Long Non-Coding RNAs in Insects
Abstract
Simple Summary
Abstract
1. Introduction
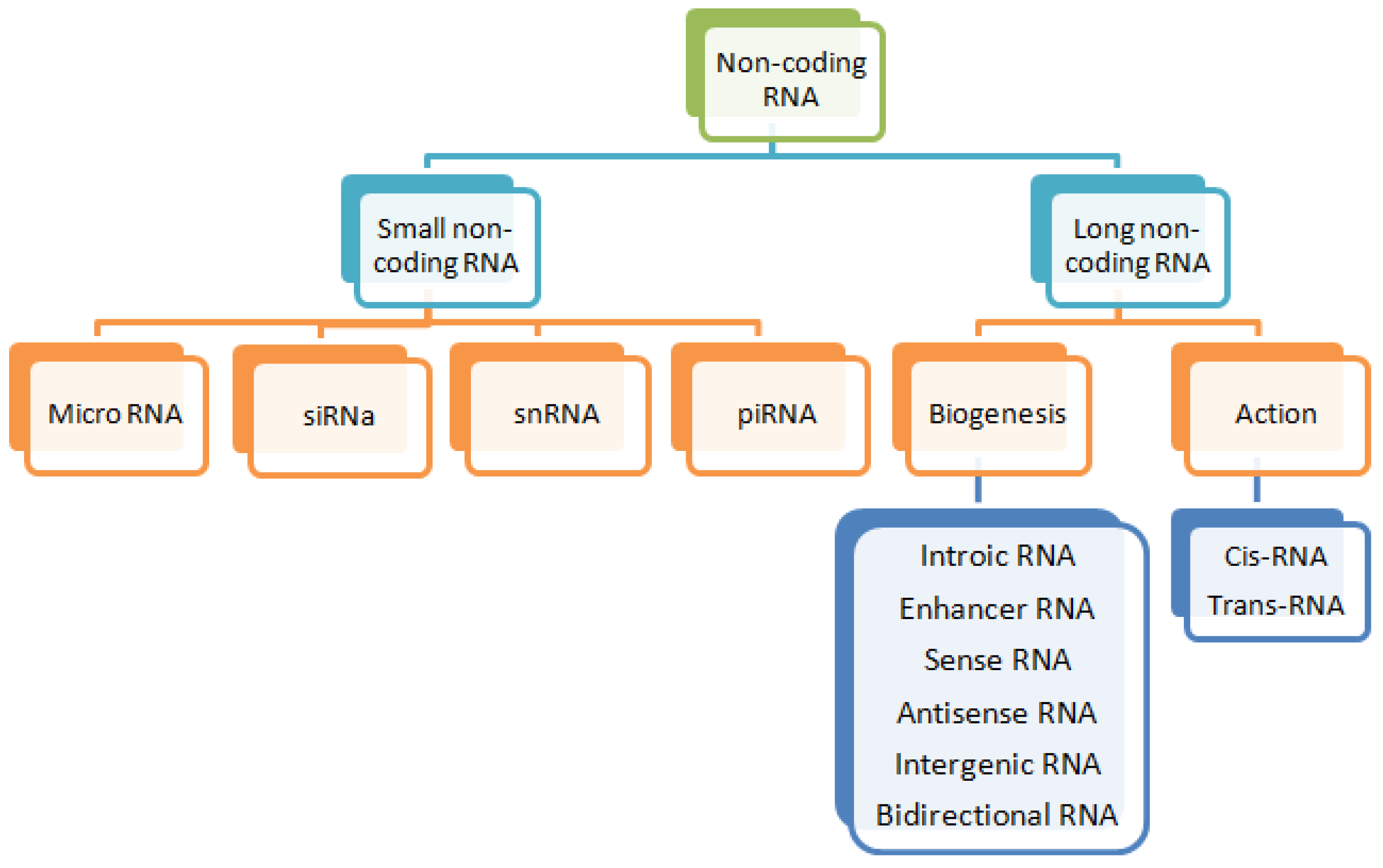
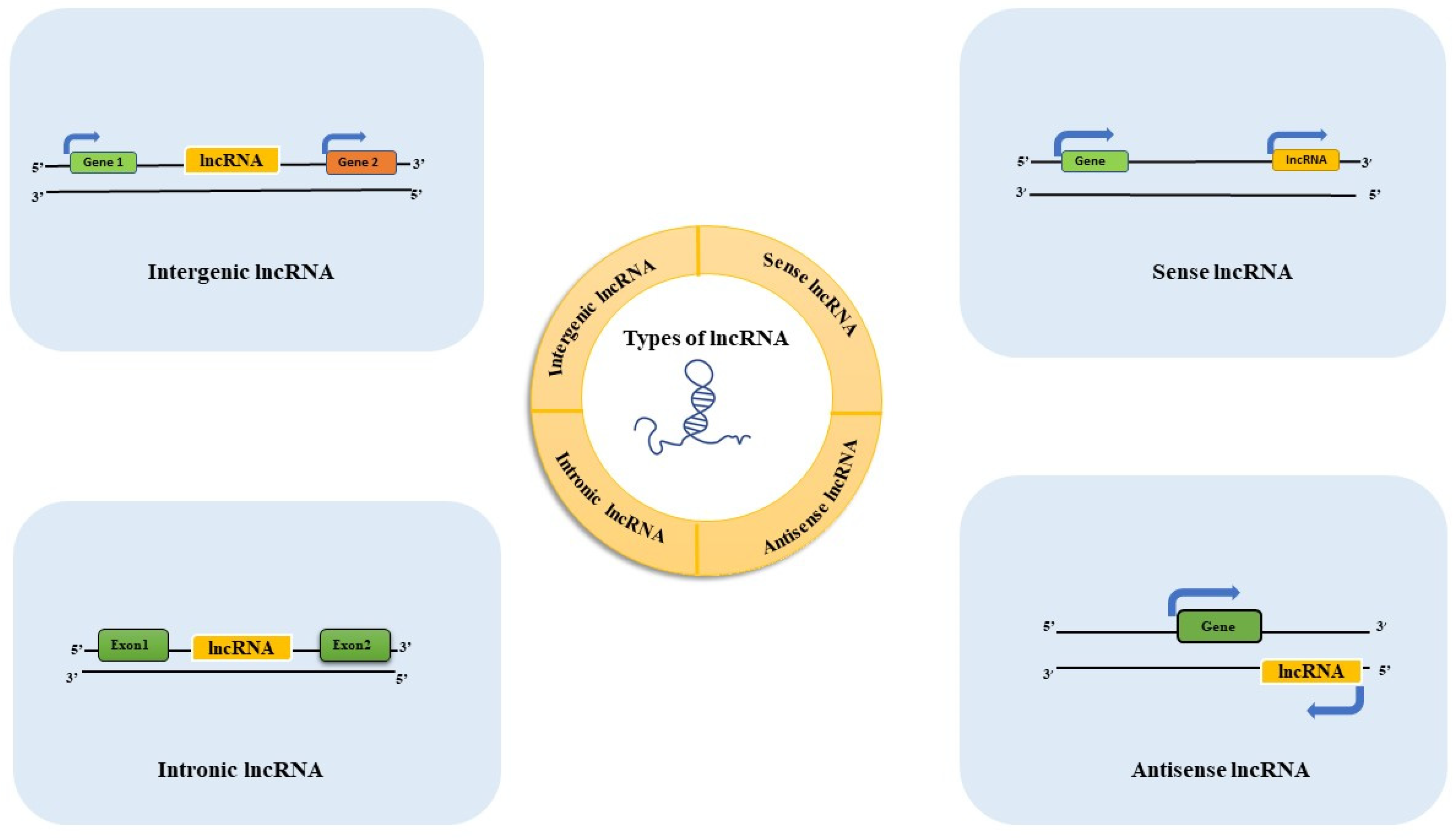
2. Long Non-Coding RNAs (lncRNAs)
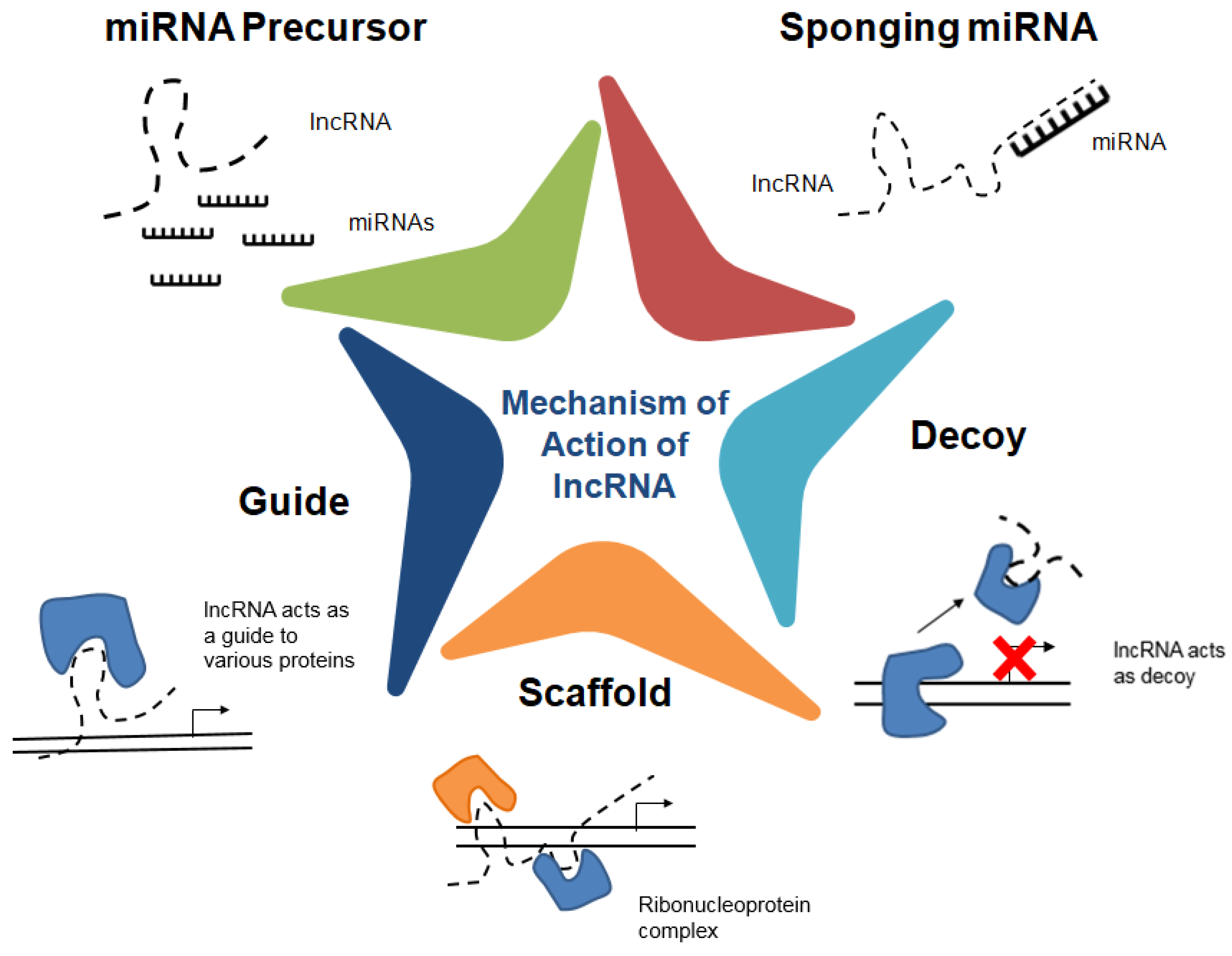
3. Identification and Functional Characterization of lncRNAs
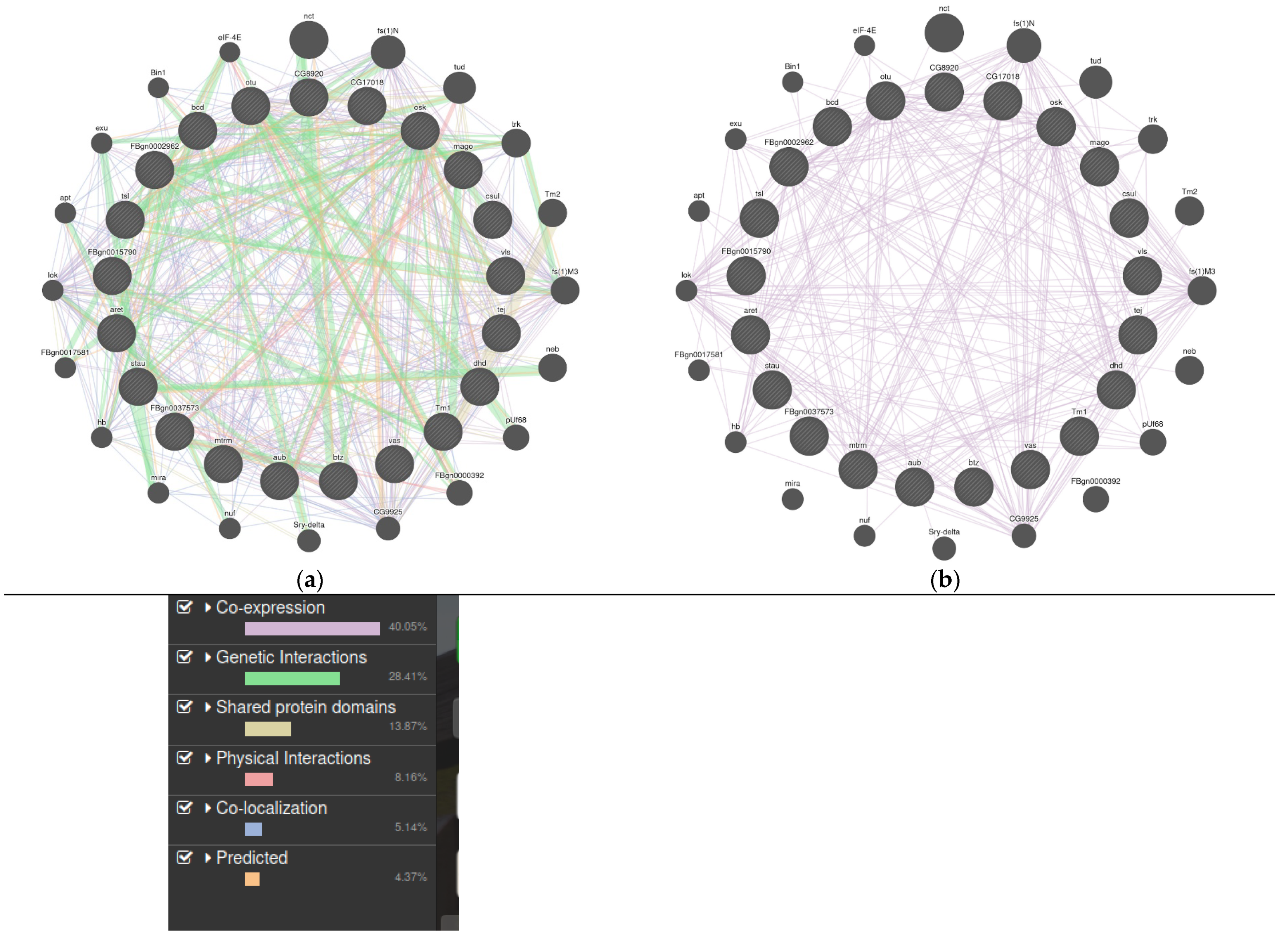
3.1. LncRNAs in Drosophila Melanogaster
3.2. LncRNAs in Apis Mellifera
3.3. LncRNAs in Aedes Aegypti
3.4. LncRNAs in Anopheles Gambiae
3.5. LncRNAs in Bombyx Mori
3.6. LncRNAs in Plutella Xylostella
3.7. LncRNA in Planthopper Insects
4. Challenges and Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of Alternative Splicing Through Coupling with Transcription and Chromatin Structure. Annu. Rev. Biochem. 2015, 84, 165–198. [Google Scholar] [CrossRef]
- Rana, T.M. Illuminating the Silence: Understanding the Structure and Function of Small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the Classification of Long Non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is Functional and What is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long Non-coding RNA: Classification, Biogenesis and Functions in Blood Cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Ma, X.; Shao, C.; Jin, Y.; Wang, H.; Meng, Y. Long Non-coding RNAs. RNA Biol. 2014, 11, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.-L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Liu, C.; Bai, B.; Skogerbø, G.; Cai, L.; Deng, W.; Zhang, Y.; Bu, D.; Zhao, Y.; Chen, R. NONCODE: An Integrated Knowledge Database of Non-coding RNAs. Nucleic Acids Res. 2005. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.P.; Clark, M.B.; Gascoigne, D.K.; Dinger, M.E.; Mattick, J.S. LncRNAdb: A Reference Database for Long Noncoding RNAs. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Deb, A.; Maji, R.K.; Saha, S.; Ghosh, Z. LncRBase: An Enriched Resource for lncRNA Information. PLoS ONE 2014. [Google Scholar] [CrossRef]
- Zheng, L.L.; Li, J.H.; Wu, J.; Sun, W.J.; Liu, S.; Wang, Z.L.; Zhou, H.; Yang, J.H.; Qu, L.H. DeepBase v2.0: Identification, Expression, Evolution and Function of Small RNAs, LncRNAs and Circular RNAs from Deep-sequencing Data. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef]
- Jenkins, A.M.; Waterhouse, R.M.; Muskavitch, M.A.T. Long Non-coding RNA Discovery across the Genus Anopheles Reveals Conserved Secondary Structures within and beyond the Gambiae Complex. BMC Genom. 2015. [Google Scholar] [CrossRef]
- Jayakodi, M.; Jung, J.W.; Park, D.; Ahn, Y.-J.; Lee, S.-C.; Shin, S.-Y.; Shin, C.; Yang, T.-J.; Kwon, H.W. Genome-wide Characterization of Long Intergenic Non-coding RNAs (lincRNAs) Provides New Insight into Viral Diseases in Honey Bees Apis cerana and Apis mellifera. BMC Genom. 2015, 16, 680. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, T.; Liu, C.; Liu, D.; Zhang, Q.; Long, R.; Zhao, P.; Xia, Q. Systematic Identification and Characterization of Long Non-coding RNAs in the Silkworm, Bombyx mori. PLoS ONE 2016. [Google Scholar] [CrossRef]
- Ali, A.; Abd El Halim, H.M. Re-thinking Adaptive Immunity in the Beetles: Evolutionary and Functional Trajectories of lncRNAs. Genomics 2020. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y.; Lu, Y.; Li, B.; Chen, K.; Li, C. Genome-wide Identification and Characterization of Long Non-coding RNAs in Tribolium castaneum. Insect Sci. 2020, 1744–7917.12867. [Google Scholar] [CrossRef]
- Kashi, K.; Henderson, L.; Bonetti, A.; Carninci, P. Discovery and Functional Analysis of lncRNAs: Methodologies to Investigate an Uncharacterized Transcriptome. Biochim. Biophys. Acta Gene Regul. Mech. 2016. [Google Scholar] [CrossRef]
- Etebari, K.; Furlong, M.J.; Asgari, S. Genome Wide Discovery of Long Intergenic Non-coding RNAs in Diamondback Moth (Plutella xylostella) and Their Expression in Insecticide Resistant Strains. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Asad, S.; Zhang, G.; Asgari, S. Identification of Aedes aegypti Long Intergenic Non-coding RNAs and Their Association with Wolbachia and Dengue Virus Infection. PLoS Negl. Trop. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Lin, J.D. Long Noncoding RNAs: A New Regulatory Code in Metabolic Control. Trends Biochem. Sci. 2015, 40, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Satyavathi, V.; Ghosh, R.; Subramanian, S. Long Non-Coding RNAs Regulating Immunity in Insects. Noncoding RNA 2017, 3, 14. [Google Scholar] [CrossRef]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene Regulation by the Act of Long Non-coding RNA Transcription. BMC Biol. 2013. [Google Scholar] [CrossRef]
