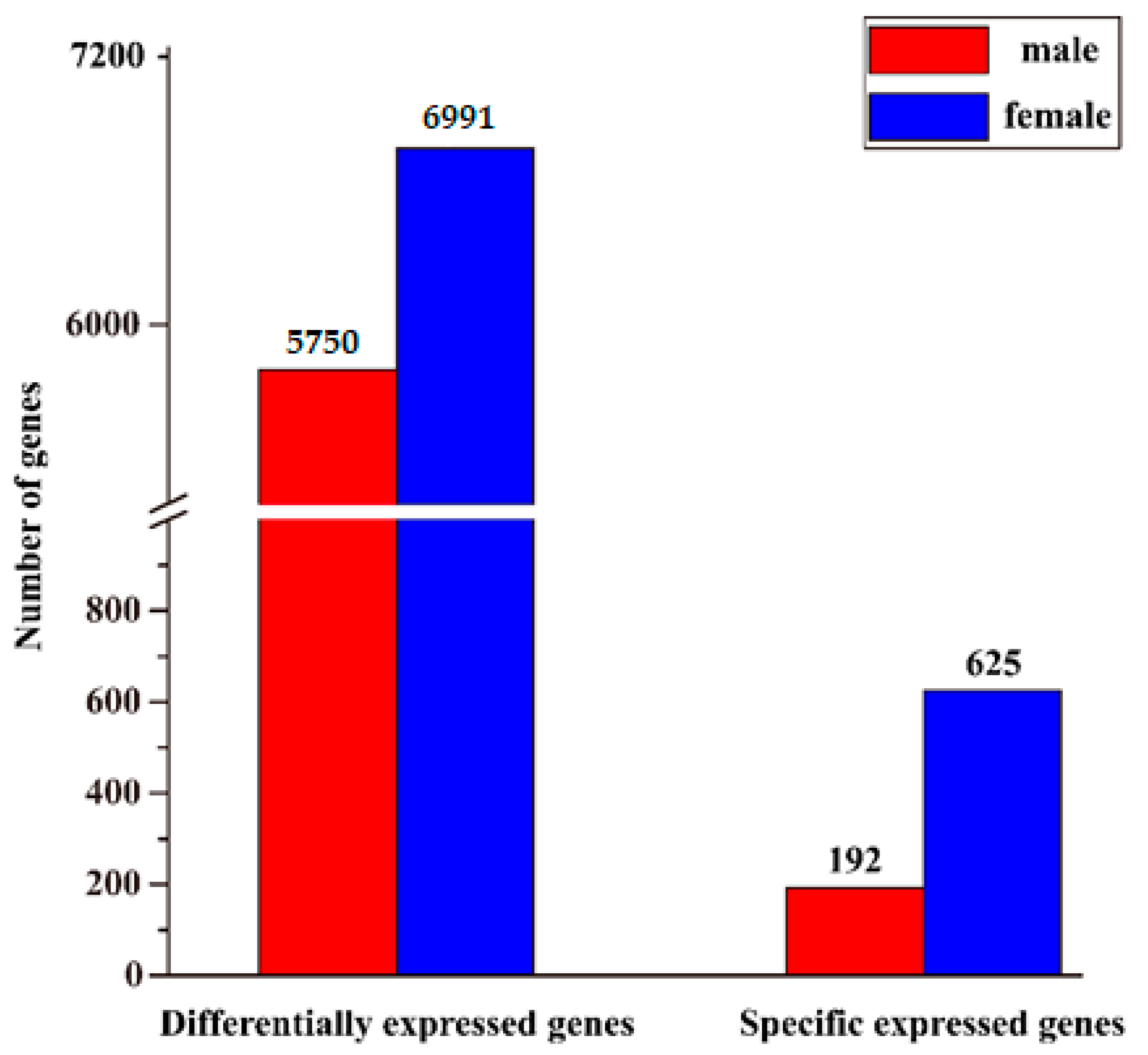
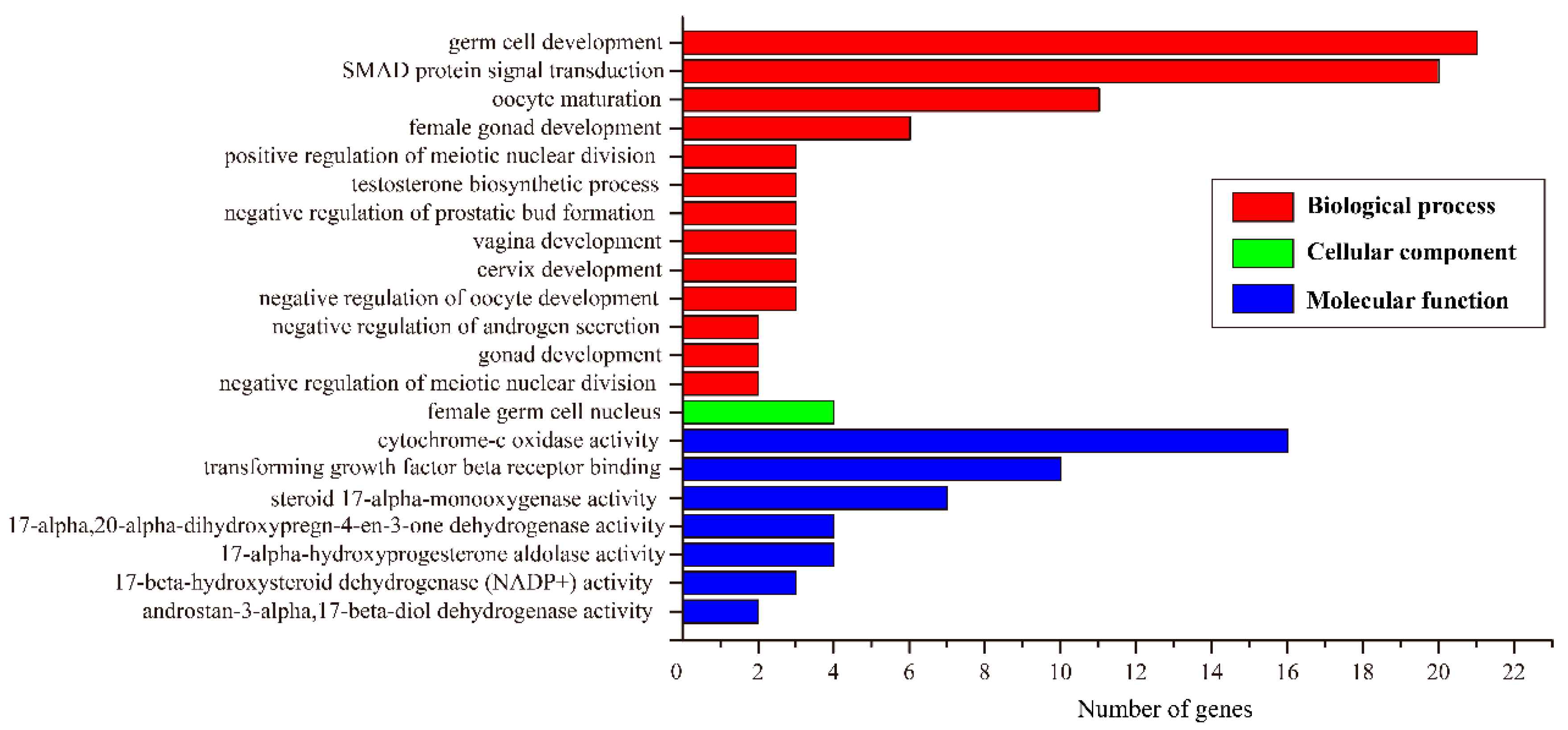
3.4. Identification of Sex-Biased Genes and Enrichment Analysis
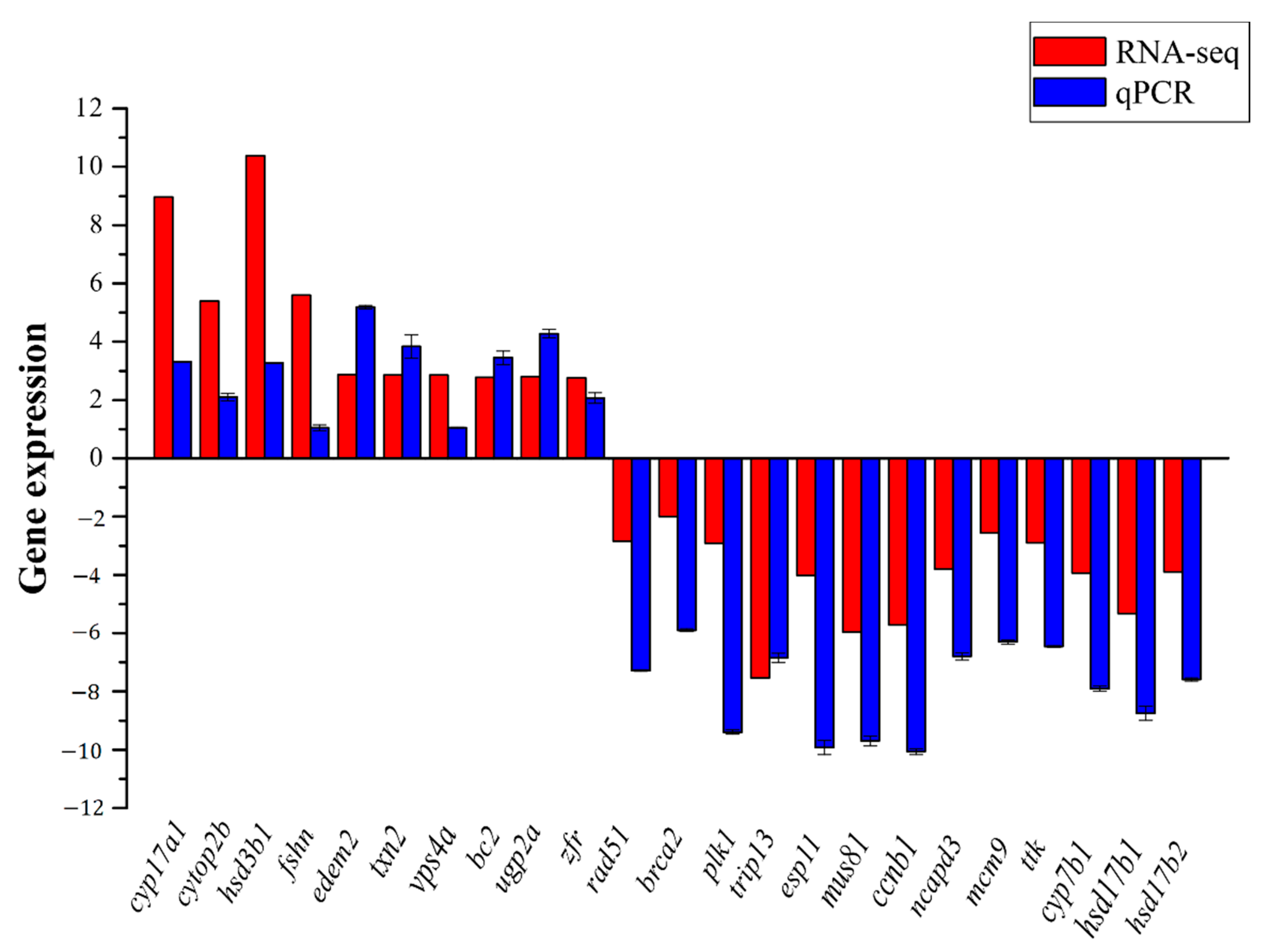
3.5. Validation of RNA-seq Results by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Dean, R.; Mank, J.E. The role of sex chromosomes in sexual dimorphism: Discordance between molecular and phenotypic data. J. Evol. Biol. 2014, 27, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Li, J.; Deng, S.-P.; Tian, Y.-S.; Wang, Q.-Y.; Zhuang, Z.-M.; Sha, Z.-X.; Xu, J.-Y. Isolation of Female-Specific AFLP Markers and Molecular Identification of Genetic Sex in Half-Smooth Tongue Sole (Cynoglossus semilaevis). Mar. Biotechnol. 2007, 9, 273–280. [Google Scholar] [CrossRef]
- Beardmore, J.; Mair, G.; Lewis, R. Monosex male production in finfish as exemplified by tilapia: Applications, problems, and prospects. Aquaculture 2001, 197, 283–301. [Google Scholar] [CrossRef]
- Naylor, R.L.; Goldburg, R.J.; Primavera, J.H.; Kautsky, N.; Beveridge, M.C.M.; Clay, J.; Folke, C.; Lubchenco, J.; Mooney, H.; Troell, M. Effect of aquaculture on world fish supplies. Nat. Cell Biol. 2000, 405, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guan, B.; Xu, J.; Hou, C.; Tian, H.; Chen, H. Genetic Manipulation of Sex Ratio for the Large-Scale Breeding of YY Super-Male and XY All-Male Yellow Catfish (Pelteobagrus fulvidraco (Richardson)). Mar. Biotechnol. 2012, 15, 321–328. [Google Scholar] [CrossRef]
- Ahmed, G.; Sultana, N.; Shamsuddin, M.; Hossain, M.B. Growth and Production Performance of Monosex Tilapia (Oreochromis niloticus) Fed with Homemade Feed in Earthen Mini Ponds. Pak. J. Biol. Sci. 2013, 16, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Pongthana, N.; Penman, D.J.; Baoprasertkul, P.; Hussain, M.G.; Islam, M.S.; Powell, S.F.; McAndrew, B.J. Monosex female production in the silver barb (Puntius gonionotus Bleeker). Aquaculture 1999, 173, 247–256. [Google Scholar] [CrossRef]
