Genomic Prediction for Twin Pregnancies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources for Genetic Evaluation
2.2. Data Editing and Genetic Trait Definition for Genetic Evaluation
2.3. Statistical Models for Genetic Evaluation
2.4. Inclusion of Twinning Prediction in a Multitrait Selection Index
2.5. Demonstration of Evaluation Efficacy
3. Results
3.1. Data Characteristics for Genetic Evaluation
3.2. Variance Components and Summary Statistics for Genetic Evaluation
3.3. Correlations of Twinning with Other Traits
3.4. Demonstration of Evaluation Efficacy
4. Discussion
4.1. Variance Components and Genetic (omic) Correlations
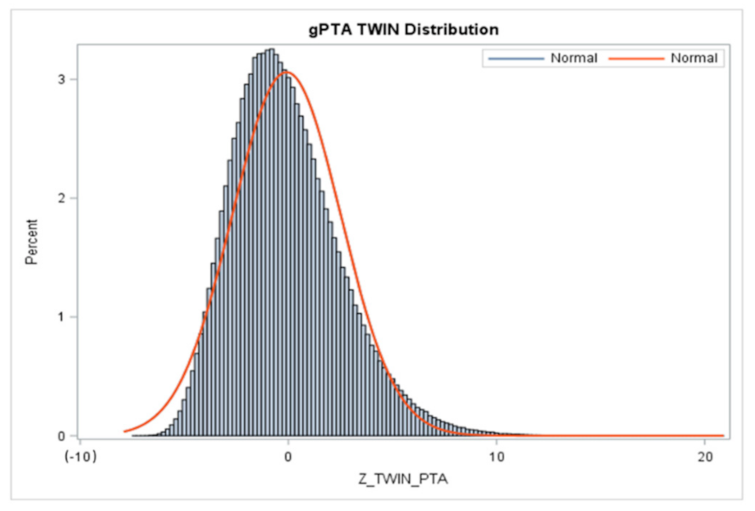
4.2. (g)PTAs and (g)STAs of Twinning
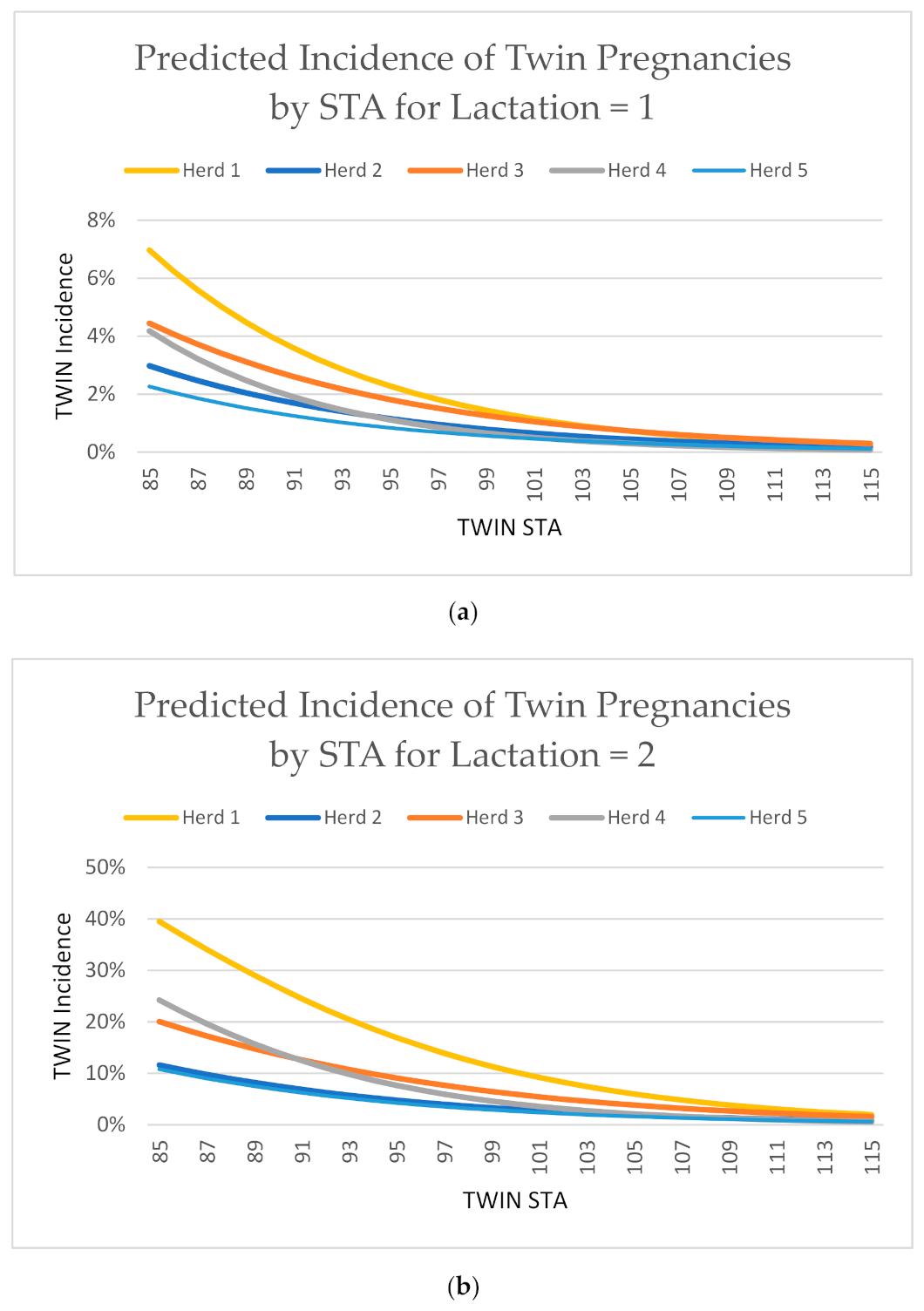
4.3. Demonstration of Evaluation Efficacy
4.4. Inclusion in Index & Breeding Strategies to Complement Current Management Strategies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Trait | Expected Response to Selection |
|---|---|
| Fat (lbs) | 15 |
| Protein (lbs) | 10 |
| Milk (lbs) | 218 |
| Productive Life (mo.) | 1.44 |
| Cow Livability (%) | 0.90 |
| Somatic Cell Score (log) | −0.05 |
| Body Size Composite (pts) | −0.22 |
| Udder Composite (pts) | 0.21 |
| Feet & Leg Composite (pts) | 0.10 |
| Daughter Pregnancy Rate (%) | 0.27 |
| Heifer Conception Rate (%) | 0.32 |
| Cow Conception Rate (%) | 0.52 |
| Calving Ability ($) | 9.47 |
| Zoetis Mastitis (STA) | 2.44 |
| Zoetis Metritis (STA) | 1.98 |
| Zoetis Retained Placenta (STA) | 0.80 |
| Zoetis Displaced Abomasum (STA) | 1.14 |
| Zoetis Ketosis (STA) | 2.04 |
| Zoetis Lameness (STA) | 1.22 |
| Zoetis Calf Respiratory (STA) | 1.16 |
| Zoetis Calf Scours (STA) | 1.16 |
| Zoetis Calf Livability (STA) | 1.46 |
| Zoetis Cow Respiratory (STA) | 1.35 |
| Zoetis Cystic Ovary (STA) | 0.26 |
| Zoetis Twinning (STA) | 0.81 |
| Zoetis Cow Abortion (STA) | 0.55 |
References
- Kinsel, M.; Marsh, W.; Ruegg, P.; Etherington, W. Risk Factors for Twinning in Dairy Cows. J. Dairy Sci. 1998, 81, 989–993. [Google Scholar] [CrossRef]
- Nielen, M.; Schukken, Y.; Scholl, D.; Wilbrink, H.; Brand, A. Twinning in dairy cattle: A study of risk factors and effects. Theriogenology 1989, 32, 845–862. [Google Scholar] [CrossRef]
- Ryan, D.; Boland, M. Frequency of twin births among Holstein-Friesian cows in a warm dry climate. Theriogenology 1991, 36, 1–10. [Google Scholar] [CrossRef]
- Labhsetwar, A.P.; Tyler, W.J.; Casida, L.E. Analysis of Variation in Some Factors Affecting Multiple Ovulations in Holstein Cattle. J. Dairy Sci. 1963, 46, 840–842. [Google Scholar] [CrossRef]