- Han, P.; Chang, C.P. Long Non-coding RNA and Chromatin Remodeling. RNA Biol. 2015. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-coding RNAs: Multi-tasking Molecules in the Cell. Int. J. Mol. Sci. 2013, 14, 16010–16039. [Google Scholar] [CrossRef]
- Liu, F.; Shi, T.; Qi, L.; Su, X.; Wang, D.; Dong, J.; Huang, Z.Y. LncRNA Profile of Apis melliferaand Its Possible Role in Behavioural Transition from Nurses to Foragers. BMC Genom. 2019. [Google Scholar] [CrossRef]
- Stork, N.E. How Many Species of Insects and Other Terrestrial Arthropods Are There on Earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Stork, N.E.; McBroom, J.; Gely, C.; Hamilton, A.J. New Approaches Narrow Global Species Estimates for Beetles, Insects, and Terrestrial Arthropods. Proc. Natl. Acad. Sci. USA 2015, 112, 7519–7523. [Google Scholar] [CrossRef] [PubMed]
- Legeai, F.; Derrien, T. Identification of Long Non-coding RNAs in Insects Genomes. Curr. Opin. Insect. Sci. 2015, 7, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Zimmer-Bensch, G. Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 2019, 8, 1399. [Google Scholar] [CrossRef]
- Mallory, A.C.; Shkumatava, A. LncRNAs in Vertebrates: Advances and Challenges. Biochimie 2015, 117, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.H.; Pan, X.; Liu, M.; Wang, S.; Huang, T.; Cai, Y.D. Tissue Expression Difference between Mrnas and Lncrnas. Int. J. Mol. Sci. 2018, 3416. [Google Scholar] [CrossRef]
- Perry, R.B.-T.; Ulitsky, I. The Functions of Long Noncoding RNAs in Development and Stem Cells. Development 2016, 143, 3882–3894. [Google Scholar] [CrossRef]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef]
- Quinn, J.J.; Zhang, Q.C.; Georgiev, P.; Ilik, I.A.; Akhtar, A.; Chang, H.Y. Rapid Evolutionary Turnover Underlies Conserved lncRNA-genome Interactions. Genes Dev. 2016. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Tan, J.; Deng, P.; Li, T.; He, H.; Bian, J.; Wu, L. Pesticide Application Has Little Influence on Coding and Non-coding Gene Expressions in Rice. BMC Genom. 2019. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhou, L.; Ge, M.; Zhang, B.; Yang, X.; Xiong, X.; Fu, G.; Zhang, J.; Nie, X.; Li, H.; et al. Whole Exome Sequencing Identifies lncRNA GAS8-AS1 and LPAR4 as Novel Papillary Thyroid Carcinoma Driver Alternations. Hum. Mol. Genet. 2016. [Google Scholar] [CrossRef]
- Gupta, S.; Gupta, N.; Tiwari, P.; Menon, S.; Mathur, P.; Kothari, S.; Nallapeta, S.; Medicherla, K.; Suravajhala, P. Lnc-EPB41-Protein Interactions Associated with Congenital Pouch Colon. Biomolecules 2018, 8, 95. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018. [Google Scholar] [CrossRef]
- Tripathi, R.; Chakraborty, P.; Varadwaj, P.K. Unraveling Long Non-coding RNAs through Analysis of High-throughput RNA-sequencing Data. Non Coding RNA Res. 2017, 2, 111–118. [Google Scholar] [CrossRef]
- Abugessaisa, I.; Ramilowski, J.A.; Lizio, M.; Severin, J.; Hasegawa, A.; Harshbarger, J.; Kondo, A.; Noguchi, S.; Yip, C.W.; Ooi, J.L.C.; et al. FANTOM Enters 20th year: Expansion of Transcriptomic Atlases and Functional Annotation of Non-coding RNAs. Nucleic Acids Res. 2021, 49, D892–D898. [Google Scholar] [CrossRef] [PubMed]
- Martianov, I.; Ramadass, A.; Serra Barros, A.; Chow, N.; Akoulitchev, A. Repression of the Human Dihydrofolate Reductase Gene by a Non-coding Interfering Transcript. Nature 2007. [Google Scholar] [CrossRef]
- Grote, P.; Herrmann, B.G. The long non-coding RNA Fendrr Links Epigenetic Control Mechanisms to Gene Regulatory Networks in Mammalian Embryogenesis. RNA Biol. 2013, 10, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Tafer, H.; Hofacker, I.L. RNAplex: A Fast Tool for RNA-RNA Interaction Search. Bioinformatics 2008. [Google Scholar] [CrossRef] [PubMed]