- Kikuchi, K.; Hamaguchi, S. Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dyn. 2013, 242, 339–353. [Google Scholar] [CrossRef]
- Nagahama, Y. Molecular mechanisms of sex determination and gonadal sex differentiation in fish. Fish Physiol. Biochem. 2005, 31, 105–109. [Google Scholar] [CrossRef]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.-F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar] [CrossRef]
- Young, M.J.A.; Fast, A.W. Temperature and photoperiod effects on ovarian maturation in the Chinese catfish (Clarias fuscus: Lacepede). J. Aquac. Trop. 1990, 5, 19–30. [Google Scholar]
- Szyper, J.P.; Tamaru, C.S.; Howerton, R.D.; Hopkins, K.D.; Fast, A.W.; Weidenbach, R.P. Maturation, Hatchery, and Nursery Techniques for Chinese Catfish, Clarias fuscus, in Hawaii. Aquac. Ext. Bull. 2001, 1–8. [Google Scholar]
- Deng, S.P.; Wang, J.J.; Wu, T.L.; Zhu, C.H.; Li, G.L. cDNA cloning and expression analysis of dmrt1 in Clarias fuscus. Acta Hydrobiol. Sin. 2012, 36, 610–617. [Google Scholar]
- Deng, S.P.; Zhu, C.H.; Sun, J.; Wang, W.D.; Wu, T.L.; Chen, H.P.; Shi, S.L.; Li, G.L. Foxl2 of the Hong Kong catfish (Clari-as fuscus): cDNA cloning, tissue distribution and changes in gene expression towards methyltestosterone, estradiol and letro-zole exposure of the fries during gonadal differentiation. Genes Genom. 2015, 37, 669–677. [Google Scholar] [CrossRef]
- Sun, J.; Li, G.; Zhu, C.; Wu, T.; Deng, S. Molecular cloning and expression of Cyp19a1b cDNA in Clarias fuscus. J. Fish. Sci. China 2012, 19, 408–415. [Google Scholar] [CrossRef]
- Martsikalis, P.V.; Gkafas, G.A.; Palaiokostas, C.; Exadactylos, A. Genomics era on breeding aquaculture stocks. In Organic Aquaculture: Impacts and Future Developments; Lembo, G., Mente, E., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 65–77. [Google Scholar] [CrossRef]
- Exadactylos, A.; Arvanitoyannis, I.S. Aquaculture Biotechnology for enhanced fish production for human consumption. In Microbial Biotechnology in Agriculture and Aquaculture; Ray, R.C., Ed.; Science Publishers Inc.: Enfield, NH, USA, 2006; pp. 453–510. [Google Scholar]