- Fricke, P.M.; Wiltbank, M.C. Effect of milk production on the incidence of double ovulation in dairy cows. Theriogenology 1999, 52, 1133–1143. [Google Scholar] [CrossRef]
- Bendixen, P.; Oltenacu, P.; Andersson, L. Case-referent study of cystic ovaries as a risk indicator for twin calvings in dairy cows. Theriogenology 1989, 31, 1059–1066. [Google Scholar] [CrossRef]
- Gregory, K.E.; Echternkamp, S.; Dickerson, G.E.; Cundiff, L.V.; Koch, R.M.; Van Vleck, L.D. Twinning in cattle: I. Foundation animals and genetic and environmental effects on twinning rate. J. Anim. Sci. 1990, 68, 1867–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, K.E.; Bennett, G.L.; Van Vleck, L.D.; Echternkamp, S.E.; Cundiff, L.V. Genetic and environmental parameters for ovulation rate, twinning rate, and weight traits in a cattle population selected for twinning. J. Anim. Sci. 1997, 75, 1213–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rose, E.P.; Wilton, J.W. Productivity and profitability of twin births in beef cattle. J. Anim. Sci. 1991, 69, 3085–3093. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Martinez, P.; Dickerson, G.E.; Anderson, G.B.; Green, R.D. Embryo-transfer twinning and performance efficiency in beef production1. J. Anim. Sci. 1990, 68, 4039–4050. [Google Scholar] [CrossRef]
- López-Gatius, F.; Andreu-Vázquez, C.; Mur-Novales, R.; Cabrera, V.E.; Hunter, R.H.F. The dilemma of twin pregnancies in dairy cattle. A review of practical prospects. Livest. Sci. 2017, 197, 12–16. [Google Scholar] [CrossRef]
- Fricke, P. Twinning in Dairy Cattle. Prof. Anim. Sci. 2001, 17, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Cady, R.A.; Van Vleck, L.D. Factors Affecting Twinning and Effects of Twinning in Holstein Dairy Cattle. J. Anim. Sci. 1978, 46, 950–956. [Google Scholar] [CrossRef] [Green Version]
- Foote, R. Factors affecting gestation length in dairy cattle. Theriogenology 1981, 15, 553–559. [Google Scholar] [CrossRef]
- Rutledge, J.J. Twinning in Cattle. J. Anim. Sci. 1975, 40, 803–815. [Google Scholar] [CrossRef]
- Johanson, J.; Berger, P.; Kirkpatrick, B.; Dentine, M. Twinning Rates for North American Holstein Sires. J. Dairy Sci. 2001, 84, 2081–2088. [Google Scholar] [CrossRef]
- Day, J.D.; Weaver, L.D.; E Franti, C. Twin pregnancy diagnosis in Holstein cows: Discriminatory powers and accuracy of diagnosis by transrectal palpation and outcome of twin pregnancies. Can. Veter J. La Rev. Veter-Can. 1995, 36, 93. [Google Scholar]