- Mückstein, U.; Tafer, H.; Hackermüller, J.; Bernhart, S.H.; Stadler, P.F.; Hofacker, I.L. Thermodynamics of RNA-RNA Binding. Bioinformatics 2006. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Richter, A.S.; Backofen, R. IntaRNA: Efficient Prediction of Bacterial sRNA Targets Incorporating Target Site Accessibility and Seed Regions. Bioinformatics 2008. [Google Scholar] [CrossRef] [PubMed]
- Suresh, V.; Liu, L.; Adjeroh, D.; Zhou, X. RPI-Pred: Predicting ncRNA-protein Interaction Using Sequence and Structural Information. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef]
- Lu, Q.; Ren, S.; Lu, M.; Zhang, Y.; Zhu, D.; Zhang, X.; Li, T. Computational Prediction of Associations between Long Non-coding RNAs and Proteins. BMC Genom. 2013. [Google Scholar] [CrossRef]
- Muppirala, U.K.; Honavar, V.G.; Dobbs, D. Predicting RNA-Protein Interactions Using Only Sequence Information. BMC Bioinform. 2011. [Google Scholar] [CrossRef]
- Wang, L.; Brown, S.J. BindN: A Web-based Tool for Efficient Prediction of DNA and RNA Binding Sites in Amino Acid Sequences. Nucleic Acids Res. 2006, 34, W243–W248. [Google Scholar] [CrossRef]
- Li, Q.; Cao, Z.; Liu, H. Improve the Prediction of RNA-Binding Residues Using Structural Neighbours. Protein Pept. Lett. 2010, 17, 287–296. [Google Scholar] [CrossRef]
- Ahmad, S.; Sarai, A. Analysis of Electric Moments of RNA-binding Proteins: Implications for Mechanism and Prediction. BMC Struct. Biol. 2011, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.B.; Boley, N.; Eisman, R.; May, G.E.; Stoiber, M.H.; Duff, M.O.; Booth, B.W.; Wen, J.; Park, S.; Suzuki, A.M.; et al. Diversity and Dynamics of the Drosophila transcriptome. Nature 2014. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; Van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The Developmental Transcriptome of Drosophila melanogaster. Nature 2011. [Google Scholar] [CrossRef]
- Li, K.; Tian, Y.; Yuan, Y.; Fan, X.; Yang, M.; He, Z.; Yang, D. Insights into the Functions of LncRNAs in Drosophila. Int. J. Mol. Sci. 2019, 20, 4646. [Google Scholar] [CrossRef] [PubMed]
- Schor, I.E.; Bussotti, G.; Maleš, M.; Forneris, M.; Viales, R.R.; Enright, A.J.; Furlong, E.E.M. Non-coding RNA Expression, Function, and Variation during Drosophila Embryogenesis. Curr. Biol. 2018. [Google Scholar] [CrossRef]
- Maeda, R.K.; Sitnik, J.L.; Frei, Y.; Prince, E.; Gligorov, D.; Wolfner, M.F.; Karch, F. The lncRNA Male-specific Abdominal Plays a Critical Role in Drosophila Accessory Gland Development and Male Fertility. PLoS Genet. 2018. [Google Scholar] [CrossRef]
- Li, M.Z.; Xiao, H.M.; Kang, H.; Li, F. Progress and Prospects of Noncoding RNAs in Insects. J. Integr. Agric. 2019. [Google Scholar] [CrossRef]
- Vedelek, V.; Bodai, L.; Grézal, G.; Kovács, B.; Boros, I.M.; Laurinyecz, B.; Sinka, R. Analysis of Drosophila melanogaster Testis Transcriptome. BMC Genom. 2018. [Google Scholar] [CrossRef]
- Wen, K.; Yang, L.; Xiong, T.; Di, C.; Ma, D.; Wu, M.; Xue, Z.; Zhang, X.; Long, L.; Zhang, W.; et al. Critical Roles of Long Noncoding RNAs in Drosophila Spermatogenesis. Genome Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Sansom, S.N.; Lee, S.; Chalei, V.; Kong, L.; Cooper, S.E.; Oliver, P.L.; Ponting, C.P. The Long Non-coding RNA Paupar Regulates the Expression of Both Local and Distal Genes. EMBO J. 2014. [Google Scholar] [CrossRef]
- Goff, L.A.; Groff, A.F.; Sauvageau, M.; Trayes-Gibson, Z.; Sanchez-Gomez, D.B.; Morse, M.; Martin, R.D.; Elcavage, L.E.; Liapis, S.C.; Gonzalez-Celeiro, M.; et al. Spatiotemporal Expression and Transcriptional Perturbations by Long Noncoding RNAs in the Mouse Brain. Proc. Natl. Acad. Sci. USA 2015. [Google Scholar] [CrossRef]
- Soshnev, A.A.; Ishimoto, H.; Mcallister, B.F.; Li, X.; Wehling, M.D.; Kitamoto, T.; Geyer, P.K. A Conserved Long Noncoding RNA Affects Sleep Behavior in Drosophila. Genetics 2011. [Google Scholar] [CrossRef]
- Li, M.; Liu, L. Neural Functions of Long Noncoding RNAs in Drosophila. J. Comp. Physiol. A 2015, 201, 921–926. [Google Scholar] [CrossRef]
- García-Bellido, A.; de Celis, J.F. The Complex Tale of the Achaete-scute Complex: A Paradigmatic Case in the Analysis of Gene Organization and Function during Development. Genetics 2009, 182, 631–639. [Google Scholar] [CrossRef]