- Zhang, X.; Zhou, J.; Li, L.; Huang, W.; Ahmad, H.I.; Li, H.; Jiang, H.; Chen, J. Full-length transcriptome sequencing and comparative transcriptomic analysis to uncover genes involved in early gametogenesis in the gonads of Amur sturgeon (Acipenser schrenckii). Front. Zool. 2020, 17, 1–21. [Google Scholar] [CrossRef]
- Zeng, D.; Chen, X.; Peng, J.; Yang, C.; Peng, M.; Zhu, W.; Xie, D.; He, P.; Wei, P.; Lin, Y.; et al. Single-molecule long-read sequencing facilitates shrimp transcriptome research. Sci. Rep. 2018, 8, 16920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, C.; He, M.; Xiang, Z.; Yin, Y.; Liu, S.; Zhuang, Z. A full-length transcriptome of Sepia esculenta using a combination of single-molecule long-read (SMRT) and Illumina sequencing. Mar. Genom. 2019, 43, 54–57. [Google Scholar] [CrossRef]
- Melo, R.M.C.; Arantes, F.P.; Sato, Y.; Dos Santos, J.E.; Rizzo, E.; Bazzoli, N. Comparative morphology of the gonadal structure related to reproductive strategies in six species of neotropical catfishes (Teleostei: Siluriformes). J. Morphol. 2011, 272, 525–535. [Google Scholar] [CrossRef]
- Van Dyk, J.C.; Pieterse, G.M. A histo-morphological study of the testis of the sharptooth catfish (Clarias gariepinus) as reference for future toxicological assessments. J. Appl. Ichthyol. 2008, 24, 415–422. [Google Scholar] [CrossRef]
- Liu, S.Y. The Occurrence and Development of the Testis and the Sperm Characteristics of Silurus Asotus; Henan Normal University: Xinxiang, China, 2017. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Apweiler, R. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.G.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Goseq: Gene Ontology testing for RNA-seq datasets. Genome Biol. 2012, 11, R14. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Matzuk, M.M. Smad5 is required for mouse primordial germ cell development. Mech. Dev. 2001, 104, 61–67. [Google Scholar] [CrossRef]