- Hossein-Zadeh, N.G.; Nejati-Javaremi, A.; Miraei-Ashtiani, S.; Kohram, H. An Observational Analysis of Twin Births, Calf Stillbirth, Calf Sex Ratio, and Abortion in Iranian Holsteins. J. Dairy Sci. 2008, 91, 4198–4205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Gregory, K.; E Echternkamp, S.; Cundiff, L.V. Effects of twinning on dystocia, calf survival, calf growth, carcass traits, and cow productivity. J. Anim. Sci. 1996, 74, 1223–1233. [Google Scholar] [CrossRef]
- Echternkamp, S.E.; Gregory, K.E. Effects of twinning on postpartum reproductive performance in cattle selected for twin births. J. Anim. Sci. 1999, 77, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echternkamp, S.E.; Gregory, K.E. Effects of twinning on gestation length, retained placenta, and dystocia. J. Anim. Sci. 1999, 77, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markusfeld, O. Periparturient Traits in Seven High Dairy Herds. Incidence Rates, Association with Parity, and Interrelationships Among Traits. J. Dairy Sci. 1987, 70, 158–166. [Google Scholar] [CrossRef]
- Andreu-Vázquez, C.; Garcia-Ispierto, I.; Ganau, S.; Fricke, P.; López-Gatius, F. Effects of twinning on the subsequent reproductive performance and productive lifespan of high-producing dairy cows. Theriogenology 2012, 78, 2061–2070. [Google Scholar] [CrossRef]
- Buoen, L.C.; Zhang, T.Q.; Weber, A.F.; Ruth, G.R. Non-Freemartin Rate in Holstein Heterosexual Twins. American Association of Bovine Practitioners. Proc. Annu. Conf. 1992, 1, 300–303. [Google Scholar] [CrossRef]
- Padula, A. The freemartin syndrome: An update. Anim. Reprod. Sci. 2005, 87, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Beerepoot, G.M.M.; Dykhuizen, A.A.; Nielen, Y.; Schukken, Y.H. The Economics of Naturally Occurring Twinning in Dairy Cattle. J. Dairy Sci. 1992, 75, 1044–1051. [Google Scholar] [CrossRef]
- Mur-Novales, R.; Lopez-Gatius, F.; Fricke, P.M.; Cabrera, V.E. An economic evaluation of management strategies to mitigate the negative effect of twinning in dairy herds. J. Dairy Sci. 2018, 101, 8335–8349. [Google Scholar] [CrossRef]
- Eddy, R.G.; Davies, O.; David, C. An economic assessment of twin births in British dairy herds. Veter. Rec. 1991, 129, 526–529. [Google Scholar]
- Lett, B.M.; Kirkpatrick, B.W. Short communication: Heritability of twinning rate in Holstein cattle. J. Dairy Sci. 2018, 101, 4307–4311. [Google Scholar] [CrossRef]
- Fricke, P.M. Double vision: Management of twinning in dairy cows; University of Wisconsin—Madison: American Association of Bovine Practitioners. In Proceedings of the Annual Conference, New Orleans, LA, USA, 17–19 September 2015. [Google Scholar]
- López-Gatius, F. Twins in Dairy Herds. Is It Better to Maintain or Reduce a Pregnancy? Animals 2020, 10, 2006. [Google Scholar] [CrossRef] [PubMed]