- Marcellini, S.; Gibert, J.M.; Simpson, P. Achaete, but not Scute, is Dispensable for the Peripheral Nervous System of Drosophila. Dev. Biol. 2005. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coolon, J.D.; McManus, C.J.; Stevenson, K.R.; Graveley, B.R.; Wittkopp, P.J. Tempo and Mode of Regulatory Evolution in Drosophila. Genome Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Faucillion, M.L.; Larsson, J. RNA-on-X 1 and 2 in Drosophila melanogasterFulfill Separate Functions in Dosage Compensation. PLoS Genet. 2018. [Google Scholar] [CrossRef]
- Ilik, I.; Akhtar, A. roX RNAs: Non-coding Regulators of the Male X Chromosome in Flies. RNA Biol. 2009, 6, 113–121. [Google Scholar] [CrossRef]
- Deng, X.; Koya, S.K.; Kong, Y.; Meller, V.H. Coordinated Regulation of Heterochromatic Genes in Drosophila melanogaster Males. Genetics 2009. [Google Scholar] [CrossRef] [PubMed]
- Penalva, L.O.F.; Sánchez, L. RNA Binding Protein Sex-Lethal (Sxl) and Control of Drosophila Sex Determination and Dosage Compensation. Microbiol. Mol. Biol. Rev. 2003. [Google Scholar] [CrossRef]
- Mulvey, B.B.; Olcese, U.; Cabrera, J.R.; Horabin, J.I. An Interactive Network of Long Non-coding RNAs Facilitates the Drosophila Sex Determination Decision. Biochim. Biophys. Acta Gene Regul. Mech. 2014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Davie, K.; Janssens, J.; Koldere, D.; De Waegeneer, M.; Pech, U.; Kreft, Ł.; Aibar, S.; Makhzami, S.; Christiaens, V.; Bravo González-Blas, C.; et al. A Single-Cell Atlas of the Aging Drosophila Brain. Cell 2018. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Rajendra, T.K.; Lal, A.K.; Lakhotia, S.C. Omega Speckles–A Novel Class of Nuclear Speckles Containing hnRNPs Associated with Noncoding hsr-omega RNA in Drosophila. J. Cell. Sci. 2000, 113, 3485–3497. [Google Scholar]
- Singh, A.K.; Lakhotia, S.C. Dynamics of hnRNPs and Omega Speckles in Normal and Heat Shocked Live Cell Nuclei of Drosophila melanogaster. Chromosoma 2015. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, S.C.; Sharma, A. The 93D (hsr-omega) Locus of Drosophila: Non-coding Gene with House-keeping Functions. Genetica 1996, 97, 339–348. [Google Scholar] [CrossRef]
- Lakhotia, S.C.; Mallik, M.; Singh, A.K.; Ray, M. The Large Noncoding hsrω-n Transcripts are Essential for Thermotolerance and Remobilization of hnRNPs, HP1 and RNA Polymerase II during Recovery from Heat Shock in Drosophila. Chromosoma 2012. [Google Scholar] [CrossRef]
- Lakhotia, S.C. Forty Years of the 93D Puff of Drosophila melanogaster. J. Biosci. 2011. [Google Scholar] [CrossRef]
- Johnson, T.K.; Carrington, L.B.; Hallas, R.J.; McKechnie, S.W. Protein Synthesis Rates in Drosophila Associate with Levels of the hsr-omega Nuclear Transcript. Cell Stress Chaperones 2009. [Google Scholar] [CrossRef]
- Barciszewski, J.; Erdmann, V.A.; Lakhotia, S.C. hsrω ω ω ω ω Gene of Drosophila melanogaster Integrates Post-Transcriptional Processing of Other Nuclear Transcripts; Kluwer Academic Publishers-Plenum Publishers: Dodrecht, The Netherlands, 2003. [Google Scholar]
- Geisler, S.; Coller, J. RNA in Unexpected Places: Long Non-coding RNA Functions in Diverse Cellular Contexts. Nat. Rev. Mol. Cell. Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Lo Piccolo, L.; Yamaguchi, M. RNAi of arcRNA hsrω Affects Sub-cellular Localization of Drosophila FUS to Drive Neurodiseases. Exp. Neurol. 2017. [Google Scholar] [CrossRef]
- Lehmann, R. Germ-plasm Formation and Germ-cell Determination in Drosophila. Curr. Opin. Genet. Dev. 1992. [Google Scholar] [CrossRef]
- Jenny, A.; Hachet, O.; Závorszky, P.; Cyrklaff, A.; Weston, M.D.J.; St Johnston, D.; Erdélyi, M.; Ephrussi, A. A Translation-independent Role of Oskar RNA in Early Drosophila Oogenesis. Development 2006. [Google Scholar] [CrossRef] [PubMed]
- Micklem, D.R.; Adams, J.; Grünert, S.; St Johnston, D. Distinct Roles of Two Conserved Staufen Domains in Oskar mRNA Localization and Translation. EMBO J. 2000. [Google Scholar] [CrossRef] [PubMed]
- Zimyanin, V.; Lowe, N.; St Johnston, D. An Oskar-Dependent Positive Feedback Loop Maintains the Polarity of the Drosophila Oocyte. Curr. Biol. 2007. [Google Scholar] [CrossRef]
- Kim-Ha, J.; Kerr, K.; Macdonald, P.M. Translational Regulation of Oskar mRNA by Bruno, an Ovarian RNA-binding Protein, is Essential. Cell 1995, 81, 403–412. [Google Scholar] [CrossRef]