- Cao, G.L.; Jiang, Y.L.; Chu, M.X. Progress on Lin28 in the Regulation of Mammal Reproduction. J. Agric. Biotechnol. 2019, 27, 534–541. [Google Scholar] [CrossRef]
- West, J.A.; Viswanathan, S.R.; Yabuuchi, A.; Cunniff, K.; Takeuchi, A.; Park, I.-H.; Sero, J.E.; Zhu, H.; Perez-Atayde, A.; Frazier, A.L.; et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009, 460, 909–913. [Google Scholar] [CrossRef]
- Ramasamy, S.; Wang, H.; Quach, H.N.B.; Sampath, K. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev. Biol. 2006, 292, 393–406. [Google Scholar] [CrossRef]
- Choi, Y.; Ballow, D.J.; Xin, Y.; Rajkovic, A. Lim Homeobox Gene, Lhx8, Is Essential for Mouse Oocyte Differentiation and Survival1. Biol. Reprod. 2008, 79, 442–449. [Google Scholar] [CrossRef]
- Qiu, Y.; Sun, S.; Charkraborty, T.; Wu, L.; Sun, L.; Wei, J.; Nagahama, Y.; Wang, D.; Zhou, L. Figla Favors Ovarian Differentiation by Antagonizing Spermatogenesis in a Teleosts, Nile Tilapia (Oreochromis niloticus). PLoS ONE 2015, 10, e0123900. [Google Scholar] [CrossRef]
- Qin, M.; Zhang, Z.; Song, W.; Wong, Q.W.-L.; Chen, W.; Shirgaonkar, N.; Ge, W. Roles of Figla/figla in Juvenile Ovary Development and Follicle Formation During Zebrafish Gonadogenesis. Endocrinology 2018, 159, 3699–3722. [Google Scholar] [CrossRef]
- Fu, L.; Koganti, P.P.; Wang, J.; Wang, L.; Wang, C.-L.; Yao, J. Lhx8 interacts with a novel germ cell-specific nuclear factor containing an Nbl1 domain in rainbow trout (Oncorhynchus mykiss). PLoS ONE 2017, 12, e0170760. [Google Scholar] [CrossRef]
- Dranow, D.B.; Hu, K.; Bird, A.M.; Lawry, S.T.; Adams, M.T.; Sanchez, A.; Amatruda, J.F.; Draper, B.W. Bmp15 Is an Oocyte-Produced Signal Required for Maintenance of the Adult Female Sexual Phenotype in Zebrafish. PLoS Genet. 2016, 12, e1006323. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumaran, B. Pesticide- and sex steroid analogue-induced endocrine disruption differentially targets hypothalamo–hypophyseal–gonadal system during gametogenesis in teleosts–A review. Gen. Comp. Endocrinol. 2015, 219, 136–142. [Google Scholar] [CrossRef]