- Van Vleck, L.D.; E Gregory, K.; E Echternkamp, S. Ovulation rate and twinning rate in cattle: Heritabilities and genetic correlation. J. Anim. Sci. 1991, 69, 3213–3219. [Google Scholar] [CrossRef] [Green Version]
- Echternkamp, S.E.; Gregory, K.E. Reproductive, growth, feedlot, and carcass traits of twin vs single births in cattle. J. Anim. Sci. 2002, 80, 1–10. [Google Scholar] [CrossRef]
- Syrstad, O. Genetic Aspects of Twinning in Dairy Cattle. Acta Agric. Scand. 1974, 24, 319–322. [Google Scholar] [CrossRef]
- Ron, M.; Ezra, E.; Weller, J. Genetic analysis of twinning rate in Israeli Holstein cattle. Genet. Sel. Evol. 1990, 22, 349–359. [Google Scholar] [CrossRef]
- Karlsen, A.; Ruane, J.; Klemetsdal, G.; Heringstad, B. Twinning rate in Norwegian cattle: Frequency, (co)variance components, and genetic trends. J. Anim. Sci. 2000, 78, 15–20. [Google Scholar] [CrossRef]
- Hossein-Zadeh, N.G.; Nejati-Javaremi, A.; Miraei-Ashtiani, S.; Kohram, H. Estimation of variance components and genetic trends for twinning rate in Holstein dairy cattle of Iran. J. Dairy Sci. 2009, 92, 3411–3421. [Google Scholar] [CrossRef] [Green Version]
- Johansson, I.; Lindhé, B.; Pirchner, F. Causes of variation in the frequency of monozygous and dizygous twinning in various breeds of cattle. Hered 2009, 78, 201–234. [Google Scholar] [CrossRef] [PubMed]
- Moioli, B.; Steri, R.; Marchitelli, C.; Catillo, G.; Buttazzoni, L. Genetic parameters and genome-wide associations of twinning rate in a local breed, the Maremmana cattle. Animal 2017, 11, 1660–1666. [Google Scholar] [CrossRef]
- Van Vleck, L.D.; E Gregory, K. Genetic trend and environmental effects in a population of cattle selected for twinning. J. Anim. Sci. 1996, 74, 522–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.; Golik, M.; Seroussi, E.; Ron, M.; Ezra, E. Detection of Quantitative Trait Loci Affecting Twinning Rate in Israeli Holsteins by the Daughter Design. J. Dairy Sci. 2008, 91, 2469–2474. [Google Scholar] [CrossRef] [Green Version]
- Allan, M.F.; Kuehn, L.A.; Cushman, R.A.; Snelling, W.M.; Echternkamp, S.E.; Thallman, R.M. Confirmation of quantitative trait loci using a low-density single nucleotide polymorphism map for twinning and ovulation rate on bovine chromosome 51,2. J. Anim. Sci. 2009, 87, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.-S.; Shi, X.; Cobanoglu, O.; Weigel, K.; Berger, P.J.; Kirkpatrick, B.W. Refined mapping of twinning-rate quantitative trait loci on bovine chromosome 5 and analysis of insulin-like growth factor-1 as a positional candidate gene1. J. Anim. Sci. 2009, 87, 835–843. [Google Scholar] [CrossRef] [Green Version]