- Ríos-Barrera, L.D.; Gutiérrez-Pérez, I.; Domínguez, M.; Riesgo-Escovar, J.R. Acal is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure. PLoS Genet. 2015. [Google Scholar] [CrossRef]
- Hardiman, K.E.; Brewster, R.; Khan, S.M.; Deo, M.; Bodmer, R. The Bereft Gene, a Potential Target of the Neural Selector Gene Cut, Contributes to Bristle Morphogenesis. Genetics 2002. [Google Scholar] [CrossRef]
- Petruk, S.; Sedkov, Y.; Riley, K.M.; Hodgson, J.; Schweisguth, F.; Hirose, S.; Jaynes, J.B.; Brock, H.W.; Mazo, A. Transcription of bxd Noncoding RNAs Promoted by Trithorax Represses Ubx in cis by Transcriptional Interference. Cell 2006. [Google Scholar] [CrossRef]
- Pease, B.; Borges, A.C.; Bender, W. Noncoding RNAs of the Ultrabithorax domain of the Drosophila bithorax complex. Genetics 2013. [Google Scholar] [CrossRef] [PubMed]
- Gummalla, M.; Maeda, R.K.; Alvarez, J.J.; Gyurkovics, H.; Singari, S.; Edwards, K.A.; Karch, F.; Bender, W. Abd-A Regulation by the iab-8 Noncoding RNA. PLoS Genet. 2012. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wen, S.; Guo, X.; Bai, B.; Gong, Z.; Liu, X.; Wang, Y.; Zhou, Y.; Chen, X.; Liu, L.; et al. The Novel Long Non-coding RNA CRG Regulates Drosophila Locomotor Behavior. Nucleic Acids Res. 2012. [Google Scholar] [CrossRef]
- Calderone, N.W. Temporal Division of Labor in the Honey Bee, Apis mellifera: A Developmental Process or the Result of Environmental Influences? Can. J. Zool. 1995. [Google Scholar] [CrossRef]
- Kolmes, S.A. A Quantitative Study of the Division of Labour among Worker Honey Bees. Z. Tierpsychol. 1985. [Google Scholar] [CrossRef]
- Heylen, K.; Gobin, B.; Billen, J.; Hu, T.T.; Arckens, L.; Huybrechts, R. Amfor Expression in the Honeybee Brain: A Trigger Mechanism for Nurse-forager Transition. J. Insect Physiol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Tadano, H.; Yamazaki, Y.; Takeuchi, H.; Kubo, T. Age- and Division-of-labour-dependent Differential Expression of a Novel Non-coding RNA, Nb-1, in the Brain of Worker Honeybees, Apis mellifera L. Insect Mol. Biol. 2009, 18, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Sawata, M.; Yoshino, D.; Takeuchi, H.; Kamikouchi, A.; Ohashi, K.; Kubo, T. Identification and Punctate Nuclear Localization of a Novel Noncoding RNA, Ks-1, from the Honeybee Brain. RNA 2002. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, G.M.; Robinson, G.E.; Gibbs, R.A.; Worley, K.C.; Evans, J.D.; Maleszka, R.; Robertson, H.M.; Weaver, D.B.; Beye, M.; Bork, P.; et al. Insights into Social Insects from the Genome of the Honeybee Apis mellifera. Nature 2006. [Google Scholar] [CrossRef]
- Sawata, M.; Takeuchi, H.; Kubo, T. Identification and Analysis of the Minimal Promoter Activity of a Novel Noncoding Nuclear RNA Gene, AncR-1, from the Honeybee (Apis mellifera L.). RNA 2004. [Google Scholar] [CrossRef]
- Kiya, T.; Kunieda, T.; Kubo, T. Inducible- and Constitutive-type Transcript Variants of Kakusei, a Novel Non-coding Immediate Early Gene, in the Honeybee Brain. Insect Mol. Biol. 2008. [Google Scholar] [CrossRef]
- Kiya, T.; Ugajin, A.; Kunieda, T.; Kubo, T. Identification of Kakusei, a Nuclear Non-coding RNA, as an Immediate Early Gene from the Honeybee, and Its Application for Neuroethological Study. Int. J. Mol. Sci. 2012, 5496. [Google Scholar] [CrossRef] [PubMed]
- Humann, F.C.; Tiberio, G.J.; Hartfelder, K. Sequence and Expression Characteristics of Long Noncoding RNAs in Honey Bee Caste Development–Potential Novel Regulators for Transgressive Ovary Size. PLoS ONE 2013. [Google Scholar] [CrossRef]
- Guo, Z.; Kang, S.; Zhu, X.; Xia, J.; Wu, Q.; Wang, S.; Xie, W.; Zhang, Y. The Novel ABC Transporter ABCH1 is a Potential Target for RNAi-based Insect Pest Control and Resistance Management. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Z.; Fang, S.M.; Zhang, Q.; Yu, Q.Y.; Zhang, Z. Identification and Comparison of Long Non-coding RNAs in the Silk Gland between Domestic and Wild Silkworms. Insect Sci. 2018. [Google Scholar] [CrossRef]