- Gennotte, V.; Akonkwa, B.; Mélard, C.; Denoël, M.; Cornil, C.A.; Rougeot, C. Do sex reversal procedures differentially affect agonistic behaviors and sex steroid levels depending on the sexual genotype in Nile tilapia? J. Exp. Zool. Part A Ecol. Integr. Physiol. 2017, 327, 153–162. [Google Scholar] [CrossRef]
- Uno, T.; Ishizuka, M.; Itakura, T. Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 2012, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hilborn, E.; Stål, O.; Jansson, A. Estrogen and androgen-converting enzymes 17β-hydroxysteroid dehydrogenase and their involvement in cancer: With a special focus on 17β-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget 2017, 8, 30552–30562. [Google Scholar] [CrossRef] [PubMed]
- Zhai, G.; Shu, T.; Xia, Y.; Jin, X.; He, J.; Yin, Z. Androgen signaling regulates the transcription of anti-Müllerian hormone via synergy with SRY-related protein SOX9A. Sci. Bull. 2017, 62, 197–203. [Google Scholar] [CrossRef]
- Zhai, G.; Shu, T.; Xia, Y.; Lu, Y.; Shang, G.; Jin, X.; He, J.; Nie, P.; Yin, Z. Characterization of Sexual Trait Development in cyp17a1-Deficient Zebrafish. Endocrinology 2018, 159, 3549–3562. [Google Scholar] [CrossRef]
- Zhang, Q.; Ye, D.; Wang, H.; Wang, Y.; Hu, W.; Sun, Y. Zebrafish cyp11c1 Knockout Reveals the Roles of 11-ketotestosterone and Cortisol in Sexual Development and Reproduction. Endocrinology 2020, 161. [Google Scholar] [CrossRef]
- Zheng, Q.; Xiao, H.; Shi, H.; Wang, T.; Sun, L.; Tao, W.; Kocher, T.D.; Li, M.; Wang, D. Loss of Cyp11c1 causes delayed spermatogenesis due to the absence of 11-ketotestosterone. J. Endocrinol. 2020, 244, 487–499. [Google Scholar] [CrossRef]