- Cruickshank, J.; Dentine, M.R.; Berger, P.J.; Kirkpatrick, B.W. Evidence for quantitative trait loci affecting twinning rate in North American Holstein cattle. Anim. Genet. 2004, 35, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Cobanoglu, O.; Berger, P.J.; Kirkpatrick, B.W. Genome screen for twinning rate QTL in four North American Holstein families. Anim. Genet. 2005, 36, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, B.W.; Morris, C.A. A Major Gene for Bovine Ovulation Rate. PLoS ONE 2015, 10, e0129025. [Google Scholar] [CrossRef] [Green Version]
- Gonda, M.G.; Arias, J.A.; Shook, G.E.; Kirkpatrick, B.W. Identification of an ovulation rate QTL in cattle on BTA14 using selective DNA pooling and interval mapping. Anim. Genet. 2004, 35, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Bierman, C.D.; Kim, E.; Weigel, K.; Berger, P.J.; Kirkpatrick, B.W. Fine-mapping quantitative trait loci for twinning rate on Bos taurus chromosome 14 in North American Holsteins1. J. Anim. Sci. 2010, 88, 2556–2564. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, B.W.; Byla, B.M.; Gregory, K.E. Mapping quantitative trait loci for bovine ovulation rate. Mamm. Genome 2000, 11, 136–139. [Google Scholar] [CrossRef] [Green Version]
- Kappes, S.M.; Bennett, G.L.; Keele, J.W.; Echternkamp, S.E.; Gregory, K.E.; Thallman, R.M. Initial results of genomic scans for ovulation rate in a cattle population selected for increased twinning rate. J. Anim. Sci. 2000, 78, 3053–3059. [Google Scholar] [CrossRef]
- Lien, S.; Karlsen, A.; Klemetsdal, G.; Våge, D.I.; Olsaker, I.; Klungland, H.; Aasland, M.; Heringstad, B.; Ruane, J.; Gomez-Raya, L. A primary screen of the bovine genome for quantitative trait loci affecting twinning rate. Mamm. Genome 2000, 11, 877–882. [Google Scholar] [CrossRef]
- Van Vleck, L.D.; Gregory, K.E.; Echternkamp, S.E. Prediction of breeding values for twinning rate and ovulation rate with a multiple trait, repeated records animal model. J. Anim. Sci. 1991, 69, 3959–3966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaddis, K.P.; Cole, J.; Clay, J.; Maltecca, C. Genomic selection for producer-recorded health event data in US dairy cattle. J. Dairy Sci. 2014, 97, 3190–3199. [Google Scholar] [CrossRef] [PubMed]
- McNeel, A.K.; Reiter, B.C.; Weigel, D.; Osterstock, J.; Di Croce, F.A. Validation of genomic predictions for wellness traits in US Holstein cows. J. Dairy Sci. 2017, 100, 9115–9124. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.; VanRaden, P. Symposium review: Possibilities in an age of genomics: The future of selection indices. J. Dairy Sci. 2018, 101, 3686–3701. [Google Scholar] [CrossRef] [Green Version]