- Xu, X.; Wang, K.; Zha, X. An Antisense lncRNA Functions in Alternative Splicing of Bmdsx in the Silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, H.; Xiang, Z.; Lu, C.; Dai, F.; Tong, X. Identification and Characterization of a New Long Noncoding RNA iab-1 in the Hox Cluster of Silkworm, Bombyx mori Identification of iab-1. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Kiya, T.; Iwami, M. Identification and Expression Analysis of Nervous Wreck, Which is Preferentially Expressed in the Brain of the Male Silkworm Moth, Bombyx mori. Insect Mol. Biol. 2011, 20, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Li, H.; Zhang, H.; An, S. Comparative Analysis of dsRNA-induced lncRNAs in Three Kinds of Insect Species. Arch. Insect Biochem. Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Meller, V.H. roX RNAs are Required for Increased Expression of X-linked Genes in Drosophila melanogaster Males. Genetics 2006. [Google Scholar] [CrossRef]
- Muraoka, Y.; Nakamura, A.; Tanaka, R.; Suda, K.; Azuma, Y.; Kushimura, Y.; Lo Piccolo, L.; Yoshida, H.; Mizuta, I.; Tokuda, T.; et al. Genetic Screening of the Genes Interacting with Drosophila FIG4 Identified a Novel Link between CMT-Causing Gene and Long Noncoding RNAs. Exp. Neurol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dai, H.; Chen, S.; Zhang, L.; Long, M. Highly Tissue Specific Expression of Sphinx Supports Its Male Courtship Related Role in Drosophila melanogaster. PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed]
- Partap, U. The Pollination Role of Honeybees. In Honeybees of Asia; Springer: Berlin/Heidelberg, Germany, 2011; ISBN 9783642164224. [Google Scholar]
- Park, D.; Jung, W.W.; Choi, B.S.; Jayakodi, M.; Lee, J.; Lim, J.; Yu, Y.; Choi, Y.S.; Lee, M.L.; Park, Y.; et al. Uncovering the Novel Characteristics of Asian Honey Bee, Apis cerana, by Whole Genome Sequencing. BMC Genom. 2015. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chen, H.; Du, Y.; Zhou, D.; Geng, S.; Wang, H.; Wan, J.; Xiong, C.; Zheng, Y.; Guo, R. Genome-wide Identification of Long Non-coding RNAS and Their Regulatory Networks Involved in Apis mellifera Ligustica Response to Nosema Ceranae Infection. Insects 2019, 245. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.; Nelson, C.B.; Picot, V.; Gubler, D.J. Arboviruses: A Global Public Health Threat. Vaccine 2020, 38, 3989–3994. [Google Scholar] [CrossRef]
- Weaver, S.C.; Charlier, C.; Vasilakis, N.; Lecuit, M. Zika, Chikungunya, and Other Emerging Vector-Borne Viral Diseases. Annu. Rev. Med. 2018. [Google Scholar] [CrossRef]
- Azlan, A.; Obeidat, S.M.; Yunus, M.A.; Azzam, G. Systematic Identification and Characterization of Aedes aegypti Long Noncoding RNAs (lncRNAs). Sci. Rep. 2019. [Google Scholar] [CrossRef]
- Akbari, O.S.; Antoshechkin, I.; Amrhein, H.; Williams, B.; Diloreto, R.; Sandler, J.; Hay, B.A. The Developmental Transcriptome of the Mosquito Aedes aegypti, an Invasive Species and Major Arbovirus Vector. G3 2013. [Google Scholar] [CrossRef]
- Azlan, A.; Halim, M.A.; Mohamad, F.; Azzam, G. Identification and Characterization of Long Noncoding RNAs and Their Association with Acquisition of Blood Meal in Culex quinquefasciatus. Insect Sci. 2020. [Google Scholar] [CrossRef]
- Jensen, T.H.; Jacquier, A.; Libri, D. Dealing with Pervasive Transcription. Mol. Cell. 2013, 52, 473–484. [Google Scholar] [CrossRef]
- Soumya, M.; Harinatha Reddy, A.; Nageswari, G.; Venkatappa, B. Silkworm (Bombyx mori) and Its Constituents: A Fascinating Insect in Science and Research. J. Entomol. Zool. Stud. 2017, 5, 1701–1705. [Google Scholar]
- Taguchi, S.; Iwami, M.; Taketoshi, K. Identification and Characterization of a Novel Nuclear Noncoding RNA, Fben-1, Which is Preferentially Expressed in the Higher Brain Center of the Female Silkworm Moth, Bombyx mori. Neurosci. Lett. 2011. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.Z.; Sun, X.; Zhang, B.; Pu, J.; Jiang, Z.Y.; Li, M.; Fan, Y.J.; Xu, Y.Z. Alternative Splicing Regulation of Doublesex Gene by RNA-binding Proteins in the Silkworm Bombyx mori. RNA Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Shukla, J.N.; Nagaraju, J. Two Female-specific DSX Proteins are Encoded by the Sex-specific Transcripts of dsx, and are Required for Female Sexual Differentiation in Two Wild Silkmoth Species, Antheraea assama and Antheraea mylitta (Lepidoptera, Saturniidae). Insect Biochem. Mol. Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Shukla, J.N.; Nagaraju, J. Doublesex: A Conserved Downstream Gene Controlled by Diverse Upstream Regulators. J. Genet. 2010, 89, 341–356. [Google Scholar] [CrossRef]