- Puranen, T.; Poutanen, M.; Ghosh, D.; Vihko, P.; Vihko, R. Characterization of Structural and Functional Properties of Human 17?-Hydroxysteroid Dehydrogenase Type 1 Using Recombinant Enzymes and Site-Directed Mutagenesis. Mol. Endocrinol. 1997, 11, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Sharp, T.V.; Xiao, Q.; Jeffrey, I.; Gewert, D.R.; Clemens, M.J. Reversal of the double-stranded-RNA-induced inhibition of protein synthesis by a catalytically inactive mutant of the protein kinase PKR. JBIC J. Biol. Inorg. Chem. 1993, 214, 945–948. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Monsivais, D.; Matzuk, M.M.; Pangas, S.A. The TGF-β Family in the Reproductive Tract. Cold Spring Harb. Perspect. Biol. 2017, 9, a022251. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.-H. The regulation of TGF signal transduction. Development 2009, 136, 3699–3714. [Google Scholar] [CrossRef]
- Mullen, A.C.; Wrana, J.L. TGF-β Family Signaling in Embryonic and Somatic Stem-Cell Renewal and Differentiation. Cold Spring Harb. Perspect. Biol. 2017, 9, a022186. [Google Scholar] [CrossRef]
- Attisano, L. Signal Transduction by the TGF-beta Superfamily. Science 2002, 296, 1646–1647. [Google Scholar] [CrossRef]
- Itman, C.; Wong, C.; Hunyadi, B.; Ernst, M.; Jans, D.A.; Loveland, K.L. Smad3 Dosage Determines Androgen Responsiveness and Sets the Pace of Postnatal Testis Development. Endocrinology 2011, 152, 2076–2089. [Google Scholar] [CrossRef]
- Li, Y.; Schang, G.; Boehm, U.; Deng, C.-X.; Graff, J.; Bernard, D.J. SMAD3 Regulates Follicle-stimulating Hormone Synthesis by Pituitary Gonadotrope Cells in Vivo. J. Biol. Chem. 2017, 292, 2301–2314. [Google Scholar] [CrossRef]
- Roh, J.-S.; Bondestam, J.; Mazerbourg, S.; Kaivo-Oja, N.; Groome, N.; Ritvos, O.; Hsueh, A.J.W. Growth Differentiation Factor-9 Stimulates Inhibin Production and Activates Smad2 in Cultured Rat Granulosa Cells. Endocrinology 2003, 144, 172–178. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Pangas, S.A.; Jorgez, C.J.; Graff, J.M.; Weinstein, M.; Matzuk, M.M. Redundant Roles of SMAD2 and SMAD3 in Ovarian Granulosa Cells In Vivo. Mol. Cell. Biol. 2008, 28, 7001–7011. [Google Scholar] [CrossRef]