- Fessenden, B.; Weigel, D.J.; Osterstock, J.; Galligan, D.T.; Di Croce, F. Validation of genomic predictions for a lifetime merit selection index for the US dairy industry. J. Dairy Sci. 2020. [Google Scholar] [CrossRef]
- Hazel, L.N. The Genetic Basis for Constructing Selection Indexes. Genetics 1943, 28, 476–490. [Google Scholar] [PubMed]
- Byrne, T.; Santos, B.; Amer, P.; Martin-Collado, D.; Pryce, J.; Axford, M. New breeding objectives and selection indices for the Australian dairy industry. J. Dairy Sci. 2016, 99, 8146–8167. [Google Scholar] [CrossRef]
- VanRaden, P.M. Invited Review: Selection on Net Merit to Improve Lifetime Profit. J. Dairy Sci. 2004, 87, 3125–3131. [Google Scholar] [CrossRef] [Green Version]
- Miglior, F.; Muir, B.L.; Van Doormaal, B.J. Selection Indices in Holstein Cattle of Various Countries. J. Dairy Sci. 2005, 88, 1255–1263. [Google Scholar] [CrossRef]
- Vukasinovic, N.; Bacciu, N.; Przybyla, C.; Boddhireddy, P.; Denise, S. Development of genetic and genomic evaluation for wellness traits in US Holstein cows. J. Dairy Sci. 2017, 100, 428–438. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Chesnais, J.; Schenkel, F. FImpute-An efficient imputation algorithm for dairy cattle populations. J. Dairy Sci. 2011, 94, 421. [Google Scholar]
- Norman, H.; Waite, L.; Wiggans, G.; Walton, L. Improving Accuracy of the United States Genetics Database with a New Editing System for Dairy Records. J. Dairy Sci. 1994, 77, 3198–3208. [Google Scholar] [CrossRef]
- Gonzalez-Peña, D.; Vukasinovic, N.; Brooker, J.; Przybyla, C.; Baktula, A.; Denise, S. Genomic evaluation for wellness traits in US Jersey cattle. J. Dairy Sci. 2020, 103, 1735–1748. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T. BLUPF90 and related programs (BGF90). In Proceedings of the World Congress Genet. Appl. Livest. Prod., Montpellier, France, 19–23 August 2002; p. 743. [Google Scholar]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Aguilar, I.; Leggara, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs. University of Georgia. 2014. Available online: http://nce.ads.uga.edu/wiki/doku.php?id=documentation (accessed on 17 December 2020).
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar, I.; Misztal, I.; Johnson, D.; Legarra, A.; Tsuruta, S.; Lawlor, T. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, D.; (University of Georgia, Athens, GA, USA). Personal communication, 2016.
- VanRaden, P.M.; Cole, J.B. Net Merit as a Measure of Lifetime Profit: 2014 Revision. USDA. 2014. Available online: Aipl.arsusda.gov/reference/nmcalc-2014.htm (accessed on 17 December 2020).