- Suzuki, M.G.; Imanishi, S.; Dohmae, N.; Asanuma, M.; Matsumoto, S. Identification of a Male-Specific RNA Binding Protein That Regulates Sex-Specific Splicing of Bmdsx by Increasing RNA Binding Activity of BmPSI. Mol. Cell. Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yin, H.; Shen, M.; Huang, H.; Hou, Q.; Zhang, Z.; Zhao, W.; Guo, X.; Wu, P. Analysis of lncRNA-mediated Gene Regulatory Network of Bombyx mori in Response to BmNPV Infection. J. Invertebr. Pathol. 2020. [Google Scholar] [CrossRef]
- Chen, P.; Bao, X.Y.; Kang, T.T.; Dong, Z.Q.; Zhu, Y.; Pan, M.H.; Lu, C. Screening and Identification of Proteins Interacting with Bombyx mori IAP and Their Effects on BmNPV Proliferation. Sci. Agric. Sin. 2019. [Google Scholar] [CrossRef]
- Zhu, B.; Xu, M.; Shi, H.; Gao, X.; Liang, P. Genome-wide Identification of lncRNAs Associated with Chlorantraniliprole Resistance in Diamondback Moth Plutella xylostella (L.). BMC Genom. 2017. [Google Scholar] [CrossRef]
- Wang, L.; Tang, N.; Gao, X.; Chang, Z.; Zhang, L.; Zhou, G.; Guo, D.; Zeng, Z.; Li, W.; Akinyemi, I.A.; et al. Genome Sequence of a Rice Pest, the White-backed Planthopper (Sogatella furcifera). Gigascience 2017, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.Y.; Qu, L.Y.; Zhao, D.; Chen, L.B.; Jin, H.Y.; Xu, L.M.; Cheng, J.A.; Zhang, C.X. The Genome- and Transcriptome-wide Analysis of Innate Immunity in the Brown Planthopper, Nilaparvata lugens. BMC Genom. 2013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jiang, F.; Wang, X.; Yang, P.; Bao, Y.; Zhao, W.; Wang, W.; Lu, H.; Wang, Q.; Cui, N.; et al. Genome Sequence of the Small Brown Planthopper, Laodelphax striatellus. Gigascience 2017. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Yuan, Z.; Guo, D.; Hou, B.; Yin, C.; Zhang, W.; Li, F. Genome-wide Identification of Long Noncoding RNA Genes and Their Potential Association with Fecundity and Virulence in Rice Brown Planthopper, Nilaparvata lugens. BMC Genom. 2015. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Y.; Ye, W.Y.; Xiao, H.M.; Li, M.Z.; Cao, Z.H.; Ye, X.H.; Zhao, X.X.; He, K.; Li, F. LncRNAs are Potentially Involved in the Immune Interaction between Small Brown Planthopper and Rice Stripe Virus. J. Integr. Agric. 2019. [Google Scholar] [CrossRef]
- Chang, Z.X.; Ajayi, O.E.; Guo, D.Y.; Wu, Q.F. Genome-wide Characterization and Developmental Expression Profiling of Long Non-coding RNAs in Sogatella furcifera. Insect Sci. 2020. [Google Scholar] [CrossRef]
- Xie, X.; Tang, B.; Xiao, Y.F.; Xie, R.; Li, B.S.; Dong, H.; Zhou, J.Y.; Yang, S.M. Long Non-coding RNAs in Colorectal Cancer. Oncotarget 2016. [Google Scholar] [CrossRef]
- Cagliani, R.; Forni, D.; Clerici, M.; Sironi, M. Coding Potential and Sequence Conservation of SARS-CoV-2 and Related Animal Viruses. Infect. Genet. Evol. 2020. [Google Scholar] [CrossRef]
- Zhou, Q.; Huang, X.R.; Yu, J.; Yu, X.; Lan, H.Y. Long Noncoding RNA Arid2-IR is a Novel Therapeutic Target for Renal Inflammation. Mol. Ther. 2015. [Google Scholar] [CrossRef]
- Damas, N.D.; Fossat, N.; Scheel, T.K.H. Functional Interplay between RNA Viruses and Non-Coding RNA in Mammals. Noncoding RNA 2019, 5, 7. [Google Scholar] [CrossRef]
| S.No. | Organism | Name of lncRNA | Function | Size (bp) | Site of Expression | Interaction with Other RNA/Protein | Reference |
|---|---|---|---|---|---|---|---|
| Order: Hymenoptera | |||||||
| 1 | Apis mellifera | Nb-1 (Nurse bee brain-selective gene-1) | Shows varying expression in the brain of honey bees relative to the age of the colony. | 599 | Brain | Octopamine and Juvenile hormone | [109] |
| 2 | KS-1 (Kenyon cell/small-type preferential gene-1) | Expresses in the mushroom body of Kenyon cells in the honey bee brain and is accumulated in the nucleus. It is also involved in neuronal functions. | 17,525 | Kenyon cells | - | [110,112] | |
| 3 | AncR-1 | Exhibits spatial expression in the brain, sexual tissues, and in some secretory organs. It is also involved in neuronal functions. | 6861 | Sexual tissues and secretory organs | - | [112] | |