- Xu, J.; Oakley, J.; McGee, E.A. Stage-Specific Expression of Smad2 and Smad3 During Folliculogenesis1. Biol. Reprod. 2002, 66, 1571–1578. [Google Scholar] [CrossRef]
- Hardy, K.; Mora, J.M.; Dunlop, C.; Carzaniga, R.; Franks, S.; Fenwick, M.A. Nuclear exclusion of SMAD2/3 in granulosa cells is associated with primordial follicle activation in the mouse ovary. J. Cell Sci. 2018, 131, jcs218123. [Google Scholar] [CrossRef]
- Rodriguez, A.; Tripurani, S.K.; Burton, J.C.; Clementi, C.; Larina, I.; Pangas, S.A. SMAD Signaling Is Required for Structural Integrity of the Female Reproductive Tract and Uterine Function During Early Pregnancy in Mice. Biol. Reprod. 2016, 95, 44. [Google Scholar] [CrossRef]
- Liu, Z.W. Effects of GDF9 and BMP15 during the Developmental Process of Oocyte of Gibel Carp (Carassius Auratus Gibelio); Shanghai Ocean University: Shanghai, China, 2012. [Google Scholar]
| Item | Full-Length Transcriptome |
|---|---|
| Number of CCS | 290,291 |
| Read bases of CCS | 727,649,017 |
| Average read length of CCS | 2506 |
| Number of FLNC reads | 248,408 |
| Number of consensus isoforms | 69,148 |
| Number of high-quality isoforms | 66,958 |
| Number of unigenes | 37,305 |
| Group | Clean Read Number | Clean Base Number | Q30 (%) | Q20 (%) | GC Content (%) | Mapping Rate (%) |
|---|---|---|---|---|---|---|
| Female1 | 23,589,409 | 7,063,060,762 | 95.02 | 98.27 | 50.11 | 88.30 |
| Female2 | 26,686,421 | 7,990,222,868 | 95.01 | 98.28 | 49.97 | 87.40 |
| Female3 | 23,801,164 | 7,125,984,844 | 95.15 | 98.34 | 49.94 | 88.13 |
| Male1 | 21,172,626 | 6,329,873,382 | 94.36 | 97.82 | 48.59 | 74.74 |
| Male2 | 25,166,743 | 7,532,229,636 | 94.29 | 97.82 | 48.54 | 75.58 |
| Male3 | 24,181,267 | 7,231,220,526 | 94.66 | 97.97 | 48.14 | 74.17 |
| Annotation Database | Number of Unigenes |
|---|---|
| GO annotation | 26,710 |
| KEGG annotation | 22,010 |
| KOG annotation | 25,866 |
| Pfam annotation | 29,797 |