- Weigel, K.; Hoffman, P.; Herring, W.; Lawlor, T. Potential gains in lifetime net merit from genomic testing of cows, heifers, and calves on commercial dairy farms. J. Dairy Sci. 2012, 95, 2215–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwald, N.R.; Weigel, K.A.; Chang, Y.M.; Welper, R.D.; Clay, J.S. Genetic Selection for Health Traits Using Producer-Recorded Data. I. Incidence Rates, Heritability Estimates, and Sire Breeding Values. J. Dairy Sci. 2004, 87, 4287–4294. [Google Scholar] [CrossRef] [Green Version]
- Mrode, R. Linear Models for the Prediction of Animal Breeding Values; CABI: Oxfordshire, UK, 2005. [Google Scholar] [CrossRef]
- Misztal, I.; Legarra, A.; Aguilar, I. Computing procedures for genetic evaluation including phenotypic, full pedigree, and genomic information. J. Dairy Sci. 2009, 92, 4648–4655. [Google Scholar] [CrossRef] [Green Version]
- Misztal, I.; Aggrey, S.E.; Muir, W.M. Experiences with a single-step genome evaluation1. Poult. Sci. 2013, 92, 2530–2534. [Google Scholar] [CrossRef]
- Berry, D. Symposium review: Breeding a better cow—Will she be adaptable? J. Dairy Sci. 2018, 101, 3665–3685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, H.; Caraviello, D.; Satter, L.; Fricke, P.; Wiltbank, M. Relationship Between Level of Milk Production and Multiple Ovulations in Lactating Dairy Cows. J. Dairy Sci. 2005, 88, 2783–2793. [Google Scholar] [CrossRef]
- McGovern, S.; Purfield, D.; Ring, S.; Carthy, T.; Graham, D.; Berry, D. Candidate genes associated with the heritable humoral response to Mycobacterium avium ssp. paratuberculosis in dairy cows have factors in common with gastrointestinal diseases in humans. J. Dairy Sci. 2019, 102, 4249–4263. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Event | Remark | Standardized Event |
|---|---|---|
| ABORT | TWIN, TW M, TW F, TW=, TWF, TWDOA, TWM, BULHFRTW, TW2BULL, TWNB, TWNH, BULLSTW, TWNHFRS, TWBULL, TW (and remark is only two letters long) | TWIN |
| RP | TWIN | |
| MISC | TWIN | |
| DRYOFF | TWIN | |
| ILL | TWIN | |
| OK | TWIN | |
| REMARK | TWIN | |
| SOLD | TWIN | |
| FRESH | TWIN |
| Item | Count |
|---|---|
| Pedigree records total | 3,687,609 |
| Phenotypic records total | 3,528,053 |
| Animals with phenotypes | 1,800,296 |
| Animals with genotypes | 1,145,323 |
| Animals with genotypes and phenotypes | 78,509 |
| Incidence of twinning | 3.25% |
| Trait | σ2g | σ2pe | σ2hys | σ2e | h2 | r2 |
|---|---|---|---|---|---|---|
| TWIN | 0.1315 | 0.1318 | 0.2272 | 1.0 | 0.0882 | 0.1767 |
| All Animals with Genotypes | |||||
|---|---|---|---|---|---|
| Variables * | n | Mean | Std Dev | Minimum | Maximum |
| TWIN PTA | 1,145,323 | −0.096 | 2.606 | −7.449 | 20.792 |
| TWIN STA | 1,145,323 | 100.29 | 4.636 | 63 | 113 |
| TWIN REL | 1,145,323 | 42.02 | 6.144 | 0 | 99.5 |
| Animals with genotypes (no phenotypes, no progeny) | |||||
| TWIN PTA | 887,068 | −0.147 | 2.550 | −7.449 | 19.703 |
| TWIN STA | 887,068 | 100.38 | 4.537 | 65 | 113 |
| TWIN REL | 887,068 | 40.77 | 5.099 | 0 | 61.140 |
| Zoetis Wellness Traits * | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | DWP$ | DIAR | CALF_RESP | DEAD | RETP | METR | MAST | LAME | KETO | DA | MFV | ABRT | RESP | CYST |