| 4 | Lncov1 and Lncov2 | Shows differential expression in the ovarioles of queen and workers. | 1367 | Larval Ovary | [116] | [110] | |
| Order: Lepidoptera | |||||||
| 5 | Bombyx mori | dw4sg_0040 | Downregulates the antibacterial peptide, metabolic process, and oxidative response. | Silk gland | - | [117] | |
| 6 | dw4sg_0178 | It is downregulated in B. mori | - | - | |||
| 7 | dw4sg_0483 | Performs the post-transcriptional regulation of silk protein to yield silk | - | - | |||
| 8 | Bmdsx-AS1 | Promotes male specific splicing of Bmdsx by interacting with BmPSI | Silkworm testis | Hnrnph, BxRBP1 &3. | [118] | ||
| 9 | iab-1 | Involved in essential metabolic/physiological processes. | ~>1000 | Nervus and epidermis | Interacts with Hox gene- BmUbx,, Bmabd-A, Bmabd-B | [119] | |
| 10 | Fben-1 | Developmental/sexual functions in females. | ~2000 | Female brain (mushroom body) | - | [120] | |
| 11 | Plutella xylostella | TCONS_00186426 | Co-expresses with glycoprotein, Abd-5, which is important for cuticle formation in insects. | - | - | Abd-5 | [121] |
| 12 | TCONS_0002929 | Indirectly involved in fruit fly development as it expresses nearby C-roughest protein rst, which has a role in the development of fruit fly | - | - | - | ||
| 13 | TCONS_00008658 | Located in an intergenic region near JHEJ. JHEJ is known to activate insect JH. | - | - | - | ||
| Order: Diptera | |||||||
| 14 | Drosophila melanogaster | roX1 & roX2 | Regulates dosage compensation and formation of MSL ribonucleoprotein complex. | 4832 & 1368 respectively | roX1 expressed in Nuclei of all body parts. roX1 and roX2 expressed in the CNS of male brain. | Interacts with roX1: Clamp, Unr, mle, mof, MSL2,3. roX2: Male less (MLE), Male Sex Lethal (MSL1,2,3), mof, Unr. | [81,122] |
| 15 | hsrw | Regulates the development of neuromuscular junctions. | 21,216 | Expresses in almost every stage. | Hrb87F, Hrb98DE, Iswi, Mtor, Pep, Saf-B, TBPH, caz, sqd, hrp-40. | [89,94,123] | |
| 16 | yar | Regulates the sleep behavior and the circadian rhythm. | 1569 | Embryo and cytoplasm | - | [74] | |
| 17 | Sphinx | Directs male courtship behavior mediated by olfactory neurons. | 644,454 | - | Or92a. | [124] | |
| 18 | acal | Involved in sealing the dorsal gap during embryonic development. | 2386 | Embryonic/larval: CNS, epidermis | aop, bsk, raw | [100] | |
| 19 | oskar | Works along with Staufen to regulate oogenesis. | 2335 | Germplasm | Stau, vls, nos, SmD3, bwk, etc. | [96] | |
| 20 | bereft | Role in development of extra sensory organs (interommatidial bristles). | ~7000 | Peripheral nervous system, non -neuronal epidermis. | Krn, Nach, hid, ppk16, ppk28, ppk6, spi, upd1-3, vn | [101] | |
| 21 | msa | Responsible for male fertility and development of accessory glands. | >6500 | Secondary cells of drosophila male accessory glands | - | [68] | |
| 22 | bxd | Associated with repression of ultrabithorax (ubx) and also regulates growth and development. | 1755 | - | - | [103] | |
| 22 | iab-8 | Inhibits expression of homeotic gene abd-A. Knockdown of iab-8 causes sterility in both sexes. | 92,000 | - | - | [68,104] | |
| 23 | SxlPe-R1 and R2 | Facilitates sex determination. | 480 | - | - | [83] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choudhary, C.; Sharma, S.; Meghwanshi, K.K.; Patel, S.; Mehta, P.; Shukla, N.; Do, D.N.; Rajpurohit, S.; Suravajhala, P.; Shukla, J.N. Long Non-Coding RNAs in Insects. Animals 2021, 11, 1118. https://doi.org/10.3390/ani11041118
Choudhary C, Sharma S, Meghwanshi KK, Patel S, Mehta P, Shukla N, Do DN, Rajpurohit S, Suravajhala P, Shukla JN. Long Non-Coding RNAs in Insects. Animals. 2021; 11(4):1118. https://doi.org/10.3390/ani11041118
Chicago/Turabian StyleChoudhary, Chhavi, Shivasmi Sharma, Keshav Kumar Meghwanshi, Smit Patel, Prachi Mehta, Nidhi Shukla, Duy Ngoc Do, Subhash Rajpurohit, Prashanth Suravajhala, and Jayendra Nath Shukla. 2021. "Long Non-Coding RNAs in Insects" Animals 11, no. 4: 1118. https://doi.org/10.3390/ani11041118
APA StyleChoudhary, C., Sharma, S., Meghwanshi, K. K., Patel, S., Mehta, P., Shukla, N., Do, D. N., Rajpurohit, S., Suravajhala, P., & Shukla, J. N. (2021). Long Non-Coding RNAs in Insects. Animals, 11(4), 1118. https://doi.org/10.3390/ani11041118