| Swissprot annotation | 34,144 |
| eggNOG annotation | 33,442 |
| NR annotation | 34,221 |
| All annotated | 34,342 |
| Pathway ID | Pathway Term | p-Value |
|---|---|---|
| ko03010 | Ribosome | 1.20 × 10−14 |
| ko03460 | Fanconi anemia pathway | 0.001289 |
| ko00061 | Fatty acid biosynthesis | 0.002237 |
| ko04110 | Cell cycle | 0.002476 |
| ko04512 | ECM-receptor interaction | 0.003232 |
| ko00900 | Terpenoid backbone biosynthesis | 0.004637 |
| ko03030 | DNA replication | 0.008156 |
| ko00062 | Fatty acid elongation | 0.008802 |
| ko00230 | Purine metabolism | 0.010252 |
| ko00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 0.010259 |
| ko01212 | Fatty acid metabolism | 0.014225 |
| ko00670 | One carbon pool by folate | 0.018081 |
| ko00604 | Glycosphingolipid biosynthesis—ganglio series | 0.027686 |
| ko00910 | Nitrogen metabolism | 0.030108 |
| ko04270 | Vascular smooth muscle contraction | 0.044731 |
| ko00600 | Sphingolipid metabolism | 0.049852 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Zhou, D.; Zhang, X.; Li, G.; Zhang, Y.; Huang, C.; Zhang, Z.; Tian, C. A First Insight into the Gonad Transcriptome of Hong Kong Catfish (Clarias fuscus). Animals 2021, 11, 1131. https://doi.org/10.3390/ani11041131
Lin X, Zhou D, Zhang X, Li G, Zhang Y, Huang C, Zhang Z, Tian C. A First Insight into the Gonad Transcriptome of Hong Kong Catfish (Clarias fuscus). Animals. 2021; 11(4):1131. https://doi.org/10.3390/ani11041131
Chicago/Turabian StyleLin, Xinghua, Dayan Zhou, Xiaomin Zhang, Guangli Li, Yulei Zhang, Cailin Huang, Zhixin Zhang, and Changxu Tian. 2021. "A First Insight into the Gonad Transcriptome of Hong Kong Catfish (Clarias fuscus)" Animals 11, no. 4: 1131. https://doi.org/10.3390/ani11041131
APA StyleLin, X., Zhou, D., Zhang, X., Li, G., Zhang, Y., Huang, C., Zhang, Z., & Tian, C. (2021). A First Insight into the Gonad Transcriptome of Hong Kong Catfish (Clarias fuscus). Animals, 11(4), 1131. https://doi.org/10.3390/ani11041131