| TWIN | 0.115 | 0.014 | 0.021 | 0.065 | 0.239 | 0.123 | 0.025 | −0.025 | 0.045 | 0.024 | 0.029 | 0.252 | 0.013 | 0.056 |
| CDCB Traits ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | Milk | Fat | Prot | PL | LIV | SCS | DPR | HCR | CCR |
| TWIN | 0.011 | −0.038 | 0.025 | −0.055 | −0.029 | 0.029 | 0.014 | −0.031 | −0.011 |
| Herd | Protocol | Region | No. Females | No. Records | Average TWIN STA | Minimum TWIN STA | Maximum TWIN STA | 1st Lactation TWIN Incidence (%) | 3rd Lactation TWIN Incidence (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | No synchronization | West | 477 | 1483 | 100 | 88 | 108 | 1.48 | 14.00 |
| 2 | Presynch/ovsynch | Mid-West | 1105 | 3050 | 101 | 77 | 108 | 0.74 | 9.09 |
| 3 | Presynch/ovsynch | Mid-West | 2157 | 5434 | 100 | 86 | 110 | 1.25 | 12.93 |
| 4 | Double Ovsynch | Mid-West | 3200 | 8721 | 100 | 79 | 110 | 0.64 | 7.79 |
| 5 | Double Ovsynch | North West | 1280 | 3592 | 100 | 79 | 115 | 0.56 | 6.95 |
| Herd | STA Genetic Group | TWIN Incidence | SEM | p-Value | TWIN Cost Per Case ($) |
|---|---|---|---|---|---|
| 1 | Bottom 33% | 0.19 | 0.027 | <0.0001 | 31 |
| 34–66% | 0.14 | 0.024 | 23 | ||
| Top 33% | 0.10 | 0.022 | 16 | ||
| 2 | Bottom 33% | 0.09 | 0.013 | <0.0001 | 14 |
| 34–66% | 0.06 | 0.010 | 10 | ||
| Top 33% | 0.04 | 0.009 | 6 | ||
| 3 | Bottom 33% | 0.12 | 0.011 | <0.0001 | 19 |
| 34–66% | 0.09 | 0.009 | 14 | ||
| Top 33% | 0.06 | 0.008 | 10 | ||
| 4 | Bottom 33% | 0.09 | 0.008 | <0.0001 | 14 |
| 34–66% | 0.07 | 0.006 | 11 | ||
| Top 33% | 0.03 | 0.004 | 5 | ||
| 5 | Bottom 33% | 0.09 | 0.012 | <0.0001 | 14 |
| 34–66% | 0.06 | 0.008 | 10 | ||
| Top 33% | 0.02 | 0.006 | 3 |
| Herd | Fixed Effect | df | F-Value | p-Value |
|---|---|---|---|---|
| 1 | TWIN STA 2013 | 1 | 7.68 | 0.0060 |
| Conception Season | 3 | 0.18 | 0.9129 | |
| Peak Lactation | 1 | 0.02 | 0.8849 | |
| 2 | TWIN STA 2013 | 1 | 2.50 | 0.1147 |
| Conception Season | 3 | 0.54 | 0.6563 | |
| Peak Lactation | 1 | 2.75 | 0.0981 | |
| 3 | TWIN STA 2013 | 1 | 7.64 | 0.0058 |
| Conception Season | 3 | 0.77 | 0.5105 | |
| Peak Lactation | 1 | 3.87 | 0.0495 | |
| 4 | TWIN STA 2013 | 1 | 8.66 | 0.0033 |
| Conception Season | 3 | 1.29 | 0.2779 | |
| Peak Lactation | 1 | 0.46 | 0.4981 | |
| 5 | TWIN STA 2013 | 1 | 4.73 | 0.0300 |
| Conception Season | 3 | 0.58 | 0.6295 | |
| Peak Lactation | 1 | 3.42 | 0.0650 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McGovern, S.P.; Weigel, D.J.; Fessenden, B.C.; Gonzalez-Peña, D.; Vukasinovic, N.; McNeel, A.K.; Di Croce, F.A. Genomic Prediction for Twin Pregnancies. Animals 2021, 11, 843. https://doi.org/10.3390/ani11030843
McGovern SP, Weigel DJ, Fessenden BC, Gonzalez-Peña D, Vukasinovic N, McNeel AK, Di Croce FA. Genomic Prediction for Twin Pregnancies. Animals. 2021; 11(3):843. https://doi.org/10.3390/ani11030843
Chicago/Turabian StyleMcGovern, Shaileen P., Daniel J. Weigel, Brenda C. Fessenden, Dianelys Gonzalez-Peña, Natascha Vukasinovic, Anthony K. McNeel, and Fernando A. Di Croce. 2021. "Genomic Prediction for Twin Pregnancies" Animals 11, no. 3: 843. https://doi.org/10.3390/ani11030843






